Abstract
Staphylococcus aureus and Staphylococcus epidermidis form communities (called biofilms) on inserted medical devices, leading to infections that affect many millions of patients worldwide and cause substantial morbidity and mortality. As biofilms are resistant to antibiotics, device removal is often required to resolve the infection. Thus, there is a need for new therapeutic strategies and molecular data that might assist their development. Surface proteins S. aureus surface protein G (SasG) and accumulation-associated protein (S. epidermidis) promote biofilm formation through their “B” regions. B regions contain tandemly arrayed G5 domains interspersed with approximately 50 residue sequences (herein called E) and have been proposed to mediate intercellular accumulation through Zn2+-mediated homodimerization. Although E regions are predicted to be unstructured, SasG and accumulation-associated protein form extended fibrils on the bacterial surface. Here we report structures of E–G5 and G5–E–G5 from SasG and biophysical characteristics of single and multidomain fragments. E sequences fold cooperatively and form interlocking interfaces with G5 domains in a head-to-tail fashion, resulting in a contiguous, elongated, monomeric structure. E and G5 domains lack a compact hydrophobic core, and yet G5 domain and multidomain constructs have thermodynamic stabilities only slightly lower than globular proteins of similar size. Zn2+ does not cause SasG domains to form dimers. The work reveals a paradigm for formation of fibrils on the 100-nm scale and suggests that biofilm accumulation occurs through a mechanism distinct from the “zinc zipper.” Finally, formation of two domains by each repeat (as in SasG) might reduce misfolding in proteins when the tandem arrangement of highly similar sequences is advantageous.
Keywords: device infection, protein biophysics, X-ray crystallography, protein domains
Health care-associated infections affect many millions of patients every year worldwide, causing substantial morbidity and mortality and high costs to health services (1). Health care-associated infections are a particular problem in adult (2) and neonatal intensive care units (3), and frequently arise as a result of formation of biofilms on the surfaces of indwelling medical devices (4). Staphylococcus aureus and Staphylococcus epidermidis are a common cause of such infections (2). Device-related staphylococcal infections are difficult to eradicate and treat clinically (5) because bacteria in the biofilm are protected from antimicrobial agents and the host immune system (6). Prolonged antibiotic therapy and device removal can be required to resolve the infection (5).
A biofilm is a functional multilayered community of microorganisms, adhering to a surface and organized within a self-produced exopolymeric matrix (4). Initiation of biofilm formation is a two-step process involving attachment (in which bacteria adhere to a surface) and subsequent maturation (when a 3D structure evolves). The maturation phase requires intercellular aggregation, during which bacteria divide and accumulate (6). Staphylococci can mediate cell-to-cell adhesion using two types of exopolymers: the polysaccharide intercellular adhesin and proteins (7). Polysaccharide intercellular adhesin is also known as poly-N-acetyl-glucosamine and is synthesized by enzymes encoded on the icaADBC operon (7). Accumulation can occur independently of ica, instead relying on the expression of surface proteins such as accumulation-associated protein (Aap) (8) in S. epidermidis and S. aureus surface protein G (SasG) (9) in S. aureus.
Aap and SasG have a domain organization that is typical of the LPXTG protein family, comprising an N-terminal secretion signal and a C-terminal sorting peptide, which is essential for covalent linkage to wall peptidoglycan (10) (Fig. 1A). They contain an N-terminal A domain, followed by a stretch of tandemly arrayed “B” repeats. The A domains of SasG and Aap have been implicated in adhesion of bacteria to desquamated epithelial cells (10) and B repeat regions are responsible for cell-to-cell accumulation during biofilm formation (11, 12). Although annotations in the literature differ (13, 14), the Pfam database (15) recognizes a B-repeat sequence as containing a G5 domain (∼80 residues) (14) followed by an approximately 50-residue sequence (herein called E; Fig. 1A). The formation of Aap-mediated biofilm requires proteolytic cleavage of the N-terminal A domain (11). A similar processing pathway was proposed for SasG (9). However, recent findings indicated that the full-length protein exposed on the bacterial surface undergoes limited processing within B repeats (12). The current model of protein-mediated intercellular adhesion promoted by Aap and SasG in staphylococci is known as a zinc zipper, in which Zn2+-mediated self-association events occur between stretches of B repeats on opposing Aap or SasG molecules (12, 13).
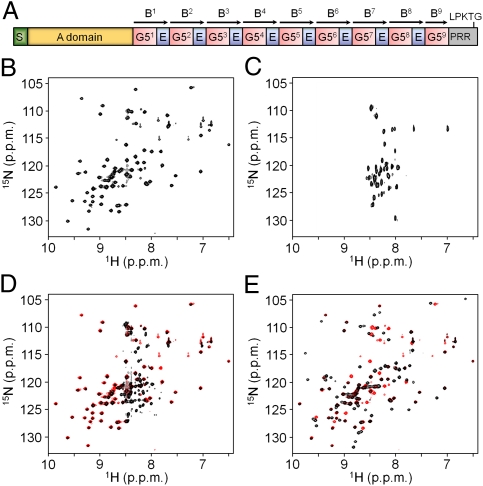
Fig. 1.
SasG from S. aureus NCTC 8325 (UniProt no. Q2G2B2). (A) Schematic representation of the domain arrangement showing nine 78-residue G5 domains and eight 50-residue E segments. The location of the signal sequence (S), A domain, proline-rich region (PRR), and sorting peptide (LPKTG) are also presented. B repeats [according to the previous annotation (16)] are indicated with arrows. HSQC spectra (1H-15N) of G52 (B) and E (C). (D) Superimposed 1H-15N HSQC spectra of G52-E (black) and G52 (red). (E) Superimposed 1H-15N HSQC spectra of E-G52 (black) and G52 (red).
SasG and Aap have sequence identities of approximately 34% and approximately 50% for G5 and E repeats, respectively, and contain a variable number of repeats [three to 10 in SasG (16) and four to 17 in Aap (17)], dependent on the strain. High DNA sequence identity results in pairwise protein sequence similarity between B repeats within SasG (Fig. S1A) and within Aap of 90% to 100% and 82% to 91%, respectively. It has been shown recently that tandemly arrayed domains with high sequence identity are prone to misfolding events (18). This is likely to be a particular problem for long-lived proteins, or those which undergo shear stresses, and might explain the apparent evolutionary pressure for sequences of adjacent domains to have less than 40% sequence identity (19). Thus, the SasG and Aap biofilm-forming region also raises the question of how misfolding is avoided when sequence identity between repeats is otherwise advantageous.
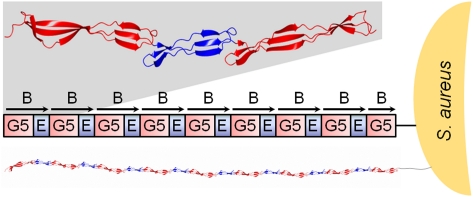
The minimum number of SasG B repeats required for biofilm formation is five. SasG variants with more than four repeats blocked the binding of S. aureus surface adhesins to their ligands; for example, the clumping factor B binding to cytokeratin and fibrinogen (9). It is therefore possible that a minimum number of repeats is required for projection of the biofilm forming domains beyond other surface proteins. EM shows that SasG and Aap form highly elongated fibrils on the bacterial surface. S. aureus strains expressing SasG with eight full-length B repeats form peritrichous fibrils of varying density and a mean length of 53 ± 3 nm (9). Aap-expressing strains produce localized tufts of fibrillar appendages, usually in a lateral position in relation to the septum. The mean estimated length of the fibrils formed by an Aap variant with 12 full-length B repeats was 122 ± 11 nm for S. epidermidis NCTC 11047 and 159 ± 35 nm for RP62A (20). Although it was proposed that fibrils correspond to a single molecule of Aap (20), individual fibrils could not be distinguished as a result of very close packing.
Here we show that the biofilm-forming region of SasG has an elongated, contiguous structure formed by folded E and G5 domains connected by mutually stabilizing interfaces in head-to-tail fashion. The high resolution crystal structures of E-G5 (1.7 Å) and G5-E-G5 (1.85 Å) reveal that the domain structures are composed of flat, single-layer β-sheets and thus lack a compact hydrophobic core. Interdomain interfaces form interlocking connections between G5s and Es, leading to extended rod-like structures, which explain the appearance of the SasG fibrils on the bacterial surface.
Results
Structural Annotation of SasG Repeat Region.
The SasG B repeat region of S. aureus strain NCTC 8325 contains nine 78-residue G5 domains and eight 50-residue segments (herein called E; Fig. 1A and Fig. S1A). Although there is one G5 domain structure (21) in the Protein Data Bank (PDB; with low ∼15% sequence identity to SasG G5 domains), the E segments are predicted to be disordered (with high probability) by algorithms such as PONDR (22) and IUPred (23). The 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectrum of G52 shows that most peaks have a wide distribution in the 1H dimension, indicating that, as expected, it has a stable fold (Fig. 1B). In addition, as predicted by the sequence analysis tools, the shorter E segment appears disordered in isolation (Fig. 1C), with peaks having a narrow range of 1H chemical shifts. The spectrum of G52-E (a B repeat in the previous annotation; Fig. 1D) shows a subpopulation of intense and poorly dispersed peaks whereas the remaining peaks are less intense but widely distributed and overlay almost exactly with the spectrum of G5 in isolation. Thus, the G5 domain retains its fold in the G5-E (B-repeat) context, E is disordered, and there is no evidence of a significant interface between G5 and E. Surprisingly, in the spectrum of E-G52 (Fig. 1E), most peaks show wide 1H chemical shift dispersion, and an overlay of the G52 and E-G52 spectra shows that, although the majority of peaks associated with G5 in isolation remain in the same position, several are shifted in the E-G52 context. This implies that E is folded and that, although the overall structure of the G5 domain is similar, there is a significant E–G5 interface. Backbone dynamics of the E segment in the context of G52-E and E-G52 were estimated by using a 1H-15N heteronuclear nuclear Overhauser effect (NOE) experiment (Fig. 2A). In the case of G52-E, the average NOE values calculated for E residues were significantly lower than those for the G52 residues, indicating the high flexibility of the E region. The NOE values obtained for E-G52 were similar for most residues, demonstrating similar backbone dynamics for both subdomains on the subnanosecond timescale, and thus that E is folded.
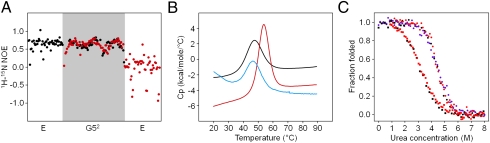
Fig. 2.
Dynamics and stability of SasG domains. (A) Heteronuclear NOE experiment (1H-15N) measuring the dynamics of backbone 15N nuclei of E-G52 (black) and G52-E (red). The G52 residues were assigned in both E-G52 and G52-E, whereas the E segment values were measured but not assigned to individual residues. (B) DSC thermograms for G52 (black), G52-E (blue), and E-G52 (red; Table S1). (C) Urea-induced equilibrium denaturation curves for G52 (■) and E-G52 (●). Folding was followed by monitoring changes in intrinsic tyrosine fluorescence starting with folded (black) or unfolded (red) protein. The purple points show unfolding of E-G52 monitored by changes in the circular dichroism at 235 nm.
To test the thermodynamic significance of the interdomain interfaces, we monitored the unfolding of a G5 domain in isolation and linked with E. Differential scanning calorimetry (DSC) thermograms (Fig. 2B and Table S1) show a significant increase in the melting temperature (Tm) of E-G52 (54 °C) compared with an isolated G52 domain (47 °C), implying a stabilizing effect of the N-terminal E. Consistent with the NMR studies, the Tm of the G52 domain was unaffected by the presence of a C-terminal E segment. Furthermore, the urea-induced equilibrium unfolding transitions (Fig. 2C) reveal that E-G52 is significantly more stable (m-value = 1.4 kcal⋅mol−1⋅M−1) than an individual G52 domain (m-value = 0.9 kcal⋅mol−1⋅M−1) and that both unfold as single cooperative units.
Crystal Structures of E-G52 and G51-E-G52 Explain the Elongated Nature of SasG Fibrils.
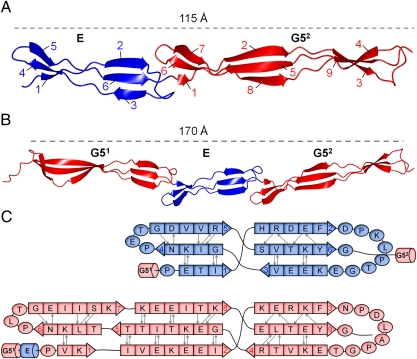
Having established the domain boundaries of stably folded segments of SasG, we sought to characterize them structurally. Crystal structures of E-G52 and G51-E-G52 were determined at a resolution of 1.70 Å and 1.87 Å, respectively (statistics are provided in Tables S2 and S3). Both structures reveal a highly extended topology, which can be depicted as a cylinder with a diameter of approximately 20 Å and lengths of 115 Å for E-G52 and 170 Å for G51-E-G52 (Fig. 3 A and B). Each structure is formed from consecutive single-layer triple-stranded β-sheets, and their unusual elongation arises from the head-to-tail arrangement of the β-sheets.
Fig. 3.
Crystal structures of SasG domains; E and G5 domains are shown in blue and red, respectively. (A) Structure of E-G52. The β-strands are numbered for E and G5 domains. (B) Structure of G51-E-G52. (C) Schematic of the secondary structure of E (Upper) and G52 (Lower), as defined by the method of Kabsch and Sanders (64).
The crystal structures, together with the NMR spectroscopy analysis, reveal that G5 domains and E segments are the building blocks of the B repeat region. G5 and E share 24% sequence identity (Fig. S1B) and show similar overall topology (Figs. 3 and 4B and C). They are each composed of triple-stranded single-layer β-sheets connected by an intertwined motif, which leads to a strand switch. Such a supersecondary structural motif was denoted by Ruggiero et al. as a β–triple helix–β and was first described for the G5 domain from RpfB from Mycobacterium tuberculosis (21). The SasG E segment is approximately 45 Å in length and is composed of two β-sheets: an N-terminal antiparallel β-sheet and a C-terminal β-sheet with a mixed parallel/antiparallel arrangement of β-strands (Fig. 3). The G5 domain extends to approximately 70 Å and is assembled from two (G51) or three (G52) triple-stranded β-sheets. The N-terminal antiparallel β-sheet of G51 is longer than the C-terminal sheet and corresponds to the two most N-terminal β-sheets in the G52 structure, which are also antiparallel. The C-terminal β-sheets of both G5 domains are nearly identical and show a mixed parallel/antiparallel arrangement of β-strands (Fig. 3). SasG G51 and G52 are nearly identical (rmsd = 1.34 Å; Fig. 4A) and show significant structural homology to the previously reported structure of the G5 domain. However, the RpfB G5 is composed of two β-sheets that are bent with respect to each other, providing the molecule with an overall arch-like structure, whereas SasG G5 domains are planar (Fig. 4 D and E).
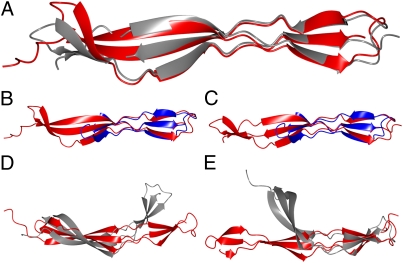
Fig. 4.
Structural superposition of G5 and E domains. Automatic secondary structure matching in CCP4mg was implemented (65): (A) G51 (red) and G52 (gray), (B) G51 (red) and E (blue), (C) G52 (red) and E (blue), (D) G51 (red) and G5 from RpfB (gray), and (E) G52 (red) and G5 from RpfB (gray).
The SasG β-sheets are exposed to the solvent on both faces, yet the exclusion of apolar side chains from aqueous solvent is considered to be a key stabilizing force in folded proteins. As expected from the appearance of the structure, the total relative accessible surface area (RSA) and the RSA for nonpolar side chains are larger for SasG domains than for globular proteins with a similar number of residues (Table S4). The ratio of the RSA calculated for apolar side chains and for all atoms is, however, comparable between the globular proteins and the extended SasG domains, suggesting a sufficient burial of hydrophobic residues. Moreover, the DSC thermograms (Fig. 2B and Table S1) clearly demonstrate a positive change in heat capacity upon thermal unfolding (and that the melting temperatures are not unusually low; Table S1). Thus, although they lack a typical compact hydrophobic core, the burial of nonpolar groups makes an important contribution to the stabilization of SasG domains. This appears to be achieved by strategic distribution of nonpolar side chains and aromatic residues throughout the SasG sequence. Bulky aromatics (tyrosines and phenylalanines) and longer hydrophobic side chains (isoleucines, leucines) are located at the interdomain interfaces, where they contribute to the formation of pseudohydrophobic cores. Smaller nonpolar residues are distributed along the single-layer β-sheets and pack against the hydrophobic moieties of long charged side chains, such as glutamates and lysines.
Interdomain Interfaces.
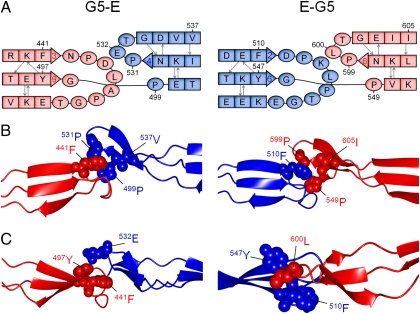
The structural similarity in the N- and C-terminal β-sheets of SasG G5 and E domains results in highly homologous interdomain interfaces (G5–E and E–G5; Fig. 5A). The interfaces are formed by equivalent residues from G5 and E domains, which are involved in similar networks of van der Waals interactions. For example, Phe441 from G51 interacts with Pro499, Pro531, and Val537 from E, which corresponds to Phe510 from E interacting with Pro549, Pro599, and Ile605 from G52 (Fig. 5 A and B). In comparison with G5–E, the E–G5 interface is additionally stabilized by the presence of Leu600 in G52, which interacts with Phe510 and Tyr547 from E (Fig. 5C). In the context of the G5–E interface, this position is occupied by Glu532, which appears to distort the packing of nonpolar side chains. These structural differences partially rationalize the NMR and DSC data.
Fig. 5.
G5–E and E–G5 interfaces in SasG. G5 and E domains are shown in red and blue, respectively. (A) Schematic representation of the interfaces indicating the similarity between the N- and C-terminal sheets of each domain. (B) Structural representation of the interfaces showing the equivalent residues involved in interdomain interactions. (C) Structural representation of the interfaces highlighting the major difference between G5–E and E–G5 (stereo images showing a portion of electron density are shown in Fig. S4).
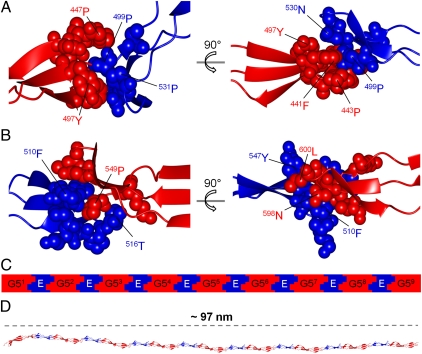
The residues involved in the E–G5 and G5–E interfaces are distributed across three strands of both domains and interdigitate (Fig. 6). Thus, the interdomain flexibility of G5s and Es is likely to be restricted, providing rigidity to multidomain SasG constructs. To test this hypothesis, we carried out sedimentation velocity analytical ultracentrifugation to estimate the overall shape of SasG constructs containing different numbers of domains. The calculated frictional ratios from the known molecular weights and sedimentation coefficients, as well as the prolate axial ratios (a/b), confirmed that all tested proteins are highly elongated monomers (Table 1). These results demonstrate that E-G52 and G51-E-G52 (Fig. 3 A and B) also adopt an extended conformation in solution. Furthermore, the length of constructs appears to be directly proportional to the number of G5 and E domains; that is, the long axis dimensions for modules of varying length are approximately additive. For example, the experimental a/b value for G51-E-G52 is 10.4, whereas the estimated axial ratio would be 10.7 if based on a/b values obtained for G51 (4.8) and E-G52 (5.9; Table 1). This additive nature of the axial ratios confirms the head-to-tail arrangement of domains and the presence of substantial rigidity in the interdomain connections.
Fig. 6.
Interlocking domain interfaces in SasG. G5 and E domains are shown in red and blue, respectively. (A) The G51–E interface highlighting the interdigitated residues. (B) The E–G52 interface highlighting the interdigitated residues. (C) Schematic representation and (D) structural model of interlocking G5 and E domains within SasG with nine G5 domains.
Table 1.
Summary of analytical ultracentrifugation data
| Protein | Theoretical MW (Da) | s* (S) | s20,w (S) | f/f0* | a/b* | f/f0 | a/b |
| G51 | 9,699.1 | 1.007 | 1.047 | 1.53 | 5.5 | 1.47 | 4.8 |
| G52 | 9,654.8 | 1.066 | 1.107 | 1.56 | 5.8 | 1.45 | 4.4 |
| E-G52 | 14,471.2 | 1.291 | 1.341 | 1.72 | 8.0 | 1.57 | 5.9 |
| G51- E-G52 | 23,729.8 | 1.472 | 1.530 | 1.89 | 10.7 | 1.87 | 10.4 |
| E-G51- E-G52 | 28,512.1 | 1.594 | 1.655 | 2.06 | 13.5 | 1.98 | 12.2 |
a/b, prolate axial ratio calculated from f/f0; a/b*, axial ratio for a prolate ellipsoid; f/f0, frictional ratio calculated from the known molecular weight and sedimentation coefficient; f/f0*, experimental frictional ratio; MW, molecular weight; s* (S), experimental sedimentation coefficient; s20,w (S), sedimentation coefficient corrected to water.
SasG Domains Are Elongated Monomers in Solution.
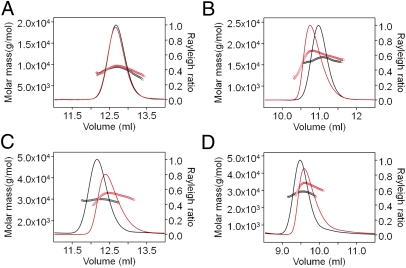
Previously, we and others showed that SasG and Aap B repeats dimerize in the presence of Zn2+; however, the structural basis of dimerization was not clear (12, 13). The crystal structure of a SasG G52 dimer (Fig. S2) reveals that Zn2+ is coordinated by an N-terminal nonnative histidine and two glutamate residues from each monomer. Mutation of the histidine abolished the Zn2+-mediated dimerization of G52 and of a previously reported B repeat (G52-E; Fig. S3). Using the new SasG domain boundaries (defined based on the crystal structure of G51-E-G52), size-exclusion chromatography with multiangle laser light scattering (SEC-MALLS) confirmed that all stably folded modules (G51, G52, E-G52, G51-E-G52, and E-G52-E-G53) and partially or fully disordered SasG modules (E, G51-E, G52-E and G51-E-G52-E) are monomeric in solution at neutral pH in the absence and presence of 5 mM Zn2+ (Fig. 7, Fig. S3, and Table S5).
Fig. 7.
SEC-MALLS analysis of the oligomeric state of SasG domains in the presence (red) and absence (black) of Zn2+ (5 mM): (A) G52, (B) E-G52, and (C) G51-E-G52, i.e., native sequence with four additional N-terminal amino acids (i.e., GPHM); and (D) E-G51-E-G52, i.e., native sequence with four additional N-terminal amino acids (i.e., GPHM). The exact molar masses and sequences are listed in Table S5.
Discussion
Bacterial biofilm matrices are heterogeneous composed mainly of polysaccharides, proteins, nucleic acids, and lipids (24). Proteins detected in biofilms are either enzymes, responsible for degradation and modification of the extracellular biopolymers, or structural proteins that are attached to the bacterial surface and are involved in the formation and stabilization of the matrix. Some nonenzymatic surface proteins, such as LecA (25) and LecB (26) of Pseudomonas aeruginosa, bind to extracellular saccharides and link the bacterial surface with the matrix. Other proteinaceous components of the biofilm matrix are functional amyloids. Examples include amyloid fibers of Gram-positive Bacillus subtilis composed of TasA (27), as well as curli (28) and Tafi (29) amyloid fibrils of Escherichia coli and Salmonella spp., composed of the highly homologous subunit proteins CsgA and AgfA, respectively. Staphylococcal biofilm-associated surface proteins, including Aap and SasG, represent another group of matrix proteins. They also form elongated fibrillar structures at the bacterial cell surface, but via a mechanism distinct from amyloid assembly.
Elongated filamentous structures on bacterial surfaces are most commonly assembled from multiple polypeptide chains. For example, the M protein from Streptococcus pyogenes forms hair-like fimbriae that extend approximately 50 nm from the cell surface (30) and are composed of dimeric parallel α-helical coiled-coil structures (31). Pili in Gram-negative bacteria are typically formed by noncovalent homopolymerization of major pilus subunit proteins (pilins) (32). Recently discovered pili in Gram-positive bacteria are formed by covalent polymerization of pilin subunits in a process that requires a specific sortase enzyme (33). The additive long axis dimensions for SasG modules of varying length (Table 1) suggest that the interdomain interfaces provide rigidity. This, taken together with the head-to-tail arrangement of the domains, implies that in vivo SasG structures (with as many as 10 G5 domains) could form the highly extended structures observed on the bacterial surface. As a single-chain structure composed of single-layer β-sheets, SasG would form an unusually thin filament on the cell surface, providing a highly efficient solution to the formation of an extended structure on this scale (50–100 nm). Thus, SasG represents a new paradigm in the production of thin, rod-like protein structures.
A striking feature of the SasG structures (Fig. 3 A and B) is the apparent lack of a compact hydrophobic core, although the Tm, free energies of unfolding, and m-values for urea denaturation of the G5 and E–G5 constructs suggest they have stabilities only slightly lower than globular domains of similar size. Typically, β-sheets, whether in extracellular or cytoplasmic proteins, are amphipathic, and their hydrophobic faces form a compact hydrophobic core with other secondary structure elements. In addition, extracellular β-sheet–containing domains are often stabilized through disulfide bond formation. Despite the increased solvent exposure of SasG domains, the burial of apolar residues appears to be sufficient to stabilize their fold. Moreover, more than 95% refolding efficiencies (even after multiple unfolding cycles) were observed in DSC experiments for structured SasG constructs, at protein concentrations at which misfolding and aggregation are typically observed.
Individual (rather than tandemly arrayed) single-layer β-sheet domains have been reported previously; for example, OspA, a surface protein of Borrelia burgdorferi, contains a three-stranded β-sheet between two globular domains (34), and WRKY4 transcription factors (35) contain a four-stranded single-layer β-sheet that is stabilized by zinc. In OspA and WRKY4, the thermodynamic stability of the sheets is suggested to depend on interactions of nonpolar side chains with the hydrophobic parts of long hydrophilic amino acids. The sequences of G5 and E domains show a high content of long charged residues (Glu, ∼15%; and Lys, ∼15%), and analysis of the E-G52 and G51-E-G52 crystal structures reveal that short nonpolar side chains (Ala, Val, Ile, Leu) form small hydrophobic clusters surrounded by large hydrophilic amino acids. In addition, the long, charged side chains form cross-strand arrays of alternating charges observed previously in antiparallel β-sheets (36).
Despite the difference in length, the SasG G5 and E domains show significant structural similarity (Figs. 3 and 4 B and C). As in the G5 domain from RpfB (21), the central “switch” region of the SasG G5 and E domains resembles the collagen triple-helical structural motif. The pseudotriple helix of SasG domains has a PPII-like conformation and is rich in proline and glycine residues. The packing of intertwined SasG strands within the switch region is tight, despite their mixed parallel/antiparallel arrangement. This feature of the β–triple helix–β fold is stabilized by two (21) of the five conserved glycines after which the G5 domain was named (14). The SasG E segment contains structurally equivalent glycine residues within its pseudotriple helix. The distribution of hydrophobic, charged, and proline residues within the E segment also resembles that of the G5 domain (Fig. 3). This suggests that E–G5 has evolved through processes of G5 duplication, mutation, fusion, and partial deletion. Despite the structural similarity between the two SasG domains, the E segment is significantly less stable than G5 (Figs. 1 B–E and 2 A and B) and dependent on the E–G5 interface for folding.
The sequences of SasG (and Aap) repeats are very similar at the DNA level (16) and vary in number dependent on the bacterial strain (16, 17). However, although sequence identity at the DNA level might be advantageous in facilitating recombination events, the resulting protein sequence identity is a potential problem from a protein folding perspective. The immediate juxtaposition of domains with identical sequence can promote misfolding events (18). This might explain the apparent evolutionary pressure to maintain sequence identity at less than 40% between adjacent domains that was revealed by an analysis of proteins containing strings of Ig and fibronectin-type III domains (19). The formation of two domains by each sequence repeat, as in SasG, could provide an elegantly simple solution to this problem. As there are very few structures of native tandem repeats of domain size in the PDB (37), this observation will assist future structural studies of such proteins, several of which [e.g., Rib and alphaC (38)] are expressed on the surface of pathogens.
The structure of a Zn2+-mediated G52 dimer (Fig. S3) and SEC-MALLS analysis of mutants (Fig. S2) show that our previously reported dimerization of a SasG B repeat at millimolar Zn2+ concentrations (12) was dependent on two glutamate residues and a single nonnative histidine in each monomer. This phenomenon is highly context-dependent, as the presence of this histidine did not result in Zn2+-dependent dimerization of other SasG constructs (Fig. 7). Notwithstanding, in vivo studies demonstrated that zinc is essential for protein-mediated biofilm formation in S. epidermidis (13) and S. aureus (12). Aap G5 domains, which contain native C-terminal histidine residues (not present in SasG), dimerize in a zinc-dependent manner (13). However, there is no experimental evidence that directly links this in vitro dimerization to the Zn2+-dependence of protein-mediated biofilm formation in S. epidermidis. In addition, free Zn2+ is normally present at much lower concentrations in vivo [estimated as low as femtomolar (39) in bacterial cells and subnanomolar in mammalian plasma (40)] than was required for Aap dimerization. Thus, our study implies that, rather than homodimerization, zinc might mediate binding to another polymeric component of the staphylococcal biofilm in ica-null strains, such as extracellular teichoic acid or DNA (41). Alternatively, the rod-like structure might play a role in maintaining bacterial separation in the biofilm. Furthermore, a recent study on Aap-mediated biofilm formation by S. epidermidis showed that anti-G5 monoclonal antibodies enhanced bacterial accumulation, whereas those with an epitope in the E segment inhibited biofilm accumulation to 60% of the maximum (42). This suggests that G5 and E domains might play different roles during bacterial accumulation, despite their structural similarity. Further studies will be required to reveal the full molecular basis of protein-mediated staphylococcal biofilm formation. The domain arrangement, high resolution structures, and lack of Zn2+-dependent dimerization of SasG domains reported here provide a very significant step toward this goal and the overall aim of informing the development of new therapeutic strategies for the treatment or prevention of staphylococcal biofilm infections.
Materials and Methods
Cloning.
DNA sequences of B1, B2, and E1G52, codon-optimized for Escherichia coli, were synthesized (GenScript) and subcloned into the pSKB2 expression vector providing an N-terminal hexahistidine tag. Other SasG modules were generated based on these three templates by using standard cloning and mutagenesis techniques.
Protein Production.
Unlabeled, 15N-labeled, and 13C-15N-labeled proteins were produced in E. coli BL21(DE3) by using Luria–Bertani, 15N-M9, and 13C-15N-M9 medium, respectively. Selenomethionylated E-G52-L17M-L103M was produced in E. coli B834(DE3) (Novagen) by using SeMet-supplemented minimal medium. Standard procedures were used. After induction with isopropyl β-d-1-thiogalactopyranoside at an OD600 of 0.6, E. coli cultures were grown at 20 °C for 24 h. SasG domains were purified by nickel-affinity purification using a HisTrap HP column (GE Healthcare). The His-tag was cleaved by using HRV 3C protease (Novagen), and removed with a HisTrap HP column. In case of insufficient purity, size exclusion chromatography was applied with a HiLoad 16/60 Superdex 75 column (GE Healthcare).
NMR Spectroscopy.
NMR spectra were acquired at 25 °C on a Bruker AVANCE II 700 MHz spectrometer equipped with a triple-resonance probe. Samples for HSQC experiments contained 15N-labeled protein (0.5 mM) in 20 mM Tris-HCl, pH 7.0, 100 mM NaCl, and 10% 2H2O. Steady-state 1H-15N NOE values were determined by recording HSQC spectra in the presence (i.e., NOE) and absence (i.e., NONOE) of 1H saturation. NOE and NONOE experiments were deconvoluted and the NOE value was calculated from the intensity (i.e., volume) of the cross peaks by using the following formula:
For the assignment of the G52 HSQC spectrum, a series of 3D experiments was recorded (HNCO, HNCACO, CBCACONH, and CBCANH) for a sample containing 13C-15N-G52 (1 mM) in 20 mM Tris-HCl, pH 7.0, 100 mM NaCl, and 10% 2H2O. Spectra were processed by using NMRPipe (43) and viewed in NMRView (44). NMR backbone resonances were assigned using CcpNmr Analysis 2.1 (45, 46).
DSC.
DSC scans were acquired on a MicroCal VP-DSC calorimeter for protein samples at a concentration of 1 mg/mL in 20 mM Tris-HCl, pH 7.4, 150 mM NaCl. Scans of degassed buffer and proteins were recorded for temperatures ranging from 10 to 90 °C at a scan rate of 90 °C/h. Data analysis was performed using MicroCal Origin 7.0. A progress baseline was used for the sample baseline correction before area integration or fitting of the unfolding endotherm. Data points between 20 and 90 °C were used for fitting. The reversibility of thermal unfolding was verified by repetitive scans on the same sample.
Folding Studies.
Protein stability was determined by using urea-induced equilibrium denaturation. Fluorescence measurements were performed on a Cary Eclipse fluorescence spectrophotometer. CD measurements were performed on an Applied Photophysics Chirascan CD spectrometer. The experiments were carried out by using 5 μM protein in PBS solution (pH 7.4) at 25 °C after equilibration overnight. The fluorescence excitation wavelength was 274 nm, and emission was followed at 302 nm. For CD, the ellipticity at 235 nm was followed. The data were fitted to a two-state equation (48).
Crystallography.
Crystallization trials were performed by using the sitting drop vapor diffusion method. E-G52 crystallized in 0.2 M NH4Cl and 20% (wt/vol) PEG3350, using the protein solution at a concentration of 2 mM (30 mg/mL) in 20 mM Tris, pH 7.0, 100 NaCl. Crystals of selenomethionylated E-G52-L17M-L103M were obtained in 0.05 M Tris, 0.05 M Bicine, pH 8.5, 12.5% (vol/vol) 2-methyl-2, 4-pentanediol, 12.5% (wt/vol) PEG 1000, 12.5% (wt/vol) PEG 3350, 0.02 M sodium formate, 0.02 M sodium citrate, and 0.02 M sodium oxamate, for the protein solution at a concentration of 2 mM (30 mg/mL) in 20 mM Tris, pH 7.0, 100 NaCl. G51-E-G52 crystallized in 0.2 M MgCl2, 0.1 M Bis-Tris, pH 5.5, and 25% (wt/vol) PEG 3350, using the protein solution at a concentration of 1.2 mM (27.6 mg/mL) in 20 mM Tris, pH 7.0, 100 NaCl.
Diffraction data were collected at 100 K at the Diamond Light Source on beamline I04 (native E-G52 and G51-E-G52) and I02 (SeMet-E-G52-L17M-L103M). Images recorded for E-G52 and G51-E-G52 were processed with XDS (48), and the data were scaled by using SCALA (49) from the CCP4 program suite (50). Multiple-wavelength anomalous data collected for SeMet-E-G52-L17M-L103M were processed by using the HKL2000 suite (51).
The structure of SeMet-E-G52-L17M-L103M was solved by the multiwavelength anomalous dispersion method by using SHELXC/D/E (52). Four heavy atom sites were identified, and a partial model with two molecules in the asymmetric unit was produced by Buccaneer (53). One of the molecules was used as a search model for molecular replacement for E-G52 data. The solution, produced by Phaser (54), consisted of one molecule in the asymmetric unit. A complete model of the E-G52 structure was generated in ARP/wARP (55). The structure of G51-E-G52 was also solved by molecular replacement. First, the E-G52 structure was used as a search model in Phaser, revealing two E segments and consequently two G51-E-G52 molecules in the asymmetric unit. Subsequent molecular replacement runs with MOLREP (56), followed by model building in ARP/wARP, localized the remaining four G5 domains. MOLREP was implemented because of its shorter run time. Initial refinement was carried out with REFMAC5 (57) using the “jelly body” option. Final refinement runs were performed in Phenix 1.7.1 (58) using TLS restraints [generated with TLSMD (59)] and simulated annealing. The structures were visualized and manually rebuilt in COOT (60). The stereochemistry of the final model was evaluated with MolProbity (61). Data collection and refinement statistics are shown in Tables S2 and S3. Atomic coordinates and structure factor amplitudes have been deposited to PDB under codes 3TIP and 3TIQ.
Analytical Ultracentrifugation.
Sedimentation velocity experiments were conducted on a Beckman Optima XL/I analytical ultracentrifuge using an An-60 Ti rotor at 20 °C. Protein concentrations of 2 mg/mL in 20 mM Tris, 150 mM NaCl buffer, pH 7.4, were centrifuged at 58,000 and 52,000 rpm (G51, G52, and E-G52) or 42,000 rpm (G51-E-G52, E-G52-E-G53) collecting absorbance data at 280 nm and interference data. The program SEDFIT (62) was used to determine sedimentation coefficients and frictional ratios and to convert these to axial ratios. Buffer density and viscosity and protein partial specific volumes were calculated by using SEDNTERP (63).
SEC-MALLS.
SEC-MALLS experiments were performed by using a Superdex 75 HR10/30 column (GE Healthcare) and a Shimadzu HPLC System. Protein samples (100 μL) at a concentration of 1.5 mg/mL were loaded onto a gel filtration column and eluted with one column volume (24 mL) of an appropriate running buffer at a flow rate of 0.5 mL/min. The eluting fractions were monitored using a DAWN HELEOS-II 18-angle light scattering detector (Wyatt Technologies), a SPD20A UV/Vis detector (Shimadzu), and an Optilab rEX refractive index monitor (Wyatt Technologies). Data were analyzed by using Astra (Wyatt Technologies).
Supplementary Material
Acknowledgments
We thank Garib Murshudov for assistance with refinement of the E-G52 and G51-E-G52 structures and Alexey Murzin for helpful discussions. This work was carried out with the support of the Diamond Light Source. This work was supported by British Heart Foundation nonclinical doctoral Studentship FS/08/025/24765 (D.T.G.); British Heart Foundation Senior Basic Science Research Fellowship FS/07/034 (to J.R.P.); Wellcome Trust Grant 064417 (to J.C.); Biotechnology and Biological Sciences Research Council LoLa Grant BB/G020671/1 (J.A.W.); and Science Foundation Ireland Programme Investigator Grant 08/IN.1/B1854 (to T.J.F.). J.C. is a Wellcome Trust Senior Research Fellow.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3TIP and 3TIQ).
This article is a PNAS Direct Submission. R.K. is a guest editor invited by the Editorial Board.
See Author Summary on page 6370 (volume 109, number 17).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119456109/-/DCSupplemental.
References
- 1.World Health Organization . Report on the Burden of Endemic Health Care-Associated Infection Worldwide. Geneva: World Health Organization; 2011. [Google Scholar]
- 2.Rosenthal VD, et al. International Nosocomial Infection Control Consortium Members International Nosocomial Infection Control Consortium report, data summary for 2002-2007, issued January 2008. Am J Infect Control. 2008;36:627–637. doi: 10.1016/j.ajic.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Brady MT. Health care-associated infections in the neonatal intensive care unit. Am J Infect Control. 2005;33:268–275. doi: 10.1016/j.ajic.2004.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Harris LG, Richards RG. Staphylococci and implant surfaces: A review. Injury. 2006;37(Suppl 2):S3–S14. doi: 10.1016/j.injury.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Dunne WM., Jr Bacterial adhesion: Seen any good biofilms lately? Clin Microbiol Rev. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Gara JP. ica and beyond: Biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 8.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65:519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corrigan RM, Rigby D, Handley P, Foster TJ. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153:2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 10.Roche FM, Meehan M, Foster TJ. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology. 2003;149:2759–2767. doi: 10.1099/mic.0.26412-0. [DOI] [PubMed] [Google Scholar]
- 11.Rohde H, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 12.Geoghegan JA, et al. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J Bacteriol. 2010;192:5663–5673. doi: 10.1128/JB.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conrady DG, et al. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA. 2008;105:19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bateman A, Holden MTG, Yeats C. The G5 domain: A potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics. 2005;21:1301–1303. doi: 10.1093/bioinformatics/bti206. [DOI] [PubMed] [Google Scholar]
- 15.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32(database issue):D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roche FM, et al. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology. 2003;149:643–654. doi: 10.1099/mic.0.25996-0. [DOI] [PubMed] [Google Scholar]
- 17.Rohde H, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 18.Borgia MB, et al. Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins. Nature. 2011;474:662–665. doi: 10.1038/nature10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 20.Banner MA, et al. Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation-associated protein. J Bacteriol. 2007;189:2793–2804. doi: 10.1128/JB.00952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruggiero A, et al. Crystal structure of the resuscitation-promoting factor (DeltaDUF)RpfB from M. tuberculosis. J Mol Biol. 2009;385:153–162. doi: 10.1016/j.jmb.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 22.Romero P, et al. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 24.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 25.Diggle SP, et al. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol. 2006;8:1095–1104. doi: 10.1111/j.1462-2920.2006.001001.x. [DOI] [PubMed] [Google Scholar]
- 26.Tielker D, et al. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology. 2005;151:1313–1323. doi: 10.1099/mic.0.27701-0. [DOI] [PubMed] [Google Scholar]
- 27.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci USA. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vidal O, et al. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Austin JW, Sanders G, Kay WW, Collinson SK. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol Lett. 1998;162:295–301. doi: 10.1111/j.1574-6968.1998.tb13012.x. [DOI] [PubMed] [Google Scholar]
- 30.Fischetti VA. Streptococcal M protein: Molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNamara C, et al. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science. 2008;319:1405–1408. doi: 10.1126/science.1154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geibel S, Waksman G. Crystallography and electron microscopy of chaperone/usher pilus systems. Adv Exp Med Biol. 2011;715:159–174. doi: 10.1007/978-94-007-0940-9_10. [DOI] [PubMed] [Google Scholar]
- 33.Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria - structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Dunn JJ, Luft BJ, Lawson CL. Crystal structure of Lyme disease antigen outer surface protein A complexed with an Fab. Proc Natl Acad Sci USA. 1997;94:3584–3589. doi: 10.1073/pnas.94.8.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamasaki K, et al. Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell. 2005;17:944–956. doi: 10.1105/tpc.104.026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wouters MA, Curmi PMG. An analysis of side chain interactions and pair correlations within antiparallel beta-sheets: The differences between backbone hydrogen-bonded and non-hydrogen-bonded residue pairs. Proteins. 1995;22:119–131. doi: 10.1002/prot.340220205. [DOI] [PubMed] [Google Scholar]
- 37.Jorda J, Xue B, Uversky VN, Kajava AV. Protein tandem repeats - the more perfect, the less structured. FEBS J. 2010;277:2673–2682. doi: 10.1111/j.1742-464X.2010.07684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lachenauer CS, Creti R, Michel JL, Madoff LC. Mosaicism in the alpha-like protein genes of group B streptococci. Proc Natl Acad Sci USA. 2000;97:9630–9635. doi: 10.1073/pnas.97.17.9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 40.Magneson GR, Puvathingal JM, Ray WJ., Jr The concentrations of free Mg2+ and free Zn2+ in equine blood plasma. J Biol Chem. 1987;262:11140–11148. [PubMed] [Google Scholar]
- 41.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, et al. Monoclonal antibodies against accumulation-associated protein affect EPS biosynthesis and enhance bacterial accumulation of Staphylococcus epidermidis. PLoS ONE. 2011;6:e20918. doi: 10.1371/journal.pone.0020918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Johnson BA, Blevins RA. NMR View - a computer-program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 45.Fogh RH, et al. A framework for scientific data modeling and automated software development. Bioinformatics. 2005;21:1678–1684. doi: 10.1093/bioinformatics/bti234. [DOI] [PubMed] [Google Scholar]
- 46.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 47.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W. Evaluation of single-crystal X-ray-diffraction data from a position-sensitive detector. J Appl Cryst. 1988;21:916–924. [Google Scholar]
- 49.Evans PR. Proceedings of CCP4 Study Weekend, 1993, on Data Collection and Processing. Warrington, UK: Daresbury Lab; 1993. Data reduction; pp. 114–122. [Google Scholar]
- 50.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 52.Sheldrick GM. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 54.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 56.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 57.Vagin AA, et al. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 58.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 60.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 61.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web server issue):W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, et al., editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, UK: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 64.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 65.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: The CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]