Abstract
The degradation of the plant cell wall by glycoside hydrolases is central to environmentally sustainable industries. The major polysaccharides of the plant cell wall are cellulose and xylan, a highly decorated β-1,4-xylopyranose polymer. Glycoside hydrolases displaying multiple catalytic functions may simplify the enzymes required to degrade plant cell walls, increasing the industrial potential of these composite structures. Here we test the hypothesis that glycoside hydrolase family 43 (GH43) provides a suitable scaffold for introducing additional catalytic functions into enzymes that target complex structures in the plant cell wall. We report the crystal structure of Humicola insolens AXHd3 (HiAXHd3), a GH43 arabinofuranosidase that hydrolyses O3-linked arabinose of doubly substituted xylans, a feature of the polysaccharide that is recalcitrant to degradation. HiAXHd3 displays an N-terminal five-bladed β-propeller domain and a C-terminal β-sandwich domain. The interface between the domains comprises a xylan binding cleft that houses the active site pocket. Substrate specificity is conferred by a shallow arabinose binding pocket adjacent to the deep active site pocket, and through the orientation of the xylan backbone. Modification of the rim of the active site introduces endo-xylanase activity, whereas the resultant enzyme variant, Y166A, retains arabinofuranosidase activity. These data show that the active site of HiAXHd3 is tuned to hydrolyse arabinofuranosyl or xylosyl linkages, and it is the topology of the distal regions of the substrate binding surface that confers specificity. This report demonstrates that GH43 provides a platform for generating bespoke multifunctional enzymes that target industrially significant complex substrates, exemplified by the plant cell wall.
Keywords: biofuels, biotechnology, protein engineering, structural biology
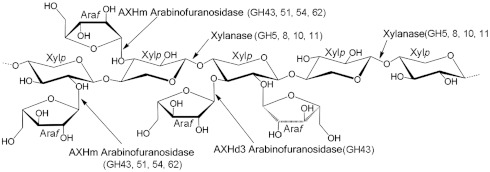
Enzymes that degrade the plant cell wall, typically glycoside hydrolases, are of increasing industrial significance, particularly in the environmentally relevant bioenergy and bioprocessing sectors. The plant cell wall contains a large number of chemically complex polysaccharides (1), exemplified by xylans, which are polymers of β-1,4-xylopyranose (Xylp) residues that are extensively decorated with O2- and/or O3-linked arabinofuranose (Araf), O2-linked uronic acids and acetyl groups at either O2 or O3. In common with many polysaccharides in the plant cell wall, the decorations of the main chain of xylans hinder the access of enzymes to the backbone structures of this polymer (2). Thus, the side chains in xylan must be removed by arabinofuranosidases, esterases, and glucuronidases before the xylan backbone can be hydrolyzed by endo-acting xylanases (see ref. 2 for review). Single Araf substitutions that decorate Xylp residues are removed by a myriad of arabinofuranosidases (defined as AXHm2,3). By contrast, backbone sugars, decorated at both O2 and O3 with Araf units, are particularly recalcitrant to enzymatic attack; currently only two arabinofuranosidases, defined as AXHd3s, are known to cleave these linkages (3, 4) (Fig. 1). The activity displayed by AXHd3s is of biotechnological significance. Indeed, the AXHd3 from Humicola insolens, designated hereafter as HiAXHd3, is a component of the Novozyme product Ultraflo L, which is used in the brewing industry (3).
Fig. 1.
Schematic of the different linkages in arabinoxylan. The structure of a model arabinoxylan and the enzymes that hydrolyze the polysaccharide. Araf indicates arabinofuranose residues and Xylp depicts xylopyranose units in the xylan backbone. The major glycoside hydrolases families that contain these enzyme activities are shown in parentheses.
Glycoside hydrolases are grouped into sequence-based families (GHs) in which specificity may vary (5). The mode of action of these enzymes reflects the topology of the active site/substrate binding region. Thus, endo-acting glycoside hydrolases, which cleave internal linkages, display a substrate binding cleft open at both ends, whereas the active site of exo-acting enzymes that target the termini of polysaccharide backbones display a substrate binding groove blocked at one end by extended loops. Endo and backbone exo-acting enzymes are closely related; in nature, short loop extensions have led to the conversion of endo-glycanases into exo-glycanases that target the polysaccharide backbone (6–8). The active site of enzymes that cleave sugars decorating the polysaccharide backbone displays a pocket topology in which the target side chain is positioned (9, 10). In contrast to enzymes that target the backbone of complex carbohydrates, the introduction of endo activity into a glycoside hydrolase that removes polysaccharide side chains has not been reported. It is believed that opening up the catalytic pocket, such that it can accommodate the polysaccharide backbone, disrupts the integrity of the active site and compromises its capacity to make productive interactions with the substrate.
Glycoside hydrolase family 43 (GH43) enzymes display a five-bladed β-propeller fold (11) in which the active site may feature a tight pocket, exemplified by side chain cleaving arabinofuranosidases (AXHm2,3s and AXHd3s), exo-acting arabinanases, and β-xylosidases (9, 12, 13), or endo-acting arabinanases (11, 14) and xylanases. AXHm2,3 arabinofuranosidases target single substitutions, presumably because the active site is enclosed, and are thus likely to make steric clashes with substrates in which the backbone sugar is decorated at both O2 and O3. By contrast, the mechanism by which AXHd3 arabinofuranosidases not only accommodate a double substitution but also display absolute specificity for such structures is unknown. A distinctive feature of GH43 is that the active site of the β-xylosidases and arabinofuranosidases are highly conserved (10) (Fig. S1), consistent with the observation that β-xylosidases and some arabinofuranosidases are capable of also cleaving α-arabinofuranosyl and β-xylopyranosyl linkages, respectively (12, 15) . It would seem, therefore, that substrate binding regions distal to the active site contribute to the specificity of GH43 enzymes. Thus, GH43 provides an excellent model system for testing the hypothesis that endo catalytic functions can be introduced into exo-acting enzymes that target polysaccharide side chains.
To explore the potential use of rational design to engineer the specificity of AXHd3 enzymes, the crystal structure of HiAXHd3 was determined. The data showed that the active site of HiAXHd3 comprises a deep pocket that houses the catalytic apparatus and a shallower adjacent pocket that confers specificity for double substitutions by binding to the O2-linked Araf component of these structures. Interactions with the xylan backbone play a key role in directing the O3-linked Araf into the active site and the O2 Araf into the distal site. Significantly, alteration of the active site, by the mutation of tyrosine 166 to alanine, generated an enzyme that not only displays exo-arabinofuranosidase activity but also, uniquely, endo-xylanase activity. The crystal structure of the Y166A mutant revealed the molecular basis for the dual endo- and exo-activities displayed by the HiAXHd3 variant. The incorporation of additional catalytic functions into the active site of an arabinofuranosidase demonstrates how the GH43 fold provides a platform for generating multifunctional enzymes.
Results
HiAXHd3 Targets Arabinoxylan and Arabinan.
Purified, recombinant HiAXHd3, in addition to releasing Araf from arabinoxylan, was active against sugar beet arabinan, which contains Araf side chains, but did not hydrolyze linear arabinan, in which the α-L-1,5-Araf-containing backbone is not decorated (Table 1 and Table S1). Arabinan-derived oligosaccharides, which are substrates for HiAXHd3, were subjected to 2D NMR analysis before and after treatment with the enzyme. The chemical shifts corresponding to the anomeric protons of Araf units substituted at both O2 and O3 were lost after treatment with HiAXHd3 (Fig. S2). Similarly, chemical shifts were lost for the anomeric proton from the respective α-L-Araf residues attached to O2 of 2,3,5-linked Araf (a backbone Araf unit that is substituted at both O2 and at O3). The chemical shift peak heights increased for the anomeric protons of α-L-Araf residues that contain a single O2 substitution, and Araf units linked O2 to the arabinan backbone. These NMR data reveal that HiAXHd3 displays specificity for the α-1,3-Araf linkages in arabinose moieties in the arabinan backbone, which contain substitutions at both O2 and O3. Thus, HiAXHd3 targets the same linkage in both arabinan and arabinoxylan.
Table 1.
The catalytic activity of wild-type HiAXHd3, Y166A, and its variants
| Wheat arabinoxylan (arabinose release) | Sugar beet arabinan (arabinose release) | Birchwood xylan (reducing sugar release) | |||||||
| kcat (min-1) | KM (M) | kcat/KM (min-1 M-1) | kcat (min-1) | KM (M) | kcat/KM (min-1 M-1) | kcat (min-1) | KM (mg ml-1) | kcat/KM (min-1 mg-1 ml) | |
| Wild type | 1.0( ± 0.13) × 104 | 3.5( ± 1.4) × 10-4 | 3.0 × 107 | 1.7( ± 0.1) × 103 | 1.3( ± 0.3) × 10-4 | 1.4 × 107 | Inactive | ||
| Y166A | 37.5 ± 8.0 | 2.1( ± 2.0) × 10-4 | 1.8 × 105 | 3.0 ± 0.8 | 6.2( ± 0.6) × 10-5 | 4.8 × 104 | 15.8 ± 3.7 | 3.5 ± 2.7 | 4.5 |
| Y166A/N184A | 26.3 ± 1.5 | 1.0( ± 2.9) × 10-4 | 2.6 × 105 | 12.5 ± 2.0 | 7.1( ± 3.2) × 10-4 | 1.8 × 104 | 78.4 ± 16 | 2.2 ± 1.7 | 35.5 |
| Y166A/H272A | Inactive | Inactive | 11.6 ± 2.7 | 0.9 ± 0.7 | 13.5 | ||||
| Y166A/P234A | 4.95 ± 0.8 | 3.0( ± 1.7) × 10-4 | 1.6 × 104 | 1.7 ± 0.3 | 8.3( ± 4.0) × 10-4 | 2.0 × 103 | 30.2 ± 8.2 | 6.8 ± 4.7 | 4.4 |
| Y166A/F493A | 121.5 ± 40 | 4.9( ± 2.4) × 10-4 | 2.5 × 105 | 50.4 ± 10.6 | 9.4( ± 4.5) × 10-4 | 5.4 × 104 | 98.0 ± 21 | 5.4 ± 3.3 | 18.1 |
| Y166A/F289A | 11.8 ± 1.8 | 1.9( ± 0.7) × 10-3 | 6.3 × 103 | 2.2 ± 0.70 | 7.2( ± 5.8) × 10-4 | 3.1 × 103 | 26.8 ± 4.5 | 4.0 ± 2.1 | 6.7 |
| Y166A/F270A | 16.6 ±1.7 | 2.7( ± 0.9) × 10-4 | 6.1 × 104 | 2.2 ± 0.2 | 2.3( ± 0.6) × 10-4 | 9.4 × 103 | 16.8 ± 4.0 | 4.2 ± 1.8 | 4.0 |
| Y166A/G183S | 41.5 ± 6.7 | 8.8( ± 1.1) × 10-5 | 4.7 × 105 | Not determined | 123 ± 16 | 4.8 ± 0.1 | 25.6 | ||
| Y166A/N184G | 150.6 ± 9.5 | 1.0( ± 0.2) × 10-4 | 1.5 × 106 | Not determined | 437 ± 31 | 4.2 ± 0.10 | 104 | ||
| Y166A/F493T | 52.6 ± 4.2 | 1.3( ± 0.2) × 10-4 | 4.0 × 105 | Not determined | 76 ± 12 | 2.6 ± 0.0 | 29 | ||
| Y166A/G183S/N184G/F493T | 1.5( ± 0.8) × 103 | 5.6( ± 0.8) × 10-4 | 2.7 × 106 | Not determined | 1,145 ± 140 | 7.5 ± 1.1 | 243 | ||
| Y166G/G183S/N184G/F493T | 1.1( ± 0.2) × 103 | 1.4( ± 0.1) × 10-4 | 7.9 × 106 | Not determined | 3267±356 | 6.9±0.7 | 473 | ||
| Y166A/D43A | Inactive | Inactive | Inactive | ||||||
| Y166A/D167A | Inactive | Inactive | Inactive | ||||||
| Y166A/E216A | Inactive | Inactive | Inactive | ||||||
Crystal Structure of HiAXHd3.
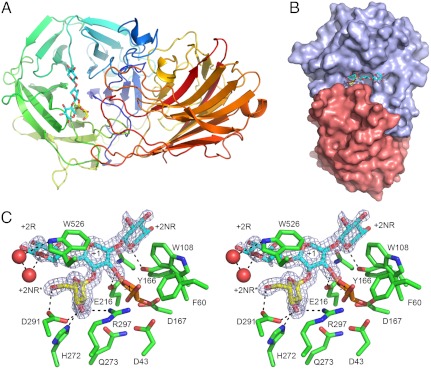
To visualize the substrate specificity of HiAXHd3 in atomic detail, its crystal structure was solved by single wavelength anomalous dispersion and refined to 1.8 Å resolution (Table S2). HiAXHd3 consists of two distinct domains that are linked by the sequence extending from Val330 to Gly339, which has relatively high B-factors compared to the rest of the protein, indicating a degree of conformational flexibility, Fig. 2A. The N-terminal domain, consisting of residues 28–329, displays a five-bladed β-propeller fold, typical of GH43 enzymes (11). The C-terminal domain, extending from amino acids 340–558, displays a canonical β-sandwich fold. A phylogenetic analysis of GH43 revealed a cohort of β-xylosidases and α-arabinofuranosidases, including the Bifidobacterium AXHd3, which also contain a C-terminal β-sandwich domain (16). The two domains in HiAXHd3 bury some 2,100 Å2 of the total protein surface at the domain interface (Fig. 2B), indicating that the stable folding of each domain is dependent upon the other. A solvent-exposed shallow cleft is presented at the interface between the two domains (Fig. 2 B and C). The N-terminal β-propeller domain, primarily blades 4 and 5 and their associated loops, provides one side of the cleft. The other face of the cleft is presented by the loops connecting β-strands 9 and 10, and β-strands 10 and 11, which are both located in the C-terminal domain. In contrast to HiAXHd3, in the other two-domain GH43 enzymes, for which crystal structures are available, the β-sandwich domains do not contribute to an interdomain cleft. In the central region of the HiAXHd3 cleft is a depression that houses the active site pocket (see below).
Fig. 2.
Structure of HiAXHd3. (A) The overall structure of HiAXHd3 in complex with Araf-Xylp3. The protein is shown as a cartoon, color-ramped from the N terminus (blue) to C terminus (red); the sugar is displayed as sticks, with xylose units depicted in cyan and arabinose in yellow. (B) The solvent accessible surface of HiAXHd3 in complex with Araf-Xylp3. The β-propeller domain is colored slate and the β-sandwich in salmon. (C) Wall-eyed stereo view of the active site, showing protein residues and the sugar as sticks with the maximum likelihood/σa-weighted 2mFobs-DFcalc electron density map for the carbohydrate shown in blue and contoured at 1σ (0.54 e-/Å3). The experimental model for the Araf-Xylp3 is shown in yellow and blue for the arabinose and xylose, respectively; arabinose has been modeled into the -1 subsite based on the positions of crystallographic waters and is shown in orange. The dotted lines show polar interactions between the ligand and the protein. This figure, and other structure figures, were drawn with PyMol (DeLano Scientific; http://pymol.sourceforge.net/).
Crystal Structure of HiAXHd3 in Complex with Ligands.
To explore the mechanism of substrate recognition in HiAXHd3, crystals of the inactive mutant, D43A (Asp43 is the catalytic base, see below), were soaked with arabinoxylan-derived oligosaccharides. These ligands were partially purified (size fractionation) from xylanase digestion of wheat arabinoxylan and contained several oligosaccharides including substrates containing a xylose decorated at O2 and O3 with Araf. The generation of these molecules is described in Supporting Information. In the subsequent crystal structure, clear electron density was observed corresponding to a xylotriose with a single α-Araf side chain, linked O2 to the central Xylp (defined as Xyl-2). Xylan chains are pseudosymmetrical and fit equally well into electron density in the two opposite orientations. The correct orientation was chosen in this case based on the observed hydrogen bonds to solvent and protein from O5 atoms (whose equivalent would be C5 in the reverse orientation), and the requirement for O3 to point into the active site for catalysis to occur. The xylotriose unit, and by inference the xylan backbone, is housed in the central region of the cleft. Weak electron density extends beyond both the reducing and nonreducing ends of the xylotriose molecule, suggesting that the bound ligand consists of a backbone that is longer, at least a xylopentaose unit, although the terminal saccharide units could not be modeled satisfactorily. Using the standard nomenclature for identifying substrate binding subsites (17), in which the scissile glycosidic bond links the sugars at the -1 and +1 subsites, in HiAXHd3 the xylotriose unit occupies subsites +2NR to +2R (the subsites that extend from the nonreducing and reducing end of the +1 sugar, respectively) and the Araf is positioned at subsite +2NR∗, which accommodates sugar decorations appended to the +1 Xylp (Figs. 2C and 3A). Despite numerous changes to the soaking procedures, no Araf moiety was observed in the -1 subsite, which comprises the active site, suggesting that the mutant displays trace catalytic activity or that a reaction product contaminant has bound preferentially to the enzyme due to its greater abundance.
Fig. 3.
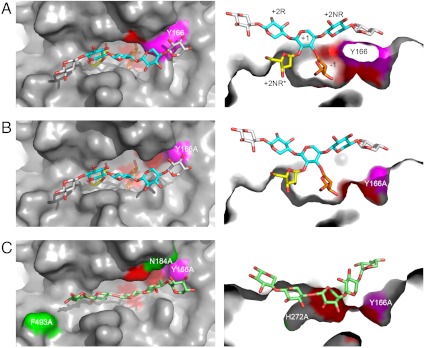
The active site cleft of HiAXHd3. (A) Orthogonal views of the active site cleft of the wild-type HiAXHd3 protein shown as a molecular surface with the sugar ligand shown as sticks. The experimental structure of Araf-Xylp3 is shown in yellow and blue, respectively, for arabinose and xylose; a modeled Ara f is shown in orange in the -1 subsite; two additional xylose residues are modeled onto the sugar backbone, shown in gray. The position of Y166 is shown in magenta and the active site residues D43, D167, and E216 in red. Left is a top view and Right is a cut-away view through the active cleft rotated 90° into the plane of the paper. (B) The active site cleft of the Y166A/E216A mutant shown as for A; the removal of the Y166 side chain increases the accessibility of the active site residues to sugar ligands. (C) Mutations that increase the xylanase activity of the Y166A HiAXHd3 mutant are shown highlighted on the surface of the protein in green (N184A and F493A; H272A is not visible from the bird’s eye view). Right is a view of the surface of the Y166A mutant with the modeled mutations showing how a Xyl5 backbone (light-green sticks), can be accommodated in the active cleft made by these mutations.
The Active Site of HiAXHd3.
The active site of HiAXHd3, located in the center of the xylotriose binding cleft, contains a constellation of carboxylate residues, Asp43, Asp167, and Glu216, which are invariant within GH43. The equivalent residues have been shown to comprise the catalytic amino acids in other members of the family (7, 11–13) (Fig. S3). The observation that D43A, D167A, and E216A are inactive (Table S1) supports the view that these three acidic residues comprise the catalytic apparatus of HiAXHd3. The O3 of the +1 Xylp makes a hydrogen bond with Oϵ1 of Glu216, the glutamate is thus in an ideal position to function as the catalytic acid, donating a proton to the scissile glycosidic oxygen. The elevated pKa required of a catalytic acid is provided by the hydrogen bond with Asp167. The conservation of Asp43 with the catalytic base in other GH43 enzymes indicates that the aspartate fulfills this function in HiAXHd3. In addition to the catalytic residues, the -1 subsite of HiAXHd3 contains several amino acids that are highly conserved in GH43, which have been shown to interact with the Araf bound in the active site of the exo-α-1,5-arabinofuranosidase (13) (Fig. S1). The complete loss in activity when these residues are substituted for alanine in HiAXHd3, supports the view that these amino acids play a central role in substrate binding (Table S1).
Structural Basis for Selection of O2 and O3 Double Substituted Substrates.
A key feature of HiAXHd3 is its absolute specificity for structures in which the backbone sugar is linked at both O2 and O3 to Araf units. These double substitutions are recalcitrant to AXHm arabinofuranosidases. Thus, HiAXHd3 is important in xylan degradation, as its capacity to target doubly substituted xylans makes this abundant polysaccharide accessible to AXHm arabinofuranosidases and endo-acting xylanases. Within the context of a double substitution, the O2-linked Araf is a critical specificity determinant for HiAXHd3. The structure of the ligand complex reveals an Araf in a shallow pocket, extending from the deeper active site pocket, which therefore comprises the +2NR∗ subsite (Fig. 2C). In addition to the hydrophobic interactions with Trp526, the sugar makes several polar contacts with the enzyme. Thus, Asp291 makes bidentate hydrogen bonds with O5 and O3 and O2 makes a polar contact with His272, while O3 also makes a hydrogen bond with Nη1 of Arg297 (Fig. 2C). The extensive interactions between the O2-linked arabinose and HiAXHd3 explains why the furanose sugar is a critical specificity determinant for double substitutions. Indeed, the importance of these polar interactions is provided by the observation that the mutants R297A and D291A are completely inactive, whereas the H272A mutation causes a 1,000-fold decrease in kcat, although it had no effect on KM (Table S1).
An intriguing feature of HiAXHd3 is its specificity for O3 linkages. Araf side chains can rotate around the glycosidic linkage, and the exocyclic C5 is also free to rotate, thus it is difficult to understand how the enzyme is able to select a specific linkage. It is possible that interactions with the backbone polysaccharide orient the O3-linked arabinofuranose into the -1 subsite. The only asymmetric feature of xylan is the endocyclic oxygen. It is likely, therefore, that HiAXHd3, by targeting these ring oxygens, binds to xylan chains in a specific orientation. In the D43A mutant the xylotriose makes relatively few interactions with the enzyme, which likely aids product departure, because the cleaved Araf cannot be released from the active site until the Xylp polymer dissociates from the enzyme. The endocyclic oxygen of the Xylp at the +2NR subsite makes a hydrogen bond with the OH of Tyr166, whereas solvent-mediated hydrogen bonds link the Nϵ1 of the indole ring of Trp526 with the ring oxygen of the +2R Xylp (Fig. 2C). Mutation of either of these amino acids results in a decrease in activity (Table 1 and Table S1) consistent with the importance of the residues in substrate binding. Significantly, the 1D NMR spectrum of arabinoxylan, after incubation with the wild-type enzyme or the Y166A mutant, revealed a ratio of single O2- and O3-linked Araf of approximately 1, whereas in the untreated polysaccharide the signal for single O2-linked Araf residues is very low (Fig. S4). Thus, Tyr166 does not contribute to the capacity of the enzyme to target the O3-linked Araf. By contrast, arabinoxylan treated with the W526A mutant displayed a ratio of O2-Araf:O3-Araf of 0.47∶1.0 (Fig. S4). Thus, the W526A mutant displayed a relaxation in its substrate specificity, because it can hydrolyze either O2- or O3-linked Araf units in a double substitution background. It would appear, therefore, that Trp526, by orienting the xylan backbone, confers the observed specificity of wild-type HiAXHd3 for O3 arabinose residues within doubly substituted structures.
Altering the Specificity of HiAXHd3.
The active site of GH43 β-D-xylosidases and α-L-arabinofuranosidases are highly conserved (10), and thus it is not clear how these enzymes select β-D-xylosyl and α-L-arabinofuranosyl linkages, respectively, in natural substrates. It is possible that this flexibility in substrate recognition could be harnessed to introduce novel functionalities into the active site of selected GH43 enzymes. For example, modification of distal subsites could be exploited to direct a xylosyl linkage into the active site of an arabinofuranosidase, generating an enzyme that hydrolyses both β-D-xylosyl and α-L-arabinofuranosyl linkages. In addition to altering the chemistry of the reactions catalyzed by these enzymes, it might also be possible to modify their mode of action. The active sites of endo- and exo-acting GH43 enzymes are housed in an open cleft and a pocket, respectively. Thus, if the pocket topology of xylan-specific arabinofuranosidases was disrupted, the distal subsites of such enzymes could be used to direct the xylan chain into the active site, thereby introducing an endo-xylanase function. Tyr166, which is not conserved in GH43 β-xylosidases, is located at the periphery of the active site of HiAXHd3, orienting the xylan chain over the active site. Thus, substituting this tyrosine with alanine may introduce some flexibility into the orientation of the xylan chain in the substrate binding cleft, while the mutation may also open up the active site pocket to allow the backbone component of the polysaccharide to enter the active site. To test this hypothesis the substrate specificity of the Y166A mutant was explored. High performance anion exchange chromatography (HPAEC) analysis of wheat arabinoxylan hydrolysis by Y166A shows that, in addition to arabinose, several other larger reaction products were evident (Fig. S5). Because wheat arabinoxylan contains short side chains (predominantly monosaccharides) the large products generated are predicted to be derived from the hydrolysis of the xylan backbone. These data indicate that Y166A not only displays arabinofuranosidase activity for doubly substituted Xylp residues (retains its AXHd3 activity) but also exhibits detectable endo-xylanase activity. The Y166A mutation did not alter the temperature or pH optimum of the enzyme (55 °C and pH 7.0, respectively). To explore whether the introduced xylanase activity is dependent on the Araf decorations, the capacity of the Y166A variant to hydrolyze birchwood xylan, which contains very limited side chains of the furanose sugar and is not a substrate for the wild-type enzyme (Table 1), was assessed. HPAEC revealed a range of reaction products that were identified as xylooligosaccharides, based on their comigration with appropriate standards, Fig. S5 and by their hydrolysis to xylose by a β-xylosidase. The xylooligosaccharides had a degree of polymerization (d.p.)≥3, although, as the reaction approached completion, xylobiose was also observed. The kinetic parameters of the arabinofuranosidase and xylanase activities displayed by Y166A were broadly similar (Table 1). To further investigate the specificity and mode of action of Y166A, the capacity of this enzyme variant to hydrolyze xylooligosaccharides was investigated. HPAEC analysis showed that although the enzyme displayed no activity against oligosaccharides with a d.p. of 2–4, xylopentaose was converted to xylotriose and xylobiose, whereas xylohexaose was cleaved exclusively into xylotriose. The catalytic efficiency of the enzyme was 30-fold greater against the hexaose (3.45 × 104 min-1 M-1) compared to the pentaose (1.26 × 103 min-1 M-1). These data demonstrate that the Y166A mutation confers xylanase activity on HiAXHd3, while the enzyme variant retains its arabinofuranosidase function, albeit at a reduced level.
To determine if the xylanase and AXHd3 activity of Y166A utilize the same active site, the effect of removing the three catalytic residues on the catalytic function of the HiAXHd3 variant was assessed. The mutants Y166A/E216A, Y166A/D167A, and Y166A/D43A displayed no arabinofuranosidase or xylanase activity (Table 1) indicating that the original active site of HiAXHd3 is now uniquely capable of catalyzing the hydrolysis of the xylan backbone and the removal of the Araf side chains in the Y166A mutant. The conversion of an exo-acting arabinofuranosidase into a glycoside hydrolase that now displays both exo-arabinofuranosidase and endo-xylanase activities from the same active site is unprecedented among glycoside hydrolases, either engineered or natural. The crystal structure of the Y166A mutant (Fig. 3) showed that the mutation disrupted the lip of the active site pocket directly below the glycosidic oxygen between the +1 and +2R Xylp residues. This modification enables a modeled xylan chain to adopt a deeper position in the enzyme such that it can enter the active site and occupy the -1 subsite, perhaps explaining how the Y166A mutation has introduced xylanase activity into HiAXHd3.
Several other residues that line the substrate binding cleft of AXHd3 are likely to contribute to the engineered xylanase activity. The significance of these amino acids was explored by further mutagenesis within the Y166A background (Table 1). Mutation of His272 (Y166A/H272A) abolished arabinofuranosidase activity, consistent with its role in the recognition of O2-linked Araf, but increased the xylanase activity of the HiAXHd3 variant threefold, primarily through a reduction in KM. As discussed above, His272 is located in the shallow pocket that abuts onto the active site. Removal of the imidazole side chain from position 272 presumably provides room for the xylan chain to occupy the 2NR* subsite, facilitating the targeting of backbone Xylp residues into the active site. The introduction of the N184A and F493A mutations into the Y166A background increased the catalytic efficiency of the xylanase activity displayed by the HiAXHd3 variant eight- and fourfold, respectively, mainly through an elevation in kcat, but had no substantial influence on the arabinofuranosidase activity of the mutant (Table 1). Phe493 contributes to the floor of the distal region of the xylan binding cleft that houses the reducing end of the polymer. Removal of the bulky phenolic side chain may allow the xylan chain to sit deeper in the cleft and thus enhance the capacity of main chain Xylp units to access the active site. Asn184 is located close to the active site of the enzyme and, through Nδ2, makes a hydrogen bond with Oϵ1 of the catalytic acid, Glu216. It is possible that the loss of this polar contact may increase the capacity of the catalytic apparatus to access xylosidic bonds within the xylan backbone. To explore further the extent to which forced protein evolution can be used to improve the dual function of the enzyme, Gly183, Asn184, and Phe493 were randomized within the Y166A background. The single mutations that had the largest impact on xylanase activity were G183S (sixfold), N184G (22-fold), and F493T (sevenfold). When these three mutations were combined the catalytic efficiency increased (over the activity of Y166A) 50-fold for the xylanase activity and 15-fold for the AXHd3 activity. Finally, Tyr166 was randomized in the G183S-N184G-F493S treble mutant, and the Y166G mutation increased activity a further two- and threefold for the xylanase and AXHd3 activities, respectively. Thus, the activity of the quadruple mutant Y166G-G183S-N184G-F493T displayed an arabinofuranosidase activity approaching that of wild-type HiAXHd3 and a xylanase activity approximately 100-fold greater than the original Y166A mutant. To summarize, it is possible to significantly enhance the xylanase activity, introduced into HiAXHd3 by the Y166A mutation, and recover the AXHd3 activity, through the modification of distal subsites.
Discussion
This report describes the structure of an unusual arabinofuranosidase that displays absolute specificity for the O3-linked Araf in the context of a double substitution and shows how additional catalytic functions can be engineered into the active site of the enzyme. In contrast to the narrow active site pockets of exo-acting GH43 enzymes that target mono substitutions, HiAXHd3 features a wider catalytic center that contains an auxiliary pocket that binds to the O2-Araf, while the O3-Araf is directed into the active site. Despite containing many of the residues that are conserved in the active sites of other GH43 AXHm2,3 arabinofuranosidases that act on mono substitutions, HiAXHd3 utilizes the O2-linked Araf as an essential specificity determinant. The spatially restricted active sites of AXHm2,3 arabinofuranosidases are likely optimized for the binding and the hydrolysis of single arabinose side chains. In HiAXHd3, however, the wider active site presumably allows more rapid solvent exchange and thus the enzyme relies on the additional binding energy provided by the O2 side chain. Specificity for the O3 side chain is conferred by orienting the direction of the polysaccharide backbone through solvent-mediated polar contacts between Trp526 and the endocyclic oxygen of Xylp at the +2 NR subsite.
This report shows that an endo-xylanase function can be introduced into a HiAXHd3 variant that retains its exo-acting arabinofuranosidase activity. The mutation leading to this activity, Y166A, disrupts the lip of the active site pocket, allowing entry of a backbone Xylp into the active site of the arabinofuranosidase. The engineered enzyme is thus able to make productive interactions with L-furanose and D-pyranose pentose sugars that display different stereochemistry at C4. This has some resonance with studies showing that GH3 and GH43 β-xylosidases can display low levels of arabinofuranosidase activity (12, 15), whereas a GH51 arabinofuranosidase is also able to hydrolyze β-xylosyl linkages (18). The capacity to hydrolyze both α-L-Araf and β-D-Xylp linkages is consistent with the observation that the two sugars share considerable spatial similarity, which is particularly evident when one considers the free rotation of the Araf exocyclic C5 hydroxymethyl group. The structural similarity of the sugars, and the cross specificity of these enzymes, explain why the active sites of GH43 β-xylosidases and α-arabinofuranosidases are highly conserved, (Fig. S1). It would appear, therefore, that substrate specificity in GH43 exo-glycosidases is dominated by the topology of the distal subsites, exemplified by curved and linear substrate binding surfaces presented by arabinan- and arabinoxylan-specific arabinofuranosidases, respectively (10, 14).
The capacity of a single active site to hydrolyze glycosidic linkages in the backbone of a polysaccharide, and to remove sugar side chains from the same polysaccharide, thus displaying both an endo and exo function, has not previously been observed in nature or generated through rational design or forced protein evolution. This flexibility in substrate recognition is consistent with the range of activities displayed by GH43 enzymes that include exo, endo, and endo-processive modes of action against β-D-pyranose and α-L-furanose substrates where the stereochemistry at C4 varies. Indeed, the capacity to introduce a unique specificity into HiAXHd3, through a single amino acid substitution, indicates that the five-bladed β-propeller fold, displayed by GH43 enzymes, provides a structural scaffold that can be harnessed to bind a range of different sugars and to catalyze the hydrolysis of glycosidic bonds through distinct modes of action. The introduction of xylanase activity into an AXHd3 arabinofuranosidase provides a proof of principle for engineering additional catalytic functions into the active site of glycoside hydrolases, without the loss of the original activity of the enzyme. This work therefore provides a platform for the future generation of enzymes with multiple catalytic functions, which may simplify the enzyme cocktails used in plant cell wall degradation.
Materials and Methods
Cloning, Expression, Purification, and Mutagenesis of HiAXHd3.
Recombinant HiAXHd3 was expressed from Escherichia coli. Details of the cloning of the gene, its expression, purification, mutagenesis, and screening of the randomized libraries are provided in Supporting Information.
Crystallization and Structure Solution of HiAXHd3.
The three structures reported herein were refined to final Rcryst and Rfree of 15.13/18.47% for seleno-methionine HiAXHd3, 16.67/20.81% for the D43A mutant, and 14.16/18.70% for the Y166A/E216A mutant, at resolutions of 1.84, 1.44, and 1.87 Å, respectively. Full information on crystallization, data collection and structure solution, refinement, and statistics are available in Supporting Information and in Table S2.
Enzyme Assays and Enzyme Screening.
The sources of the polysaccharide and oligosaccharide substrates used are detailed in Supporting Information. The arabinofuranosidase activity of HiAXHd3 was determined in 50 mM Na-Hepes buffer (pH 7.5), containing 0.2 to 20 mg/mL of substrate, 1 mg/mL BSA, 1 mM NAD+, 0.03 to 0.8 μM HiAXHd3, and 2.5 units (1 unit generates 1 μmole of product/min) of galactose dehydrogenase. The reaction was incubated at 37 °C and the generation of NADH was monitored at A340nm. For AXHd3 reactions the molar concentration of polysaccharide was determined by quantifying the moles of arabinose released per milligram of polysaccharide when the reaction has gone to completion. Reaction products were identified by HPAEC as described previously (19). Xylanase activity was determined using reducing sugar assays or through substrate depletion experiments (20).
1H and 13C NMR Spectroscopy
Polysaccharides or oligosaccharides were dissolved in D2O (99.9%; Cambridge Isotope Laboratories). 1H-NMR spectra were recorded with a Varian Inova NMR spectrometer operating at 600 MHz and with a sample temperature of 298 K. All 2D spectra were recorded using standard Varian pulse programs. The COSY spectra were collected with 800 × 1,024 complex points. The data were processed with shifted squared sinebell window functions and zero filled to obtain a 2,048 × 2,048 matrix. Heteronuclear spectra were recorded with 512 × 512 complex points. These data were processed typically with zero filling to obtain a 1,024 × 1,024 matrix. Chemical shifts were measured relative to internal acetone (δH 2.225, δC 30.89). Data were processed using MestRe-C software (Universidad de Santiago de Compostela, Spain).
Supplementary Material
ACKNOWLEDGMENTS.
The authors acknowledge support from the U.K. Forestry Commission for their contribution to the European Union WoodWisdom-Net ERA-NET project FibreSurf; the U.K. Biotechnology and Biological Sciences Research Council Sustainable Bioenergy Centre (Grant BB/G016186/1 and BB/G016224/1); U.S. Department of Energy (DOE) Bioenergy Research Center (BESC) supported by the Office of Biological and Environmental Research in the DOE Office of Science. We also thank the DOE-funded Center for Plant and Microbial Complex Carbohydrates (Grant DE-FG02-93ER20097) for supporting critical infrastructure and analytical instrumentation required for this research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3ZXJ (wild-type HiAXHd3), 3ZXK (D43A/Araf-Xylp), and 3ZXL (Y166A/E216A)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117686109/-/DCSupplemental.
References
- 1.Brett CT, Waldren K. Topics in Plant Functional Biology. London: Chapman and Hall; 1996. Physiology and Biochemistry of Plant Cell Walls. [Google Scholar]
- 2.Gilbert HJ. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 2010;153:444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorensen HR, et al. A novel GH43 α-L-arabinofuranosidase from Humicola insolens: Mode of action and synergy with GH51 α-L-arabinofuranosidases on wheat arabinoxylan. Appl Microbiol Biotechnol. 2006;73:850–861. doi: 10.1007/s00253-006-0543-y. [DOI] [PubMed] [Google Scholar]
- 4.van den Broek LA, et al. Cloning and characterization of arabinoxylan arabinofuranohydrolase-D3 (AXHd3) from Bifidobacterium adolescentis DSM20083. Appl Microbiol Biotechnol. 2005;67:641–647. doi: 10.1007/s00253-004-1850-9. [DOI] [PubMed] [Google Scholar]
- 5.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartmell A, et al. The Cellvibrio japonicus mannanase CjMan26C displays a unique exo-mode of action that is conferred by subtle changes to the distal region of the active site. J Biol Chem. 2008;283:34403–34413. doi: 10.1074/jbc.M804053200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proctor MR, et al. Tailored catalysts for plant cell-wall degradation: Redesigning the exo/endo preference of Cellvibrio japonicus arabinanase 43A. Proc Natl Acad Sci USA. 2005;102:2697–2702. doi: 10.1073/pnas.0500051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yaoi K, et al. The structural basis for the exo-mode of action in GH74 oligoxyloglucan reducing end-specific cellobiohydrolase. J Mol Biol. 2007;370:53–62. doi: 10.1016/j.jmb.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 9.Vandermarliere E, et al. Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family. Biochem J. 2009;418:39–47. doi: 10.1042/BJ20081256. [DOI] [PubMed] [Google Scholar]
- 10.Cartmell A, et al. The structure and function of an arabinan-specific α-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases. J Biol Chem. 2011;286:15483–15495. doi: 10.1074/jbc.M110.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nurizzo D, et al. Cellvibrio japonicus α-L-arabinanase 43A has a novel five-blade beta-propeller fold. Nat Struct Biol. 2002;9:665–668. doi: 10.1038/nsb835. [DOI] [PubMed] [Google Scholar]
- 12.Brux C, et al. The structure of an inverting GH43 β-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. J Mol Biol. 2006;359:97–109. doi: 10.1016/j.jmb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto Z, et al. Crystal structure of an exo-1,5-α-L-arabinofuranosidase from Streptomyces avermitilis provides insights into the mechanism of substrate discrimination between exo- and endo-type enzymes in glycoside hydrolase family 43. J Biol Chem. 2010;285:34134–34143. doi: 10.1074/jbc.M110.164251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alhassid A, et al. Crystal structure of an inverting GH 43 1,5-α-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate. Biochem J. 2009;422:73–82. doi: 10.1042/BJ20090180. [DOI] [PubMed] [Google Scholar]
- 15.Jordan DB. β-D-xylosidase from Selenomonas ruminantium: Catalyzed reactions with natural and artificial substrates. Appl Biochem Biotechnol. 2008;146:137–149. doi: 10.1007/s12010-007-8064-4. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida S, Hespen CW, Beverly RL, Mackie RI, Cann IK. Domain analysis of a modular α-L-arabinofuranosidase with a unique carbohydrate binding strategy from the fiber-degrading bacterium Fibrobacter succinogenes S85. J Bacteriol. 2010;192:5424–5436. doi: 10.1128/JB.00503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hovel K, et al. Crystal structure and snapshots along the reaction pathway of a family 51 α-L-arabinofuranosidase. EMBO J. 2003;22:4922–4932. doi: 10.1093/emboj/cdg494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charnock SJ, et al. Key residues in subsite F play a critical role in the activity of Pseudomonas fluorescens subspecies cellulosa xylanase A against xylooligosaccharides but not against highly polymeric substrates such as xylan. J Biol Chem. 1997;272:2942–2951. doi: 10.1074/jbc.272.5.2942. [DOI] [PubMed] [Google Scholar]
- 20.Pell G, et al. The mechanisms by which family 10 glycoside hydrolases bind decorated substrates. J Biol Chem. 2004;279:9597–9605. doi: 10.1074/jbc.M312278200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.