Abstract
The s48/45 domain was first noted in Plasmodium proteins more than 15 y ago. Previously believed to be unique to Plasmodium, the s48/45 domain is present in other aconoidasidans. In Plasmodium, members of the s48/45 family of proteins are localized on the surface of the parasite in different stages, mostly by glycosylphosphatydylinositol-anchoring. Members such as P52 and P36 seem to play a role in invasion of hepatocytes, and Pfs230 and Pfs48/45 are involved in fertilization in the sexual stages and have been consistently studied as targets of transmission-blocking vaccines for years. In this report, we present the molecular structure for the s48/45 domain corresponding to the C-terminal domain of the blood-stage protein Pf12 from Plasmodium falciparum, obtained by NMR. Our results indicate that this domain is a β-sandwich formed by two sheets with a mixture of parallel and antiparallel strands. Of the six conserved cysteines, two pairs link the β-sheets by two disulfide bonds, and the third pair forms a bond outside the core. The structure of the s48/45 domain conforms well to the previously defined surface antigen 1 (SAG1)-related-sequence (SRS) fold observed in the SAG family of surface antigens found in Toxoplasma gondii. Despite extreme sequence divergence, remarkable spatial conservation of one of the disulfide bonds is observed, supporting the hypothesis that the domains have evolved from a common ancestor. Furthermore, a homologous domain is present in ephrins, raising the possibility that the precursor of the s48/45 and SRS domains emerged from an ancient transfer to Apicomplexa from metazoan hosts.
Keywords: Plasmodium domain evolution, Escherichia coli periplasmic expression, solution conformation
Malaria is caused by Plasmodium parasites one of which, Plasmodium falciparum, claims the lives of nearly a million children each year in Africa alone. Efforts to develop a malaria vaccine would benefit considerably from a more complete understanding of the structure and function of Plasmodium proteins that play key roles in infection. Plasmodium is transmitted to humans when infected mosquitoes inoculate sporozoites into the skin while taking a blood meal. The sporozoites travel from the skin to the liver, where they invade hepatocytes and replicate tens- of thousands-fold, differentiating into merozoites that exit the liver and invade erythrocytes. After several rounds of replication in erythrocytes, some parasites differentiate into male and female gametocytes and are subsequently taken up by an Anopheles mosquito in a blood meal. Within the mosquito, fertilization takes place and the resulting ookinetes invade the midgut epithelium, resulting in sporozoites that travel to the salivary glands and are transmitted to the human host, completing the cycle.
The 6-cysteine Plasmodium gamete-surface homology s48/45 domain originally identified by Williamson et al. (1) (partially defined by Pfam model PF07422) is found in P. falciparum proteins that are expressed in all stages of the parasite life cycle in both the human and mosquito hosts (Table S1) (1–3). The domain might occur in 1–14 copies per protein, either as the sole globular domain or in combination with β-helix–forming hexapeptide repeats, as is the case of sequestrin. Previously believed to be exclusive to Plasmodium (4–6), the domain also exists in proteins found in all members of the aconoidasidan (hematozoan) clade of Apicomplexa, which unites the haemosporidians (Plasmodium) and piroplasms. In Plasmodium, the s48/45 family has undergone a lineage-specific expansion, with 12 distinct members [Pfs230, Pfs48/45, Pfs230p, Pfs47, P52, P36, Pf41, Pf38, Pf12, P12p, Pf92 and sequestrin (not previously included) (Table S1)] encoded by the genome of P. falciparum (2, 7). Conserved homologs are present in the genomes of other Plasmodium species. Most of the proteins containing the s48/45 domain are expressed on the surface of the parasite (8–11). Pfs48/45 and Pfs230 are current targets of malaria transmission-blocking vaccine development (12, 13).
Three of the s48/45 proteins (Pfs48/45, Pfs230, and Pfs47) are involved in male/female gamete fusion in the mosquito midgut. Pfs48/45 is predicted to be glycosylphosphatidylinositol (GPI)-anchored to the gamete surface (14). Pfs230 is a soluble protein that associates with the gamete membrane by binding to Pfs48/45 (15). Deletion of either Pfs48/45 or Pfs230 greatly reduces zygote formation (16, 17). It is unknown if the role of Pfs48/45 is only to bind to Pfs230, which functions directly in gamete fusion, or if both are directly involved in facilitating fusion. Pfs48/45 has three s48/45 domains, whereas Pfs230 has 14 s48/45 domains (2, 4). Antibodies specific for either protein block transmission to the mosquito, and complement enhances the effect of antibodies specific for Pfs230 (9, 18–20). Pfs47, a duplicated and contiguous paralogue of Pfs48/45, is found on female gametes; its deletion in rodent Plasmodium berghei, but not in P. falciparum, greatly reduces fertilization in the mosquito (11, 21). The presence of multiple species-specific paralogues of this family might result in backup functions in different lineages within the genus Plasmodium.
Sporozoites express two members of the s48/45 family, P36 and P52, from contiguous, duplicated genes. The functions of these two proteins appear to be different because the deletion of either gene influences the ability of the parasite to efficiently infect hepatocytes, with no effect on their ability to invade salivary gland cells in the mosquito (22, 23). In P. berghei there is also evidence that these genes may be required for invasion of hepatocytes and to promote normal development in the liver. Deletion of Pbs36 and Pbs36p results in sporozoites that are unable to develop beyond a small round body, reflecting a defect in normal invasion (8, 24).
The asexual-stage parasites that replicate within erythrocytes express four members of the s48/45 family (Pf12, Pf38, Pf41, and Pf92), all located on the surface of merozoites. Each member has a signal sequence and three of them are GPI-anchored to the merozoite surface (10, 14). Because Pf41 has no GPI anchor signal or transmembrane domain for attachment to the membrane, it is likely to bind another merozoite surface protein. Pf41, Pf38, and Pf12 are strongly recognized by immune sera from naturally infected patients, and Pf38 and Pf92 are under balancing selection (10, 25–27). Attempts to delete Pf92 have been reportedly unsuccessful; however, no information for the deletion of the other genes is available (28). Although a specific function has not been described for any of these proteins, the possibility exists for them to have a role in red blood cell binding.
To date, no experimentally determined molecular structure for any s48/45 domain has been reported, mainly because of protein-expression challenges (29–34). However, considering the importance of structural information in the effective design of subunit vaccine candidates, we present the solution three-dimensional structure of the s48/45 domain obtained by NMR spectroscopy of the C-terminal domain of Pf12 expressed in Escherichia coli.
Results
Production of Soluble Pf12 D2.
The C-terminal s48/45 domain of Pf12 (Pf12 D2) was produced in E. coli as a soluble histidine-tagged protein by initially harmonizing the DNA sequence of Pf12 for expression in E. coli based on the procedure published by Angov et al. (35), and then targeting the protein for expression in the bacterial periplasm (Figs. S1 and S2) (35, 36). Only the protein expressed as a soluble fraction, extracted through the osmotic shock procedure as described, was used for our studies. The use of PBS as a buffer was avoided because it led to protein aggregation. Soluble expression was assessed by Western blot analysis probing with α-His antibody (Fig. S3). Following purification by gradient immobilized metal affinity chromatography, size-exclusion FPLC on a Superdex75 column resulted in the elution of a single protein peak, indicating that Pf12 D2 is a monomer in solution (Fig. S4). The protein was more than 95% pure, as judged by Gelcode blue-stained SDS/PAGE electrophoresis (Fig. S5); the expected molecular weight of cleaved Pf12 D2 was 14.5 kDa. The identity of the protein was confirmed by N-terminal sequencing and mass spectrometry. For NMR spectroscopy, the final isotopically labeled product was found to be most stable at concentrations no higher than 7 mg/mL in 25 mM sodium acetate-d3 pH 5.0.
Structural Characterization of Pf12 D2.
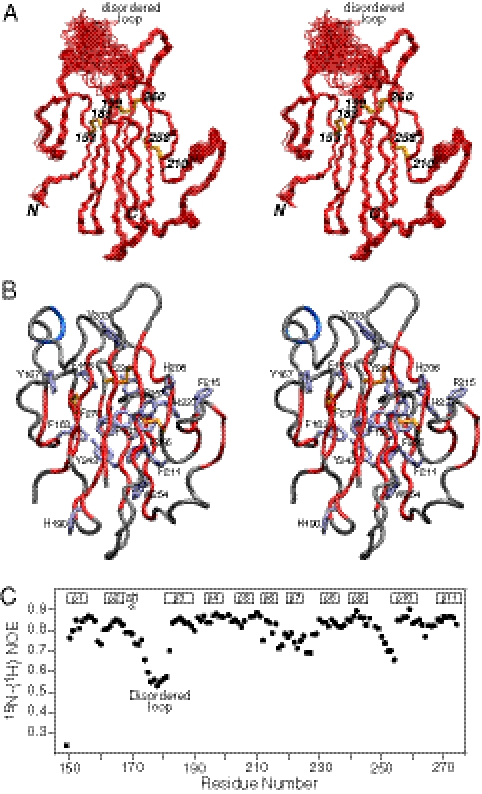
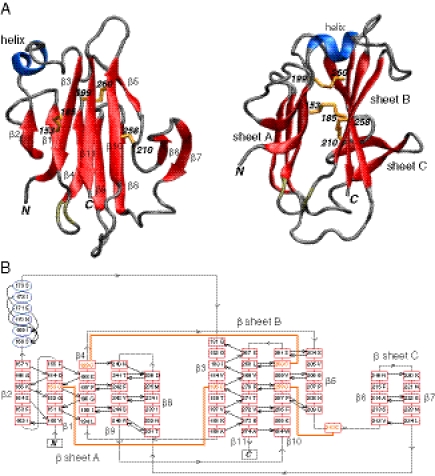
The solution NMR structure of Pf12 D2 was determined on the basis of 2,361 experimental NMR restraints, including 1,477 NOE-derived interproton distance restraints and 192 residual dipolar couplings. The latter provide long-range orientational information in the form of bond vector orientations relative to an external alignment tensor (37). A summary of the structural statistics is provided in Table S2, and a best-fit superposition of the final simulated annealing structures is shown in Fig. 1A. The three disulfide bonds are uniquely determined from the NMR structure and comprise Cys153-Cys185, Cys199–Cys260, and Cys210–Cys258. The structure comprises three β-sheets and a short helix (residues 168–173) (Fig. 2): the five-stranded β-sheet A (β2, β1, β4, β9, β8) has a 1x, −2x, −2x, 1 topology; the four-stranded β-sheet B (β3, β11, β10, β5) has a 3x, −1, −1 topology; and the two-stranded β-sheet C (β6, β7) comprises a simple antiparallel β-sheet connected by a type I turn. Disulfide bonds A (Cys153–Cys185) and B (Cys199–Cys260) bridge β-sheets A and B connecting strands β1 to β3, and β4 to β10, respectively; disulfide bond C (Cys210–Cys258) bridges the loop connecting strands β5 and β6 to strand β10 in sheet B (Fig. 2B). The core of the protein is packed by an unusually large preponderance of aromatic residues (Fig. 1B). The loop connecting the single helix with strand β3 is disordered (see, for example, the structure superpositions in Fig. 1A, and the low values of the 1H-{15N} heteronuclear NOEs in Fig. 1C).
Fig. 1.
NMR structure of Pf12 D2. (A) Stereoview of the backbone superposition of the 100 final simulated annealing structures. The backbone atoms (N, Cα, C′) are in red, and the three disulfide bonds (between residues 153–185, 199–260, and 210–258) are in yellow. (B) Stereoview of a tube presentation of the backbone (with strands colored in red, and the helix in blue) showing the large number of aromatic residues (ice blue). (C) Heteronuclear 1H-{15N}-NOE profile as a function of residue number, showing that the loop connecting the single helix with strand β3 is highly mobile and disordered.
Fig. 2.
Overall view of the structure of Pf12 D2. (A) Two approximately orthogonal views displaying a ribbon diagram of Pf12 D2 with β sheets in red, the helix in blue, and the disulfide bonds in yellow. (B) Secondary structure topology of Pf12 D2.
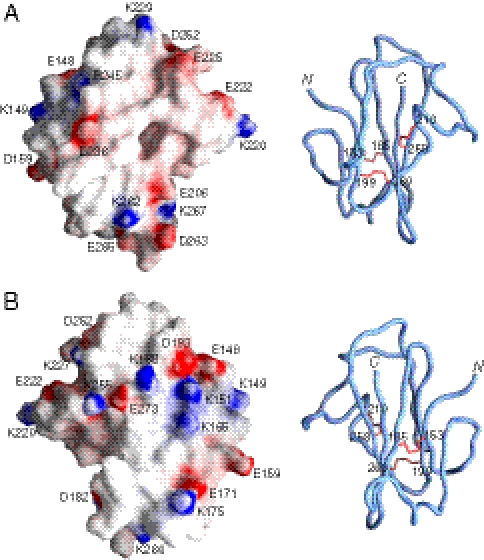
The electrostatic potential mapped on the molecular surface of Pf12 D2 is shown in Fig. 3. Although there are certainly patches of charged residues on the surface, it can also be seen that most of the protein surface is hydrophobic. In the absence of the D1 domain and knowledge of the interactions between the D1 and D2 domain, it is difficult to speculate as to the potential ligand binding site. Nevertheless, certain features of the surface of Pf12 D2 are worth noting. On the front surface in the view of Fig. 3A, the largely hydrophobic surface is surrounded by a ring of four negative charges comprising Glu206, Glu222, Glu225, and Asp236. In the center of the front face of the protein is a deep hydrophobic pocket, at the bottom of which lies Phe197 and the disulfide bridge formed by Cys210 and Cys258, bounded by Val205, Pro207, Val212, Met235, and His237. It is possible that this pocket may constitute a ligand binding site. The backside of the protein, displayed in Fig. 3B, is mildly concave and comprises a central hydrophobic region surrounded by a partial ring of charged residues comprising Lys255, Glu273, Lys188, Asp193, Glu148, Lys151, Lys165, Glu171, and Lys175. This type of surface is characteristic of many protein-interaction surfaces (38) and may therefore represent the site of interaction with another domain or protein. The surface of Pf12 D2, however, does not possess the equivalent of the basic groove or acid cap seen in the structurally related surface antigen 1 (SAG1) (39) and SporoSAG (40) proteins (see below), respectively, from Toxoplasma gondii.
Fig. 3.
Molecular surface of Pf12 D2. Two views (A and B) related by a 180° rotation displaying the molecular surface of Pf12 D2 with the electrostatic potential mapped onto the surface (Left). The corresponding panels (Right) are backbone tube representations in the same orientation as the surfaces to guide the eye. The electrostatic potential was created by the program GRASP (65).
Structural Neighbors of the s48/45 Domain and Its Relationship to the SAG1-Related Sequence Superfamily.
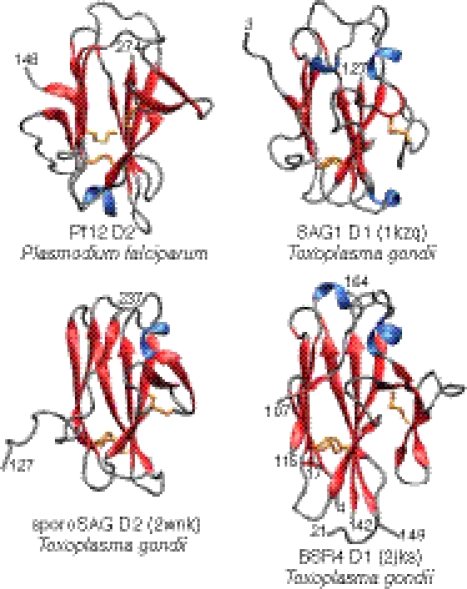
Using fold-prediction methods, it was previously shown that the s48/45 domain is likely to adopt the same fold as the SAG1-related sequence (SRS) domain, a β-sandwich found in the lineage-specifically expanded group of proteins from T. gondii, such as SAG1, SporoSAG, and BSR4 (2, 39). We confirmed this hypothesis using profile-profile comparisons with the HHpred program (41): a comparison of the PSI-BLAST–derived profile of the s48/45 family with that of the SRS superfamily yielded highly significant hits (P = 10−11; probability 94%) (Table S3). This finding was further validated by a Dali (42) search of the Protein Data Bank using the structure of Pf12 s48/45 determined in this study, which recovers the SRS domains with z > 5 (Fig. 4). Indeed, 96 residues of SAG D1, 92 residues of SporoSAG D2, and 91 residues of BSR4 D1 can be superimposed onto Pf12 D2 with Cα atomic rms differences of 2.6, 2.9, and 2.7 Å, respectively. However, the percentage sequence identities for the corresponding structurally superimposed regions are only 16%, 13%, and 12%, suggesting rather extensive sequence divergence between the SRS and s48/45 domains, despite their sharing a common structural scaffold (Fig. 4). Structure similarity searches using the DaliLite (43) program indicate that the β-sandwich fold shared by s48/45 and the SRS superfamily is further related to comparable β-sandwich domains found in a diverse array of cell-surface molecules from various organisms (z > 4 in reciprocal structure similarity searches) (44). These molecules include the cysteine protease inhibitors amoebiasin-2 (3m86) from Entamoeba histolytica and chagasin (2nnr) from Trypanosoma cruzi, ephrins (2wo3), which are animal neural signaling molecules, and the copper-binding plastocyanins (2q5b). All these β-sandwich domains are united by the pattern of crossover between the two sheets of the sandwich and the zone of variability, which is restricted to one edge of the β-sandwich (Figs. S6 and S7). The ephrins, s48/45, and SRS display a cysteine at the end of strand 3 of their respective β-sandwich domains (equivalent to the first cysteine participating in disulfide bridge B in the latter two domains) (Fig. S6). However, the partner cysteine on the opposite sheet in the ephrins is not equivalent to that seen in the s48/45 and SRS domains (Figs. S6 and S7). This structurally equivalent, unique disulfide bond B shared by s48/45 and SRS distinguishes them from the rest of the related β-sandwich domains. Disulfide bond A bridges strands β1 and β3 of the sandwich in the s48/45 domain, but in the case of SRS it bridges strand β1 to the final strand β11, which lies adjacent to the second strand in the structure (Figs. 2 and 4). Thus, one of the cysteine pairs forming disulfide bond A appears to have diverged between the s48/45 and SRS; yet, each disulfide bond mediates a comparable cross-sheet contact in both of these domains.
Fig. 4.
Comparison of the fold of Pf12 D2 with the folds of three surface antigens from T. gondii (SAG D1, sporoSAG D2, and BSR4 D1) belonging to the SRS superfamily. The ribbon diagrams show the four domains in identical orientations.
Discussion
The structure of the C-terminal domain of the protein Pf12 from P. falciparum solved by NMR defines the s48/45 domain, previously considered exclusive to the Plasmodium species. As shown in Fig. 2, this domain features an SRS fold, characterized by a sandwich formed by β-sheets resulting from a mixture of parallel and antiparallel strands (39) that positions the s48/45 domain as a homolog of the SRS family. This family of surface antigens from T. gondii is defined by the presence of an N-terminal secretion signal, a GPI anchor, and a set of conserved amino acids, among which are six cysteines involved in the formation of three disulfide bonds important in maintaining the structure of the SRS fold (40).
To date, more than 160 DNA sequences from T. gondii have been identified as belonging to the SRS superfamily (45), but only the structures for SAG1, SporoSAG, and BSR4 have been solved (39, 40, 46, 47). Although there is some structural divergence among these three proteins, and within each protein the N-terminal (D1) and C-terminal (D2) domains are also structurally distinct, the SRS fold is observed as a common feature in the domains of all three (40) (Fig. 4). Overall, comparison of the structure of Pf12 D2 to the D1 domain of SAG1, D2 of SporoSAG, and D1 of BSR4 (Fig. 4) indicates that despite having a low identity for both the overall sequence (4–9%) and the structurally matched regions (12–16%), the structure is indeed superimposable on the SRS structures with Cα rms deviations ranging from 2.6 to 2.9 Å, not a significant difference compared with the deviation reported between the superimposition of the D1 and D2 domains from the crystal structure of SAG1 (2.7 Å) (39). As seen in Fig. 4, the core structure of the SRS fold is evidently conserved in Pf12 D2, but distinct characteristics are also observed in the periphery of the core (Fig. 2) [i.e., a double-stranded β-sheet (β6, β7), a short α-helix, and a disordered loop (residues 174–180)] (Fig. 1). This last feature, indicative of extensive mobility (as demonstrated by the low 1H-{15N} NOE values), could in principle be present in the T. gondii SRS proteins for which the structures are available. However, because these features were derived from crystals, at the moment it is not clear whether this is a particular feature of Pf12, the s48/45 domain, or the SRS fold in general.
A common characteristic of both the SRS and the s48/45 domains is their tendency to occur as tandem paralogous pairs. The two tandem domains forming a pair in s48/45 proteins were respectively labeled as “A-type” and “B-type” domains in the original work defining the s48/45 domains (2, 4). The structure of Pf12 D2 reported here corresponds to the B-type or second domain in the pair.
The remarkable conservation of one of the disulfide bonds in all four structures, Pf12 D2, SAG1, SporoSAG, and BSR4 (Fig. 4 and Fig. S8), corresponding to disulfide B (Cys199–Cys260) in Pf12 D2 (Fig. 2), suggests that this structural component is essential in the stability of the SRS fold. A second bridge, disulfide A (Cys153–Cys185), although not spatially matched to the disulfide found in the three structures from T. gondii at the same position, seems to serve the equivalent purpose of keeping the main β-sheets of the sandwich together (Figs. 2 and 4, and Fig. S8). Despite structural differences, three disulfide bonds are positionally conserved within both domains of SAG1, BSR4, and SporoSAG; however, analysis of other SRS sequences predicts that the third disulfide bond might not be strictly conserved, or is even absent in some members of the family (40). In the case of Pf12 D2, the third disulfide bond (Cys210–Cys258) is positioned outside the main fold, bringing the third β-sheet closer to the core (Fig. 2); similar to the T. gondii family, not all of the s48/45 domains may require the presence of this disulfide. Analysis of the sequences from proteins Pfs230, Pfs48/45, and Pf12 by Carter et al. (4), leading to the original prediction of the disulfide bond connectivity in the s48/45 domain, showed that with the exception of the first domain of Pfs48/45, all predicted domains have an even number of cysteines, most of them anticipated to form three or two disulfide bonds, but a few would only form one bond within each domain.
The disulfide bond pattern for the s48/45 domain, where cysteines 1–2, 3–6, and 4–5 are connected, previously proposed independently by Carter et al. through visual inspection and scoring of the sequences mentioned above (4), and by Gerloff et al. through comparative structural homology models of Pfs230, Pfs48/45, Pfs47, and Pf12 based on SAG1 as a template (2), is now validated by the disulfide bonds observed between Cys153–Cys185, Cys199–Cys260, and Cys210–Cys258 in the structure of Pf12 D2 (Fig. 2).
The members of the SRS superfamily, developmentally expressed on the surface of T. gondii, are generally considered adhesins involved in host-cell attachment and invasion (45, 48). In particular, SAG1 is a highly immunogenic prototypic member of the family expressed on the invasive tachyzoite, predicted to bind polyanionic ligands and presumed to serve as an immune decoy facilitating chronic infection; anti-SAG1 antibodies reduce parasite invasion in vitro, and SAG1-null parasites showed less virulence in mice experiments (39, 49–51).
In Plasmodium, so far there are four proteins (Pf12, Pf38, Pf41, Pf92) from the s48/45 family that have been validated as merozoite surface proteins. Pf12, Pf38, and Pf41 are readily recognized by immune sera from infected patients from endemic areas (10, 25, 26); however, no function has been ascribed to any of them yet. The first step in invasion of merozoites into red blood cells is the binding of any part of the merozoite to the red blood cell surface. It is tempting to speculate that one of the s48/45 proteins could possibly be the long-anticipated initiator of the invasion cascade.
From an evolutionary perspective, β-sandwich domains with a structure comparable to the s48/45 and SRS are found in a wide range of cell-surface proteins from bacteria to eukaryotes, including versions from other parasites, such as Entamoeba and trypanosomes. This finding indicates that such domains have an ancient history of being used in extracellular interactions. However, classic members of the s48/45 and SRS family are currently only known from apicomplexan clades of coccidia and aconoidasida. These members are apparently absent in Cryptosporidium, which belongs to a more basal lineage of apicomplexans (Fig. S9), suggesting that these domains emerged in the common ancestor of the coccidians and aconoidasidans. This pattern of evolution is congruent with that of several other surface molecules [for example, the AMA1/MAEBL adhesins (44)]. Given that profile-profile searches indicate the ephrins as the next best hit after the SRS superfamily (Table S3), it is conceivable that the precursor of the s48/45 and SRS domains was derived from an ephrin-like precursor originally acquired from their metazoan hosts. Subsequent to their divergence, the SRS domain has undergone independent explosive radiations in different coccidians, but the s48/45 domains have undergone an expansion only in the Plasmodium lineage.
Despite the early recognition of Pfs230 and Pfs48/45 as transmission-blocking targets (9, 19) and subsequent identification of other members of the initially named 6-cys family (1, 5, 6), technical difficulties in recombinant protein expression have been a major hurdle in the successful production of full-length s48/45 proteins in soluble and stable conditions. The one exception has been Pfs48/45 (12). In the case of Pf12, after multiple failed attempts for the production of recombinant full length Pf12, we opted for engineering the protein for expression as separate domains with success in the production of the soluble D2 domain in the periplasm of E. coli. The conservation of this structural component is important in Plasmodium biology. Thus, further functional and structural characterization of s48/45 proteins is indispensable.
Methods
Cloning, Expression, and Purification.
Recombinant soluble Pf12 D2, the C-terminal domain of the P. falciparum protein Pf12 (PFF0615c), was produced in E. coli. Details on cloning, expression, labeling and purification are provided in SI Methods.
NMR Spectroscopy.
All NMR spectra were collected on 0.4-mM protein samples in 15 mM sodium d3-acetate at pH 5.0, and were recorded at 35 °C on Bruker DRX600, DRX800, and Avance 900 spectrometers, equipped with z-gradient cryoprobes. Spectra were processed using the program NMRPipe (52), and analyzed using the programs PIPP and CAPP (53). Sequential assignment of 1H, 15N, and 13C resonances was achieved by means of through-bond heteronuclear scalar correlations along the protein backbone and side chains (54) (55) using 3D HNCO, CBCACONH, HNCACB, (H)C(CO)NH TOCSY, and H(CCO)NH-TOCSY experiments, and a 30-ms mixing time 13C-separated NOE experiment. Interproton distance restraints were derived from 3D 15N- and 13C-separated NOE experiments at a mixing time of 120 ms. Side chain torsion angle restraints were derived from 3JNCγ(aromatic, methyl, and methylene) and 3JC'Cγ(aromatic, methyl, and methylene) scalar couplings measured by quantitative J correlation spectroscopy (56), in combination with data from a short mixing time (30 ms) 3D 13C-separated NOE spectrum recorded in H2O.
Residual 1DNH, 1DCαH, 1DNC' ,and 2DHNC' dipolar couplings (57) were measured in a liquid crystalline medium of phage pf1 (11 mg/mL) (58). The magnitudes of the axial (DaNH) and rhombic (η) components of the alignment tensor DNH were obtained by examining the distribution of the normalized residual dipolar couplings (59). Heteronuclear 15N-{1H} NOE data were collected and analyzed as described previously (60), on a 0.3 mM 15N-labeled sample in 25 mM sodium acetate, pH 5.0.
Structure Calculations.
Interproton distance restraints were classified into four distance ranges: 1.8–2.7 Å (1.8–2.9 Å for NOEs involving NH protons), 1.8–3.3 Å (1.8–3.5 Å for NOEs involving NH protons), 1.8–5.0 Å and 1.8–6.0 Å, corresponding to strong, medium, weak, and very weak NOEs, respectively (54). Next, 0.5 Å was added to the upper distance limits for NOEs involving methyl groups. Distances involving ambiguous NOEs, nonstereospecifically assigned methylene and methyl groups, and the Hδ and Hε protons of Tyr and Phe were represented as a Σ(r−6)−1/6 sum. Backbone ϕ/ψ torsion angle restraints were obtained on the basis of backbone 15N, 1HN, 13Cα, 1Hα, 13Cβ, and 13C′ chemical shifts using the program TALOS+ (61). Structures were calculated by simulated annealing in torsion angle space (62) using the program Xplor-NIH (63). The target function comprised terms for the experimental NMR restraints (NOE-derived interproton distances, backbone, and side-chain torsion angles, 13Cα/13Cβ chemical shifts, and residual dipolar couplings), terms to maintain idealized covalent geometry, a quartic van der Waals repulsion term to prevent atomic overlap, a multidimensional torsion-angle database potential of mean force, and a hydrogen bonding database potential of mean force. Structure figures were created using the programs VMD-XPLOR (64) and GRASP (65). The coordinates and experimental restraints have been deposited in the Protein Dadata Bank (PDB ID 2LOE) and the chemical-shift assignments in the Biological Magnetic Resonance Bank (BMRB ID 18210).
Supplementary Material
Acknowledgments
We thank Susan Pierce (Laboratory of Immunogenetics) and Kim Williamson and Prakash Srinivasan (Laboratory of Malaria Vector Research) for very insightful discussions. This work was supported by the intramural funds from the National Library of Medicine (to L.A.), the National Institute of Allergy and Infectious Diseases (L.H.M.), and the National Institute of Diabetes and Digestive and Kidney Diseases (G.M.C.) of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates and experimental restraints reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2LOE), and the chemical shift assignments have been deposited in the Biological Magnetic Resonance Bank, www.bmrb.wisc.edu (BMRB ID 18210).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204363109/-/DCSupplemental.
References
- 1.Williamson KC, Criscio MD, Kaslow DC. Cloning and expression of the gene for Plasmodium falciparum transmission-blocking target antigen, Pfs230. Mol Biochem Parasitol. 1993;58:355–358. doi: 10.1016/0166-6851(93)90058-6. [DOI] [PubMed] [Google Scholar]
- 2.Gerloff DL, Creasey A, Maslau S, Carter R. Structural models for the protein family characterized by gamete surface protein Pfs230 of Plasmodium falciparum. Proc Natl Acad Sci USA. 2005;102:13598–13603. doi: 10.1073/pnas.0502378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue, D1):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter R, Coulson A, Bhatti S, Taylor BJ, Elliott JF. Predicted disulfide-bonded structures for three uniquely related proteins of Plasmodium falciparum, Pfs230, Pfs48/45 and Pf12. Mol Biochem Parasitol. 1995;71:203–210. doi: 10.1016/0166-6851(94)00054-q. [DOI] [PubMed] [Google Scholar]
- 5.Templeton TJ, Kaslow DC. Identification of additional members define a Plasmodium falciparum gene superfamily which includes Pfs48/45 and Pfs230. Mol Biochem Parasitol. 1999;101:223–227. doi: 10.1016/s0166-6851(99)00066-3. [DOI] [PubMed] [Google Scholar]
- 6.Thompson J, Janse CJ, Waters AP. Comparative genomics in Plasmodium: A tool for the identification of genes and functional analysis. Mol Biochem Parasitol. 2001;118:147–154. doi: 10.1016/s0166-6851(01)00377-2. [DOI] [PubMed] [Google Scholar]
- 7.Sanders PR, et al. Identification of protein complexes in detergent-resistant membranes of Plasmodium falciparum schizonts. Mol Biochem Parasitol. 2007;154:148–157. doi: 10.1016/j.molbiopara.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Ishino T, Chinzei Y, Yuda M. Two proteins with 6-cys motifs are required for malarial parasites to commit to infection of the hepatocyte. Mol Microbiol. 2005;58:1264–1275. doi: 10.1111/j.1365-2958.2005.04801.x. [DOI] [PubMed] [Google Scholar]
- 9.Quakyi IA, et al. The 230-kDa gamete surface protein of Plasmodium falciparum is also a target for transmission-blocking antibodies. J Immunol. 1987;139:4213–4217. [PubMed] [Google Scholar]
- 10.Sanders PR, et al. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J Biol Chem. 2005;280:40169–40176. doi: 10.1074/jbc.M509631200. [DOI] [PubMed] [Google Scholar]
- 11.van Schaijk BC, et al. Pfs47, paralog of the male fertility factor Pfs48/45, is a female specific surface protein in Plasmodium falciparum. Mol Biochem Parasitol. 2006;149:216–222. doi: 10.1016/j.molbiopara.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury DR, Angov E, Kariuki T, Kumar N. A potent malaria transmission blocking vaccine based on codon harmonized full length Pfs48/45 expressed in Escherichia coli. PLoS ONE. 2009;4:e6352. doi: 10.1371/journal.pone.0006352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson KC. Pfs230: From malaria transmission-blocking vaccine candidate toward function. Parasite Immunol. 2003;25:351–359. doi: 10.1046/j.1365-3024.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 14.Gilson PR, et al. Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics. 2006;5:1286–1299. doi: 10.1074/mcp.M600035-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Kumar N. Target antigens of malaria transmission blocking immunity exist as a stable membrane bound complex. Parasite Immunol. 1987;9:321–335. doi: 10.1111/j.1365-3024.1987.tb00511.x. [DOI] [PubMed] [Google Scholar]
- 16.Eksi S, et al. Malaria transmission-blocking antigen, Pfs230, mediates human red blood cell binding to exflagellating male parasites and oocyst production. Mol Microbiol. 2006;61:991–998. doi: 10.1111/j.1365-2958.2006.05284.x. [DOI] [PubMed] [Google Scholar]
- 17.van Dijk MR, et al. A central role for P48/45 in malaria parasite male gamete fertility. Cell. 2001;104:153–164. doi: 10.1016/s0092-8674(01)00199-4. [DOI] [PubMed] [Google Scholar]
- 18.Healer J, et al. Complement-mediated lysis of Plasmodium falciparum gametes by malaria-immune human sera is associated with antibodies to the gamete surface antigen Pfs230. Infect Immun. 1997;65:3017–3023. doi: 10.1128/iai.65.8.3017-3023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vermeulen AN, et al. Sequential expression of antigens on sexual stages of Plasmodium falciparum accessible to transmission-blocking antibodies in the mosquito. J Exp Med. 1985;162:1460–1476. doi: 10.1084/jem.162.5.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vermeulen AN, et al. Plasmodium falciparum transmission blocking monoclonal antibodies recognize monovalently expressed epitopes. Dev Biol Stand. 1985;62:91–97. [PubMed] [Google Scholar]
- 21.van Dijk MR, et al. Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog. 2010;6:e1000853. doi: 10.1371/journal.ppat.1000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Schaijk BC, et al. Gene disruption of Plasmodium falciparum p52 results in attenuation of malaria liver stage development in cultured primary human hepatocytes. PLoS ONE. 2008;3:e3549. doi: 10.1371/journal.pone.0003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.VanBuskirk KM, et al. Preerythrocytic, live-attenuated Plasmodium falciparum vaccine candidates by design. Proc Natl Acad Sci USA. 2009;106:13004–13009. doi: 10.1073/pnas.0906387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dijk MR, et al. Genetically attenuated, P36p-deficient malarial sporozoites induce protective immunity and apoptosis of infected liver cells. Proc Natl Acad Sci USA. 2005;102:12194–12199. doi: 10.1073/pnas.0500925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott JF, et al. Genes for Plasmodium falciparum surface antigens cloned by expression in COS cells. Proc Natl Acad Sci USA. 1990;87:6363–6367. doi: 10.1073/pnas.87.16.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reeder JC, Wapling J, Mueller I, Siba PM, Barry AE. Population genetic analysis of the Plasmodium falciparum 6-cys protein Pf38 in Papua New Guinea reveals domain-specific balancing selection. Malar J. 2011;10:126. doi: 10.1186/1475-2875-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tetteh KK, et al. Prospective identification of malaria parasite genes under balancing selection. PLoS ONE. 2009;4:e5568. doi: 10.1371/journal.pone.0005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Bustamante PJ, et al. Differential ability of specific regions of Plasmodium falciparum sexual-stage antigen, Pfs230, to induce malaria transmission-blocking immunity. Parasite Immunol. 2000;22:373–380. doi: 10.1046/j.1365-3024.2000.00315.x. [DOI] [PubMed] [Google Scholar]
- 30.Farrance CE, et al. A plant-produced Pfs230 vaccine candidate blocks transmission of Plasmodium falciparum. Clin Vaccine Immunol. 2011;18:1351–1357. doi: 10.1128/CVI.05105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milek RL, Stunnenberg HG, Konings RN. Assembly and expression of a synthetic gene encoding the antigen Pfs48/45 of the human malaria parasite Plasmodium falciparum in yeast. Vaccine. 2000;18:1402–1411. doi: 10.1016/s0264-410x(99)00392-8. [DOI] [PubMed] [Google Scholar]
- 32.Outchkourov NS, et al. Correctly folded Pfs48/45 protein of Plasmodium falciparum elicits malaria transmission-blocking immunity in mice. Proc Natl Acad Sci USA. 2008;105:4301–4305. doi: 10.1073/pnas.0800459105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tachibana M, et al. N-terminal prodomain of Pfs230 synthesized using a cell-free system is sufficient to induce complement-dependent malaria transmission-blocking activity. Clin Vaccine Immunol. 2011;18:1343–1350. doi: 10.1128/CVI.05104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson KC, Keister DB, Muratova O, Kaslow DC. Recombinant Pfs230, a Plasmodium falciparum gametocyte protein, induces antisera that reduce the infectivity of Plasmodium falciparum to mosquitoes. Mol Biochem Parasitol. 1995;75:33–42. doi: 10.1016/0166-6851(95)02507-3. [DOI] [PubMed] [Google Scholar]
- 35.Angov E, Hillier CJ, Kincaid RL, Lyon JA. Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host. PLoS ONE. 2008;3:e2189. doi: 10.1371/journal.pone.0002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thanaraj TA, Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996;5:1594–1612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bax A, Grishaev A. Weak alignment NMR: A hawk-eyed view of biomolecular structure. Curr Opin Struct Biol. 2005;15:563–570. doi: 10.1016/j.sbi.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Williams DC, Jr, Cai M, Suh JY, Peterkofsky A, Clore GM. Solution NMR structure of the 48-kDa IIAMannose-HPr complex of the Escherichia coli mannose phosphotransferase system. J Biol Chem. 2005;280:20775–20784. doi: 10.1074/jbc.M501986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He XL, Grigg ME, Boothroyd JC, Garcia KC. Structure of the immunodominant surface antigen from the Toxoplasma gondii SRS superfamily. Nat Struct Biol. 2002;9:606–611. doi: 10.1038/nsb819. [DOI] [PubMed] [Google Scholar]
- 40.Crawford J, et al. Structural and functional characterization of SporoSAG: A SAG2-related surface antigen from Toxoplasma gondii. J Biol Chem. 2010;285:12063–12070. doi: 10.1074/jbc.M109.054866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 42.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 44.Anantharaman V, Iyer LM, Balaji S, Aravind L. Adhesion molecules and other secreted host-interaction determinants in Apicomplexa: Insights from comparative genomics. Int Rev Cytol. 2007;262:1–74. doi: 10.1016/S0074-7696(07)62001-4. [DOI] [PubMed] [Google Scholar]
- 45.Jung C, Lee CY, Grigg ME. The SRS superfamily of Toxoplasma surface proteins. Int J Parasitol. 2004;34:285–296. doi: 10.1016/j.ijpara.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 46.Crawford J, et al. Structural characterization of the bradyzoite surface antigen (BSR4) from Toxoplasma gondii, a unique addition to the surface antigen glycoprotein 1-related superfamily. J Biol Chem. 2009;284:9192–9198. doi: 10.1074/jbc.M808714200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graille M, et al. Crystal structure of the complex between the monomeric form of Toxoplasma gondii surface antigen 1 (SAG1) and a monoclonal antibody that mimics the human immune response. J Mol Biol. 2005;354:447–458. doi: 10.1016/j.jmb.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 48.Carruthers V, Boothroyd JC. Pulling together: An integrated model of Toxoplasma cell invasion. Curr Opin Microbiol. 2007;10:83–89. doi: 10.1016/j.mib.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 49.Lekutis C, Ferguson DJ, Grigg ME, Camps M, Boothroyd JC. Surface antigens of Toxoplasma gondii: Variations on a theme. Int J Parasitol. 2001;31:1285–1292. doi: 10.1016/s0020-7519(01)00261-2. [DOI] [PubMed] [Google Scholar]
- 50.Mineo JR, Kasper LH. Attachment of Toxoplasma gondii to host cells involves major surface protein, SAG-1 (P30) Exp Parasitol. 1994;79:11–20. doi: 10.1006/expr.1994.1054. [DOI] [PubMed] [Google Scholar]
- 51.Mineo JR, et al. Antibodies to Toxoplasma gondii major surface protein (SAG-1, P30) inhibit infection of host cells and are produced in murine intestine after peroral infection. J Immunol. 1993;150:3951–3964. [PubMed] [Google Scholar]
- 52.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 53.Garrett DS, Powers R, Gronenborn AM, Clore GM. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J Magn Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Clore GM, Gronenborn AM. Structures of larger proteins in solution: Three- and four-dimensional heteronuclear NMR spectroscopy. Science. 1991;252:1390–1399. doi: 10.1126/science.2047852. [DOI] [PubMed] [Google Scholar]
- 55.Bax A, Grzesiek S. Methodological advances in protein NMR. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 56.Bax A, et al. Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol. 1994;239:79–105. doi: 10.1016/s0076-6879(94)39004-5. [DOI] [PubMed] [Google Scholar]
- 57.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 58.Clore GM, Starich MR, Gronenborn AM. Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J Am Chem Soc. 1998;120:10571–10572. [Google Scholar]
- 59.Clore GM, Gronenborn AM, Bax A. A robust method for determining the magnitude of the fully asymmetric alignment tensor of oriented macromolecules in the absence of structural information. J Magn Reson. 1998;133:216–221. doi: 10.1006/jmre.1998.1419. [DOI] [PubMed] [Google Scholar]
- 60.Grzesiek S, Bax A. The importance of not saturating H2O in protein NMR—Application to sensitivity enhancement and NOE measurements. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- 61.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwieters CD, Clore GM. Internal coordinates for molecular dynamics and minimization in structure determination and refinement. J Magn Reson. 2001;152:288–302. doi: 10.1006/jmre.2001.2413. [DOI] [PubMed] [Google Scholar]
- 63.Schwieters CD, Kuszewski J, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 64.Schwieters CD, Clore GM. The VMD-XPLOR visualization package for NMR structure refinement. J Magn Reson. 2001;149:239–244. doi: 10.1006/jmre.2001.2300. [DOI] [PubMed] [Google Scholar]
- 65.Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.