Background: Antimicrobial macin proteins aggregate bacteria and exert nerve repair activities.
Results: Structures of theromacin and neuromacin were elucidated. Nerve repair activities and antimicrobial activities of macins were investigated.
Conclusion: Theromacin induces the nerve repair capacity of leech plasma. Macins seem to be proliferation factors.
Significance: The extended magnitude of macin activities demands reconsidering their potential biological impact on nerve repair/neurons in their hosts.
Keywords: Antimicrobial Peptides, Multifunctional Protein, Neurotrophic Factor, NMR, Protein Structure, Macin, Mechanism
Abstract
The tertiary structures of theromacin and neuromacin confirmed the macin protein family as a self-contained family of antimicrobial proteins within the superfamily of scorpion toxin-like proteins. The macins, which also comprise hydramacin-1, are antimicrobially active against Gram-positive and Gram-negative bacteria. Despite high sequence identity, the three proteins showed distinct differences with respect to their biological activity. Neuromacin exhibited a significantly stronger capacity to permeabilize the cytoplasmic membrane of Bacillus megaterium than theromacin and hydramacin-1. Accordingly, it is the only macin that displays pore-forming activity and that was potently active against Staphylococcus aureus. Moreover, neuromacin and hydramacin-1 led to an aggregation of bacterial cells that was not observed with theromacin. Analysis of the molecular surface properties of macins allowed confirmation of the barnacle model as the mechanistic model for the aggregation effect. Besides being antimicrobially active, neuromacin and theromacin, in contrast to hydramacin-1, were able to enhance the repair of leech nerves ex vivo. Notably, all three macins enhanced the viability of murine neuroblastoma cells, extending their functional characteristics. As neuromacin appears to be both a functional and structural chimera of hydramacin-1 and theromacin, the putative structural correlate responsible for the nerve repair capacity in leech was located to a cluster of six amino acid residues using the sequence similarity of surface-exposed regions.
Introduction
Antimicrobial peptides/proteins (AMP)3 most often target an essential bacterial structure, the bacterial cell membrane. As the elemental modification of microbial membranes to create resistance would be too complex to achieve, AMPs are instrumental in a plethora of organisms from bacteria, fungi, and protozoa to plants and animals including humans (1, 2). In general, they have remained effective for millions of years even against multidrug-resistant bacteria of today (3–6). The molecular mechanism of antimicrobial activity is mainly described by pore formation or by induction of micellization in a detergent-like manner, both of which cause the death of targeted bacteria (1). Recently, some antimicrobial peptides have been demonstrated to target specific lipid components within the bacterial membranes such as lipid II, leading to inhibition of cell wall synthesis (7). Besides being antimicrobially active, several AMPs possess further biological activities. Among others, they were found to be mediators of the adaptive immune system, participate in wound healing and angiogenesis, provide chemoattraction, modulate inflammation responses, and promote nerve repair (1, 8). To date, 1923 AMPs are deposited in the antimicrobial database (9), but for almost 80%, the tertiary structures, which are essential to better understand the molecular mechanisms of AMPs, are unknown.
The recently established macin family of antimicrobial peptides comprises three members, theromacin and neuromacin from leeches and hydramacin-1 from Hydra (10). Whereas each of the macins exerts antimicrobial activity, neuromacin also exerts a nerve repair activity. Hitherto, only the solution NMR structure of hydramacin-1 was determined, and its structure-activity relationship has been investigated (10). Hydramacin-1 adopts the knottin fold, a cysteine-stabilized α-β motif. Its mode of action was shown to be the aggregation of bacteria, which originates in its striking molecular surface property of comprising two hydrophobic hemispheres that sandwich a belt of positive charges. The corresponding mechanistic model, termed barnacle model, clearly deviates from well known mechanisms of membrane permeabilization. It suggests a dual electrostatic as well as hydrophobic peptide-membrane interaction applied in parallel to two individual bacterial cells (Fig. 1). Therefore, each bacterial cell can stick to several others, leading to the formation of huge cell aggregates. Hydramacin-1 is active against a broad range of Gram-negative and Gram-positive bacteria including multidrug-resistant strains. By contrast, the tertiary structures of theromacin and neuromacin are unknown. As the macins share high sequence identity including eight conserved cysteine residues, adoption of the same fold is likely. Nonetheless, both theromacin and neuromacin appear to act mainly against Gram-positive bacteria as no activity has been found against Gram-negative bacteria, albeit only a few strains have been tested so far (8, 11). Whereas the antimicrobial activity of neuromacin appears to involve membrane permeabilization (8), the antimicrobial mechanism of theromacin remains to be determined. Interestingly, neuromacin apparently is involved in the repair process of the central nervous system of the medicinal leech that is mediated by migration of microglia to the site of lesion and a probable cytoskeleton-driven axon outgrowth (8). However, the nerve repair mechanism of neuromacin remains elusive, and the capacity of theromacin and hydramacin-1 to promote the same is not known.
FIGURE 1.
Barnacle model exerted by hydramacin-1. Schematic representation of two negatively charged membranes (A and B) sandwiching hydramacin-1. Hydrophobic amino acid residues contributing to the molecular surface of hydramacin-1 are colored yellow, whereas cationic surface areas are colored blue.
The aim of the present study was to directly compare the three macins with each other to better understand their modes of action (antibacterial and nerve repair). Accordingly, here we have characterized the structures of the leech macins as well as the antimicrobial activities of theromacin and neuromacin and the nerve repair capacities of theromacin and hydramacin-1.
EXPERIMENTAL PROCEDURES
Cloning and Expression of Theromacin (TM) and Neuromacin (NM)
TM cDNA was cloned as follows. Total RNA from whole leech was extracted using TRIzol (Invitrogen). The RNA (3 μg) was transcribed into single-stranded cDNA using oligo(dT)18 adaptor primer, 5′-CGAGTCGACATCGATCGT18-3′ (SuperScript® kit, Invitrogen; protocol of the manufacturer). One-fourth of the reaction was amplified by PCR using the oligo(dT) primer and sense oligonucleotide 5′-atggctcctctcaacaagttgctct-3′. PCR was performed with 25 cycles using 1 unit of Taq polymerase (Go Taq, Promega) in 1.5 mm MgCl2. The cycling parameters were as follows: 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min. All PCR products were subcloned into pGEM-T easy vector (Promega), and several different cDNA clones were sequenced (GenBankTM accession number EU149766). NM cDNA was characterized in a previous study (GenBank accession number EU156754) (8).
After amplification of the sequences encoding the antimicrobially active regions of TM and NM (using the primer pairs 5′-GAAGATCTGGACGACGACGACAAGGGATGTTTCGAAGATTGG and 5′-CCCAAGCTTGTCAAGCCCAACAAGTTGG for TM and 5′-GAAGATCTGGACGACGACGACAAGGATTGCTACGAGGACTGGAGC and 5′-CCCAAGCTTGTCAGTAGCAATGACACTGTTTGTTGTTCTTGAGGACTC for NM), they were cloned into the expression vector pET-32a (Novagen) using BglII and HindIII (Fermentas). The final vectors were transformed into the Escherichia coli strain BL21(DE3). Both TM and NM were expressed as fusion proteins (FPs) comprising a thioredoxin-His6 tag N-terminally fused via an enterokinase cleavage site to the mature proteins. Uniformly 15N- and 15N/13C-labeled TM-FP was expressed in 2× M9 minimal medium supplemented with biotin and thiamine (1.5 mg/liter each) (12). When bacteria cultures reached an optical density (measured at 600 nm) of 0.5–0.6, isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 1 mm to induce protein expression. Cell growth was continued for 5 h. The TM-FP was expressed as inclusion bodies. Non-labeled TM-FP and NM-FP were generated using lysogeny broth. NM-FP was expressed as a soluble form.
Purification, Renaturation, and Cleavage of Fusion Proteins
Bacteria were harvested by centrifugation, resuspended in phosphate-buffered saline, pH 7.4 in the case of TM or Lysis-Equilibration-Wash buffer (Protino® protein purification system, Macherey-Nagel, Düren, Germany) in the case of NM, and disrupted by sonification. Soluble and insoluble cell fractions were separated by centrifugation. The TM-FP inclusion bodies were purified under denaturing conditions and renatured as described (10). The NM-FP was purified under native conditions using Protino Ni-TED 2000 columns (Macherey-Nagel) following the manufacturer's instructions. The eluates containing NM-FP were dialyzed against 50 mm Tris, pH 8. Cleavage of the FPs was performed at 37 °C overnight using EnterokinaseMaxTM (Invitrogen).
Purification of Mature Theromacin and Neuromacin
The purification of mature TM or NM was performed using reversed-phase high performance chromatography on semipreparative C18 columns. A continuous acetonitrile gradient of 0.22%/min over a period of 23 min starting at a concentration of 25% (v/v) was used to obtain isolated TM, whereas NM was isolated using a continuous acetonitrile gradient of 1.2%/min for 20 min also starting at a concentration of 25% (v/v). Fractions containing TM or NM were pooled and lyophilized. The purity of the samples was confirmed by mass spectrometry in linear mode using a 4700 Proteomics Analyzer MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Framingham, MA). Size exclusion chromatography using a HiLoad Superdex 75 prep grade (16/60) column (Amersham Biosciences) under conditions used for NMR spectroscopy confirmed the monomeric state of the mature theromacin protein (data not shown). The lyophilized samples used for activity assays were reconstituted in 0.01% (v/v) trifluoric acid, and the protein concentration was adjusted to 1 mg/ml. Aliquots of these stocks were used for investigation of activity and mode of action.
Assignment of Disulfide Bonds
Disulfide bridges of the macins were assigned by applying a combination of enzymatic digestions with different proteases (i.e. trypsin for neuromacin and sequential enzyme treatment with trypsin and Asp-N for theromacin) followed by separation of the cleavage peptides by means of ion-pairing reversed-phase liquid chromatography and analysis by electrospray mass spectrometry (LTQ-Orbitrap Velos, Thermo Fisher Scientific, Bremen, Germany). Hydramacin-14 served as a positive control because of its known disulfide isomerization state (10). Experimental details will be published elsewhere.5
Sample Preparation and NMR Experiments
Freeze-dried theromacin was dissolved in 20 mm sodium phosphate, pH 7.4, 93% (v/v) H2O, 7% (v/v) D2O. The sample was transferred into a symmetrical matched microtube (Shigemi, Inc.).
NMR measurements were performed on a Bruker Avance 600-MHz spectrometer equipped with a z-gradient triple resonance cryoprobe. Sequence-specific backbone resonance assignments of theromacin were established using the following spectra: two-dimensional total correlation spectroscopy (TOCSY) with a mixing time of 60 ms and three-dimensional HNCA, HNCO, (H)C(CO)NH, H(CCO)NH, HN(CA)CO, HN(CO)CA, HN(CO)CACB, HNCACB, 15N-edited TOCSY, and 13C-edited HCCH-TOCSY. For verification of flexible regions, (1H)-15N heteronuclear nuclear Overhauser effect (NOE) spectra were measured additionally. Two spectra with on and off resonance 1H saturation were recorded in an interleaved manner and separated before processing with the Bruker AU program split. The combined recycle and saturation delay was set to 3.2 s. Only well resolved peaks were included in the intensity (peak height) measurements and NOE ratio determination. Distance restraints were obtained from two-dimensional NOE spectroscopy (mixing time, 200 ms) and three-dimensional 15N-edited and 13C-edited NOE (mixing times, 150 ms) spectroscopy-heteronuclear single quantum correlation experiments. The spectra were acquired at 300 K and referenced to the water resonance at 4.75 ppm. The chemical shift data were deposited in the University of Wisconsin Biological Magnetic Resonance Bank database under accession number 18153. All spectra were processed with the program NMRPipe (13) and analyzed with the program NMRView (14).
Structure Calculations
Structure calculations were performed using the program CYANA (15). The calculations were based on 520 interproton constraints. Distance restraints derived from the NMR spectra were calibrated using an r̂6 function. Five disulfide bonds were defined as 15 distance restraint ranges as follows: 2.0 ≤ d(Siγ, Sjγ) ≤ 2.1 Å; 3.0 ≤ d(Ciβ, Sjγ) ≤ 3.1 Å; 3.0 ≤ d(Siγ, Cjβ) ≤ 3.1 Å. The program TALOS+ (16) was used for deriving backbone torsion angles from the NMR chemical shifts of theromacin and included in the structure calculations. 100 structures were calculated and subsequently refined in explicit solvent with the CNS program using the RECOORD protocol and parameters (17). The 10 energetically best structures were selected as the final refined structural ensemble and were deposited (Protein Data Bank code 2ln8).
Molecular Modeling of Neuromacin
Models of the three-dimensional structures of neuromacin were generated using the solution NMR structures of theromacin and hydramacin-1 (Protein Data Bank code 2k35) as templates. According to the sequence alignment, amino acid residues of neuromacin were exchanged in the templates. Insertions and deletions in the molecules were modeled using a database search approach included in the software package WHATIF (18). The model structures were finally energy-minimized using the GROMOS force field (19).
Graphic Representations
All molecular graphical representations were generated using the programs MOLMOL (20) and GRASP2 (21).
Determination of Antimicrobial Activity and Salt Tolerance
The antimicrobial activity of theromacin and neuromacin was investigated as described previously (22). Hydramacin-14 served as reference molecule and was also measured in parallel. Briefly, test strains (E. coli ATCC 35218, Staphylococcus aureus ATCC 12600, and Bacillus megaterium ATCC 14581) were grown in brain-heart infusion broth at 37 °C for 2–3 h. The cells were washed three times in 10 mm sodium phosphate buffer, pH 7.4 supplemented with 1% tryptic soy broth, and the cell number was adjusted to 104–105 cells/ml. From the prepared cell suspension, 100 μl were mixed with 10 μl of the protein stock diluted in 10 mm sodium phosphate buffer, pH 7.4, 1% tryptic soy broth and incubated at 37 °C. The final peptide concentrations tested were between 0.0125 and 100 μg/ml. After 2 h of incubation, the colony-forming units were determined. For a negative control, bacterial suspensions were supplemented with 10 μl of phosphate buffer, pH 7.4, 1% tryptic soy broth. Antimicrobial activity was tested for all strains by determination of 90% lethal dose (LD90) or minimal bactericidal concentrations (MBCs) (≥99.9% killing). The values were expressed as the median of at least three independent experiments. Reproducibility of results did not exceed one dilution step.
The salt sensitivity of the macin proteins was determined as described above using 10 mm phosphate buffer, pH 7.0 with or without supplemented sodium chloride (final concentrations of 50–150 mm). Arbitrarily, a strain was defined as susceptible to a peptide if it had an MBC or LD90 of ≤12.5 μg/ml, as intermediately susceptible if it had an MBC or LD90 of >12.5–100 μg/ml, or as resistant if it had an MBC or LD90 of >100 μg/ml as described elsewhere (22).
Assays for Permeabilization of Bacterial Membranes and for Pore-forming Activity
Bacteria with compromised membranes were detected by monitoring the fluorescence of the DNA binding dye SYTOX Green (Invitrogen) as described previously (23). B. megaterium ATCC 14581 in midlogarithmic phase were washed twice and resuspended in 10 mm HEPES, 25 mm sodium chloride, pH 7.4. A flat bottom 96-well microtiter plate (Sarstedt, Germany) was coated with 0.1% bovine serum albumin (A2153, Sigma-Aldrich) for 15 min before its use in the assay. Theromacin, neuromacin, and hydramacin-14 were 2-fold serially diluted in 10 mm HEPES, 25 mm sodium chloride, pH 7.4. Bacteria (105 colony-forming units/50 μl) were incubated with the diluted peptides (25 μl) and 2 μm fluorescent dye SYTOX Green (25 μl in 10 mm HEPES, 25 mm sodium chloride, pH 7.4) at 20 °C for different time periods. Permeabilization of the bacterial cytoplasmic membrane allows the dye to cross the membrane and to intercalate with the DNA. Excitation of the DNA-bound dye at 495 nm resulted in an increase of emitted fluorescence at 538 nm. Measurements were made in a microtiter plate reader (Fluoroskan II, Labsystems). Membrane-permeabilizing activity of the peptide was expressed as a percentage of permeabilized bacteria. For monitoring the activity at pH 5.2, 20 mm MES buffer supplemented with 25 mm sodium chloride was used. As a control, the antimicrobial peptides cecropin P1 and magainin II (Sigma-Aldrich) were used. For maximum permeabilization of the bacteria (100% value), cells were incubated with 70% ethanol for 10 min. The experiments were performed in duplicate.
Pore-forming activity of theromacin and neuromacin was determined by measuring fluorometrically the dissipation of a valinomycin-induced membrane potential in liposomes prepared from soy bean asolectin as described previously (24). Fluorescence was measured by a fluorescence spectrophotometer (model LS 50B, PerkinElmer Life Sciences) using excitation and emission wavelengths of 620 and 670 nm, respectively. Pore-forming activity was measured as the initial change in fluorescence intensity over time after the addition of the protein samples. The pore-forming peptide alamethicin (Sigma-Aldrich) served as a positive control to calibrate the assay.
Investigation of Protein-Membrane Interaction by Dynamic Light Scattering
Liposomes were prepared essentially as described by Pick (25) using defined phospholipids and 50 mm sodium phosphate buffer, pH 5.2. Initially, crude liposome samples were refined by passing them over a NAP-5 column (Amersham Biosciences). The eluate served as a stock suspension for the subsequent experiments and was stored at 4 °C. The following phospholipids were purchased from Avanti Polar Lipids Inc. (Alabaster, AL): l-α-phosphatidyl-dl-glycerol and l-α-phosphatidylcholine. Because of the net negative charge at the membrane surface, l-α-phosphatidyl-dl-glycerol liposomes serve as a very simplified model for bacterial membranes, whereas l-α-phosphatidylcholine liposomes serve as a corresponding model for eukaryotic membranes because of their electrostatically neutral surface.
The dynamic light scattering experiments were performed using a Laser-Spectroscatter 201 (RiNA GmbH, Berlin, Germany) equipped with a laser diode with a wavelength of 662 nm and an optical power of 100 milliwatts. Each experiment consisted of 10 measurements, each with a duration of 10 s. The liposome stock suspension was diluted 1:100 in the corresponding buffer (50 mm sodium phosphate with either an acidic pH of 5.2 or a pH of 7.0), and 10 μl of it were measured by dynamic light scattering. Directly after measurement, 3 μl of the protein sample (0.5 mg/ml theromacin, neuromacin, and hydramacin-14) were added and mixed thoroughly, and the sample was measured again. After 3 min, a final measurement was done without any disturbance of the sample in between.
Nerve Repair in Leech
Adult Hirudo medicinalis were purchased from Ricarimpex (Bordeaux, France) and maintained in autoclaved 1% Instant Ocean (Aquarium Systems) changed daily for 1 week before starting any experimental procedure. Leeches were anesthetized by immersion in 10% ethanol, spring water at 4 °C for 20 min. The nerve cords were removed and prepared for nerve repair assays as described previously (8). Connectives between the third and fourth nerve ganglia were hemisectioned with a thin scissor. The regenerative effect of the peptides was evaluated by adding the natural or recombinant molecule at a final concentration of 4 μm in the culture medium. Assays were performed on three different nerve cords, and the procedure was repeated three times. Photographs were taken every day for 1 week (objective, ×5) using a Leica DMIRE2 inverted microscope. Images were taken using Bioposition version 3.0 software developed on the Matrox MIL 7.5 Base Library.
Proliferation Assay and Immunocytochemistry
Murine Neuro-2A neuroblastoma cells (DSMZ) were maintained in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a humidified atmosphere with 5% CO2. 5 × 103 cells/well were plated in triplicate on a 96-well plate and incubated with 0.1 μm hydramacin-1, neuromacin, or theromacin for 48 h. Cell viability was assessed by relative cellular ATP levels using CellTiter-Glo (Promega) according to the manufacturer's instructions. Statistical significance was established at p < 0.01 by one-way analysis of variance with Bonferroni post hoc test. For immunocytochemical analysis, 104 Neuro-2A cells/well were plated on 8-well chamber slides (Nunc). After 24 h, the cells were incubated with 0.1 μm hydramacin-1, neuromacin, or theromacin for 15 min, 30 min, or 3 h. Cells were fixed with 4% warm (37 °C) paraformaldehyde at room temperature for 10 min and afterward at 4 °C for 1 h. After washing with phosphate-buffered saline (PBS) three times, with PBS, 0.12% glycine once, and with PBS once, samples were treated with 10% (v/v) fetal bovine serum in PBS for 45 min followed by 1-h incubation with the polyclonal anti-theromacin antibody at room temperature. Identical concentrations of preimmune serum were used for controls and verification of background signals. The secondary goat-anti rabbit-DyLightTM488-coupled antibody (1:300; Biomol) was applied after three washes with PBS and one wash with PBS, 10% (v/v) fetal bovine serum. After incubation at room temperature for 1 h, samples were washed four times with PBS, stained with 1 μm DAPI, and mounted with Mowiol/Dabco (Calbiochem/Sigma-Aldrich). Slides were analyzed with an FV1000 confocal microscope with a 60× lens. Image acquisition was done with FV10-ASW (version 3.0, Olympus Europa GmbH, Hamburg, Germany).
RESULTS
Tertiary Structures of Theromacin and Neuromacin
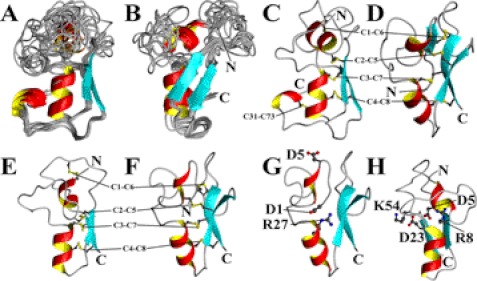
The structural statistics for the calculation of the theromacin structure are summarized in Table 1. The ensemble of the 10 energetically best theromacin structures is shown in Fig. 2, A and B. Theromacin contains two flexible loops: one is located between the N-terminal helix and the middle helix, and the second is between the two β-strands (Fig. 2, A, B, and C). The spatial positioning of the N-terminal helix and the C-terminal region also varies slightly. The flexibility of these regions is an inherent property of the protein and was verified by heteronuclear NOE experiments (supplemental Fig. SF1). Like hydramacin-1 (Fig. 2D), theromacin folds into the knottin fold, i.e. a cysteine-stabilized α-β topology. Moreover, it possesses the N-terminal helix characteristic for macins (10) but does not contain an N-terminal β-strand like hydramacin-1. Instead, its N-terminal helix is directed toward the interstrand loop. Finally, the C-terminal region of theromacin folds into a short third helix (Fig. 2C).
TABLE 1.
Structural statistics for 10 energetically best structures of theromacin
r.m.s.d., root mean square deviation.
| Distance restraints | |
| Intraresidual (|i − j| = 0) | 103 |
| Sequential (|i − j| = 1) | 180 |
| Medium range (2 ≤ |i − j| ≤ 4) | 75 |
| Long range (|i − j| ≥ 5) | 132 |
| Disulfide bonds (included) | 30 |
| Total | 520 |
| NOE restraint violations | |
| Mean number of NOE violations >0.3 Å | 0.2 |
| Maximum violation (Å) | 0.35 |
| r.m.s.d. for NOE restraints (Å) | 0.041 ± 0.003 |
| Deviation from idealized geometry | |
| Bond length (Å) | 0.0134 ± 0.0006 |
| Bond angles (°) | 1.74 ± 0.05 |
| Impropers (°) | 2.09 ± 0.14 |
| Mean global backbone r.m.s.d. to meana (Å) | 0.63 ± 0.20 |
| Mean global heavy r.m.s.d. to meana (Å) | 1.38 ± 0.29 |
| Ramachandran plot | |
| Most favored regions (%) | 71.7 |
| Additional allowed regions (%) | 24.6 |
| Generously allowed regions (%) | 2.0 |
| Disallowed regions (%) | 1.7 |
a Residues considered: 24–41 and 55–67.
FIGURE 2.
Tertiary structures of macin family members. Depicted are the ribbon presentations of an ensemble of the 10 energetically best theromacin structures (A and B), a representative theromacin structure (C), the average structure of hydramacin-1 (D; Protein Data Bank code 2k35), and the model structures of neuromacin with theromacin as template (E and H) or hydramacin-1 as template (F and G). The ensemble in B is 70° rotated around the vertical axis. Covalent cysteine residue bonds are displayed in gray and yellow balls and sticks. N and C denote the N and C termini, respectively. C1–C8 label four covalent cysteine residue bonds conserved in all macins according to their relative position in the primary structure, whereas C31-C73 labels an additional fifth cysteine residue bond in theromacin according to their absolute position in the primary structure. Further labeled residues are discussed in the text.
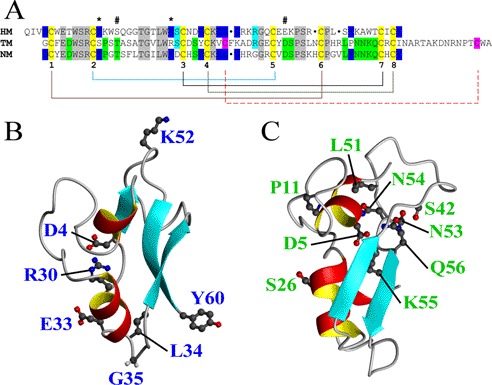
The macins possess a high sequence identity including eight conserved cysteine residues (Fig. 3A). The assignment of the disulfide bridges of theromacin and neuromacin by mass spectrometry revealed that the eight cysteine residues conserved in all three macins form four disulfide bridges: Cys1-Cys6, Cys2-Cys5, Cys3-Cys7, and Cys4-Cys8 (Fig. 2, C–F, and supplemental Table S1). Theromacin possesses two additional cysteine residues that form a fifth disulfide bond (Cys31-Cys73). The disulfide bridges identified by mass spectrometry are in agreement with the NOE pattern observed in the NMR spectra. Based on the conserved disulfide pattern and the topology of theromacin and hydramacin-1, two structural models of neuromacin were generated using either theromacin (Fig. 2E) or hydramacin-1 (Fig. 2F) as a template. Both models differ in the positioning of the N terminus. Interestingly, the residues Asp4 and Arg30, which form a salt bridge in hydramacin-1, are conserved in neuromacin (Asp1 and Arg27) and hence would allow the formation of a salt bridge in the corresponding hydramacin-1-based model as well (Fig. 2G). This would favor an interior positioning of the N-terminal region in neuromacin. In contrast, in the theromacin-based neuromacin model, the N-terminal region is directed toward the molecular surface. As a consequence, the residue Asp5 is buried inside the molecule and faces Asp23 (Fig. 2H), which would be energetically unfavorable compared with the situation in the hydramacin-1-based model where Asp5 is exposed to the surface (Fig. 2G). In the case of theromacin, the charge of the Asp5 residue is compensated by residue Lys55 or Arg8, which correspond to Lys54 and Arg8 in neuromacin (Fig. 2H). Moreover, the corresponding position 23 in theromacin is substituted by a serine residue (Fig. 3A). Therefore, an electrostatic repulsion as observed in the theromacin-based neuromacin model does not exist. Because of these charge interactions, the putative hydramacin-1-based model for neuromacin appears more likely.
FIGURE 3.
Conserved residues in macin family members. A, sequence alignment of the primary structures of TM, NM, and hydramacin-1 (HM). Lines indicate disulfide bonds between the corresponding cysteine residues (numbers 1–8) conserved in all three macins. The dashed line marks two additional cysteine residues (marked in pink) in theromacin forming a fifth disulfide bond. Residues conserved in all macins are marked in gray or yellow, respectively, whereas residues that are conserved in only two of the three macins are marked in blue, green, or light blue. Asterisks and hash marks indicate conserved residues where a conserved substitution is present in the third macin. B, representative ribbon model of the theromacin structure including residues only conserved in neuromacin. C, ribbon presentation of the hydramacin-1 average structure (Protein Data Bank code 2k35) including residues only conserved in neuromacin. Conserved residues marked by asterisks and hash marks in the sequence alignment are not shown in B and C.
The sequence alignment of the macins (Fig. 3A) depicts separately conserved residues that can be found either in neuromacin and hydramacin-1 or in neuromacin and theromacin. Interestingly, there is a clear accumulation of the majority of the separately conserved residues in two different regions of the macins (Fig. 3, B and C). In the case of hydramacin-1/neuromacin, most of the conserved residues (Glu33, Leu34, Gly35, and Tyr60) are allocated to the highly defined region of the molecule (Fig. 3B). The conserved salt bridge-forming residues Asp4 and Arg30 were already mentioned above. In contrast, most of the residues conserved exclusively between theromacin and neuromacin are localized in the more flexible region of the molecules. More precisely, these residues (Ser42, Leu51, Asn53, Asn54, Lys55, and Gln56) cluster in the inter-β loop (Fig. 3C).
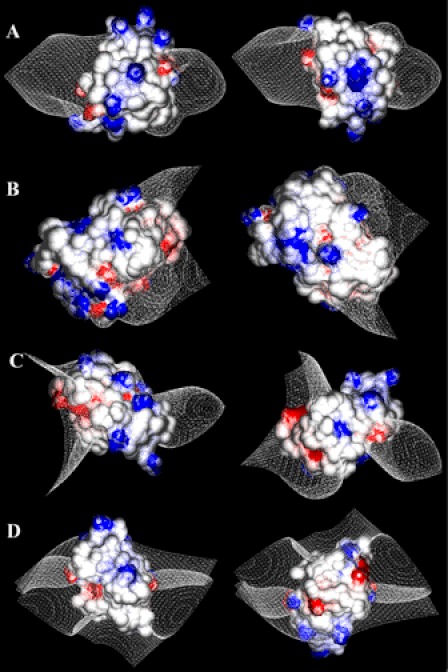
The molecular surface electrostatic potentials of the macins are shown in Fig. 4. Hydramacin-1 possesses a ringlike arrangement of cationic residues (Fig. 4A). The contour plot of the isopotential plane shows that this cationic ring is sandwiched by two hemispheres of neutral charge. By contrast, theromacin shows an accumulation of positively and negatively charged patches arranged oppositely with an isopotential plane in between (Fig. 4B). Therefore, the bipolar nature of theromacin clearly differs from the surface properties of hydramacin-1. The contour plot of the theromacin-based neuromacin model also shows a ringlike arrangement of cationic residues that is sandwiched by two isopotential planes (Fig. 4C), which is highly comparable with the situation in hydramacin-1. In the case of the hydramacin-1-based neuromacin model, the ring of cationic residues is not closed completely (Fig. 4D). As a consequence, the isopotential plane does not split the molecule completely into two hemispheres as observed for hydramacin-1. Nevertheless, both neuromacin models reveal surface properties more related to hydramacin-1 than to theromacin.
FIGURE 4.
Molecular surface properties of macin family members. The molecular surface electrostatic potentials are depicted for hydramacin-1 (A), theromacin (B), neuromacin (theromacin template) (C), and neuromacin (hydramacin-1 template) (D) with representation of the contour plot of the isopotential plane as white mesh. Cationic regions are colored blue, and anionic regions are marked in red. In the right column, molecules are presented after 180° rotation around the horizontal axis.
Antimicrobial Activity of Macins
The antimicrobial activities of the three macin proteins were measured in parallel. They showed antimicrobial activity against Gram-positive bacteria and Gram-negative bacteria in the low microgram concentration range (Table 2). The Gram-positive bacterium B. megaterium was killed by each macin in a comparable low concentration range (MBC = 0.2–0.39 μg/ml). However, significant activity differences against the Gram-positive bacterium S. aureus and the Gram-negative bacterium E. coli were observed. The latter is highly susceptible to hydramacin-1 (LD90 = 1.56 μg/ml) and, albeit 4–8 times less, also susceptible to theromacin (LD90 = 6.25 μg/ml) and neuromacin (LD90 = 12.5 μg/ml). In contrast, S. aureus appears to be resistant to hydramacin-1 (LD90 > 100 μg/ml), but it is highly susceptible to neuromacin (LD90 = 3.13 μg/ml) and intermediately susceptible to theromacin (LD90 = 25 μg/ml).
TABLE 2.
Antimicrobial activity of macins
| Strain and protein | MBCa | LD90 |
|---|---|---|
| μg/ml | μg/ml | |
| E. coli ATCC 35218 | ||
| Hydramacin-1 | 3.13 | 1.56 |
| Neuromacin | 25 | 12.5 |
| Theromacin | 25 | 6.25 |
| S. aureus ATCC 12600 | ||
| Hydramacin-1 | >100 | >100 |
| Neuromacin | 6.25 | 3.13 |
| Theromacin | 100 | 25 |
| B. megaterium ATCC 14581 | ||
| Hydramacin-1 | 0.39 | 0.1 |
| Neuromacin | 0.20 | 0.1 |
| Theromacin | 0.39 | 0.2 |
a ≥99.9% killing.
The antimicrobial activities of the macins against E. coli and S. aureus were strongly salt-dependent. The activity of theromacin and hydramacin-1 was completely lost at low concentrations of sodium chloride (50 mm NaCl) for both strains, whereas neuromacin still displayed some activity against S. aureus (Table 3). However, this activity was abolished in the presence of 100 mm sodium chloride.
TABLE 3.
Salt sensitivity of antimicrobial activities of macins
| Protein and NaCl concentration |
E. coli ATCC 35218 |
S. aureus ATCC 12600 |
||
|---|---|---|---|---|
| MBC | LD90 | MBC | LD90 | |
| μg/ml | μg/ml | μg/ml | μg/ml | |
| Hydramacin | ||||
| No NaCl | 3.13 | 1.56 | >100 | 100 |
| 50 mm NaCl | >100 | >100 | >100 | >100 |
| 100 mm NaCl | >100 | >100 | >100 | >100 |
| 150 mm NaCl | >100 | >100 | >100 | >100 |
| Neuromacin | ||||
| No NaCl | 25 | 12.5 | 6.25 | 3.13 |
| 50 mm NaCl | >100 | >100 | 100 | 50 |
| 100 mm NaCl | >100 | >100 | >100 | >100 |
| 150 mm NaCl | >100 | >100 | >100 | >100 |
| Theromacin | ||||
| No NaCl | 12.5 | 6.25 | 100 | 25 |
| 50 mm NaCl | >100 | >100 | >100 | >100 |
| 100 mm NaCl | >100 | >100 | >100 | >100 |
| 150 mm NaCl | >100 | >100 | >100 | >100 |
Membrane Permeabilization and Pore-forming Activity
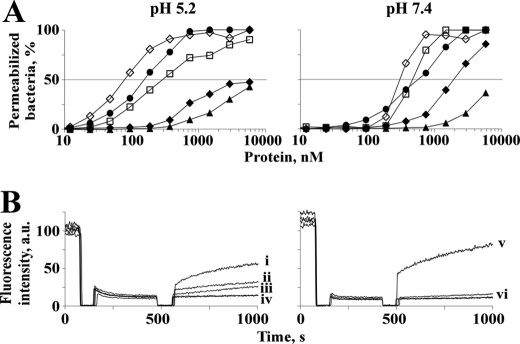
The capability of the macin proteins to permeabilize bacterial membranes was measured in parallel. They permeabilized B. megaterium cell membranes without showing a clear pH dependence (Fig. 5A). The percentage of permeabilized bacteria increased with time (data not shown) and concentration. In Fig. 5A, the permeabilization of bacteria after 5 min of incubation with the proteins at pH 5.2 and 7.4 is depicted. In the case of theromacin, 50% permeabilized bacteria were observed at a concentration of 10 μm. In comparison, hydramacin-1 led to 50% permeabilization at 2–3 times lower concentrations (2 and 6 μm). Strikingly, neuromacin was able to permeabilize 50% of the bacteria already at a concentration of 0.2–0.6 μm, which is in the range of the well known antimicrobial peptides magainin II and cecropin P1 and is 1 order of magnitude lower compared with the other two macins.
FIGURE 5.
Membrane permeabilization of viable bacteria and pore-forming activity. A, the capability of theromacin (▴), neuromacin (●), and hydramacin-1 (♦) of permeabilizing membranes of B. megaterium cells was determined at pH 5.2 (left-hand panel) and 7.4 (right-hand panel) after 5-min incubation. The fluorescence of the SYTOX Green dye was used as a measure of bacteria with compromised membranes. Magainin II (♢) and cecropin P1 (□) served as positive controls. B, the dissipation of a valinomycin-induced diffusion potential of liposomes induced by the proteins at pH 5.2 (left-hand panel) and 7.4 (right-hand panel) was monitored by fluorescence measurements. The pore-forming peptide alamethicin (0.1 μm; measurements “i” and “v”) served as a positive control, whereas the solvent of the protein sample served as a negative control. “ii” and “iii” mark measurements with 2 or 1 μm neuromacin, respectively. “iv” and “vi” summarize measurements with 2 μm theromacin and 2 μm neuromacin (pH 7.4), respectively, and negative controls, which all overlap in the diagrams. a.u., arbitrary units.
Investigation of pore-forming activities revealed that neuromacin is able to form pores only under acidic conditions (Fig. 5B). At neutral pH, pore-forming activity was not detectable up to a concentration of 2 μm. In comparison with the classical pore-forming peptide alamethicin (0.1 μm), neuromacin possesses ∼25 and 50% pore-forming activity at 10 and 20 times higher concentrations (1 and 2 μm), respectively. In contrast, theromacin did not show any pore-forming activity up to a protein concentration of 2 μm at either acidic or neutral pH. Hydramacin-1 does not form pores at any tested condition or concentration (10).
Dynamic Light Scattering
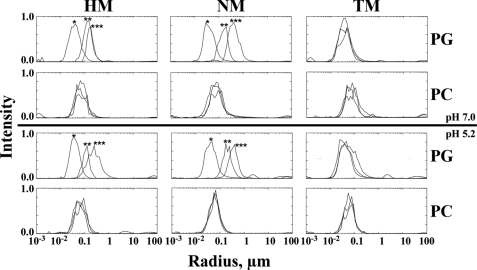
Dynamic light scattering measurements of protein-membrane interactions showed a peptide concentration-driven formation of aggregates for hydramacin-1 and neuromacin. This was independent of pH but dependent on the membrane surface charge (Fig. 6). In the case of negatively charged l-α-phosphatidyl-dl-glycerol liposomes, the measured particle radii increased within seconds from ∼30–40 to 100–200 nm after addition of the proteins to the liposome suspensions. Moreover, the radii of the aggregates progressively increased with time to reach ∼200–400 nm after 5 min of incubation. This effect was not observed for neutral l-α-phosphatidylcholine liposomes, indicating an electrostatic interaction. In contrast, theromacin did not lead to any measurable aggregation.
FIGURE 6.
Investigation of protein-membrane interaction by dynamic light scattering. Presented are the measured particle radii of l-α-phosphatidyl-dl-glycerol (PG) and l-α-phosphatidylcholine (PC) liposome suspensions in the absence (*) and presence of hydramacin-1 (HM), NM, and TM at acidic and neutral pH, respectively. Asterisks indicate measurements directly (**) or 5 min (***) after mixing liposome suspensions and protein. In the case of no apparent differences between the results of the three measurements, there are no asterisks.
Leech Nerve Cord and Murine Neuroblastoma Cells
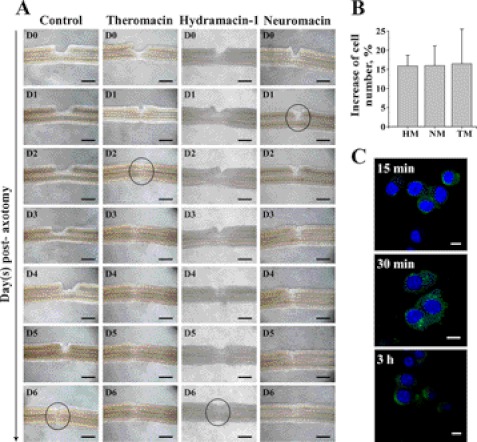
The nerve repair activities of the macin proteins were measured in parallel. In contrast to mammals, the leech central nervous system has the ability to regenerate after section, and this capacity can be visualized ex vivo (8). As shown in Fig. 7A under control conditions (without peptides), restoration of the connective nerve across the cut began at day 5 postaxotomy and was completed at day 6. As previously described for the natural neuromacin (8), the presence of the recombinantly expressed neuromacin accelerated this repair process that was then completed 24 h postaxotomy. Interestingly, theromacin also revealed a nerve repair capacity. In contrast to neuromacin, the regenerative response was slightly longer and was completed 2 days postaxotomy. Hydramacin-1 did not show any beneficial or deleterious effect on the leech nerve cord. Interestingly, all macins increased the viability of murine neuroblastoma cells. Compared with untreated controls, cell numbers were elevated by ∼15% after 48 h of incubation with the macins (Fig. 7B). After 96 h, the increase even reached more than 60% (data not shown). With a polyclonal antibody for theromacin-1 (11), we performed immunocytochemistry in murine neuroblastoma cells upon incubation with theromacin. The peptide was initially found at the membrane of the cells after 15 min (Fig. 7C). After 30 min, theromacin was localized to the cytosol, revealing an inhomogeneous distribution (Fig. 7C), whereas after 3 h, the peptide was still detectable in the cytosol but appeared distributed more homogeneously (Fig. 7C). At all recorded time points, the protein was not localized to the nuclei of the cells. Results of the corresponding negative control experiments did not show a significant green fluorescence.
FIGURE 7.
Impact of macins on damaged leech connective nerve and neuronal cells. A, after axotomy of one side of the paired connective nerve, its repair in the absence (control) and presence of recombinant theromacin, hydramacin-1, or neuromacin was studied. The repair process of the nerve cord was monitored taking sequential micrographs from the day of sectioning (D0) until day 6 (D6) after sectioning when full repair of the connective nerve had occurred. Circles mark accomplished repair. Black scale bars indicate 150 μm. B, viability of murine Neuro-2A neuroblastoma cells incubated with hydramacin-1 (HM), neuromacin, or theromacin for 48 h. Statistical significance, p < 0.01. C, immunocytochemistry of murine neuroblastoma cells incubated with theromacin for 15 min, 30 min, or 3 h, respectively. Theromacin (green) was detected using a DyLight488-coupled antibody, and cell nuclei were stained with DAPI (blue). White scale bars indicate 10 μm.
DISCUSSION
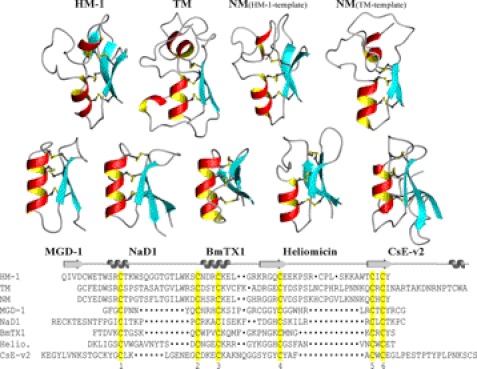
Hydramacin-1 was considered the founding member of a new family of antimicrobial proteins within the superfamily of scorpion toxin-like proteins, the macin family, which potentially also encompasses theromacin and neuromacin (10). The three-dimensional structure of theromacin established here clearly confirmed this suggestion. The tertiary structures as well as a sequential alignment of the macins and of representative members of all families of the scorpion-toxin like protein superfamily are depicted in Fig. 8. Theromacin exhibits the same structural features as hydramacin-1, in particular (i) the characteristic additional N-terminal helix and (ii) two long flexible loops separating the two helices and the two strands. Both are missing in the other five structures (Fig. 8). Because of the conserved disulfide pattern of the macins and their high sequence identity, it is very likely that neuromacin also adopts this fold. As expected, both models of neuromacin show the structural features typical for macins. For structural reasons (conserved salt bridge and Asp5 residue pointing outside), the model based on hydramacin-1 appears more likely to reflect the tertiary structure of neuromacin.
FIGURE 8.
Representatives of families of scorpion toxin-like superfamily. Displayed are the ribbon representations of the structures of hydramacin-1 (HM-1), TM, and NM either modeled on hydramacin-1 or theromacin and one representative of each of the other five families: Mediterranean mussel defensin (MGD-1) (33), Nicotiana alata plant defensin 1 (NaD1) (34), Buthus martensi toxin 1 (BmTX1) (35), heliomicin (Helio.) (36), and Centruroides sculpturatus Ewing variant 2 toxin (CsE-v2) (37). The sequence alignment of all molecules is shown below the structures. Cysteine residues conserved in the primary structure of all depicted molecules are highlighted in yellow and displayed as balls and sticks in the structures, respectively. The alignment is based on the conserved cysteine residues 1–6. Positions of α-helices and β-sheets (arrows), respectively, in the macins are depicted on top of the sequence alignment.
The most striking structural difference between hydramacin-1 and theromacin is the spatial positioning of the N-terminal region, which is located in the flexible region of the molecules. Therefore, there is the possibility for compensating the current unfavorable positioning of the mentioned residues (Asp5 and Asp23). Moreover, by comparing electrostatic surface potentials, one might favor the theromacin-based neuromacin model for functional reasons. Both hydramacin-1 and neuromacin led to aggregation of liposomes (Fig. 6) as well as Gram-negative bacteria (data not shown), whereas theromacin did not. Taking into consideration the barnacle model, the molecular surface properties of both molecules must be congruent and have to fulfill two conditions: (i) a ringlike arrangement of cationic residues and (ii) two oppositely located non-charged surface patches. The surface properties of theromacin clearly deviate from that of hydramacin-1. The bipolar character of theromacin is insufficient for aggregation according to the barnacle model. On the contrary, the surface properties of hydramacin-1 and neuromacin, regardless of the template used, are much more comparable. As the surface properties of the theromacin-based neuromacin model show a higher resemblance to that of hydramacin-1, one might favor it in the functional context. However, due to the partial flexibility of the loops, the hydramacin-1-based model might become more similar to hydramacin-1 (i.e. its surface properties). In summary, the analysis of both neuromacin models clearly shows a high probability for the adoption of hydramacin-1-like surface properties and therefore corroborates the barnacle model.
The antimicrobial character of the macins is clearly demonstrated by their ability to eradicate B. megaterium. However, noticeable differences exist in killing S. aureus and E. coli that appear analogous to the situation of human β-defensin (HBD) 2 and HBD3 (3). Actually, the sequence identity/similarity between HBD2 and HBD3 (31%/47%) is significant lower compared with that between hydramacin-1 and neuromacin (48%/61%). Therefore, it remains even more puzzling why the macins behave that differently. Nevertheless, the structural regions encoding the different activities of the HBDs were located by a rational chimeric protein design approach that may be applied to the macins to better understand their structure-activity relationships.
The macins permeabilized membranes of B. megaterium within a few minutes, suggesting the bacterial membrane as the target and its disruption as the mode of action. However, the macins showed significant differences in their mechanistic behavior. The difference in following the barnacle model was already mentioned above. Furthermore, a pore-forming activity was solely observed for neuromacin. As this was pH-dependent, it appears to be mediated through the de-/protonation of the (four) histidine residues that are missing in theromacin and hydramacin-1. The pore-forming activity of neuromacin is only moderate compared with the classical pore-forming peptide alamethicin. In contrast, the capability of neuromacin to permeabilize the membranes of B. megaterium was in the range of the antimicrobial and membrane-active peptides magainin II and cecropin. Therefore, the formation of stable pores by neuromacin is probably not the primary mechanism of its antimicrobial activity.
Besides its antimicrobial activity, neuromacin was already known to exert a nerve repair activity. Now a second leech antimicrobial peptide, namely theromacin, was for the first time demonstrated to exert this activity. In Hirudo, the production sites of the leech macins are in accordance with their regenerative effects on injured nerve cords. Indeed, theromacin is released into the blood that surrounds the nervous system, whereas neuromacin is directly produced by nerve cells and accumulates at the wounded site of the central nervous system (26). Recently, we demonstrated that the leech plasma enriched in AMPs, among them theromacin, stimulated the regeneration of the central nervous system, suggesting a nerve repair activity of at least one of these antibiotic molecules (27). The data presented here corroborate this hypothesis and support the idea of an in vivo activity of theromacin.
The neuronal repair process in leech does not include the de novo regeneration of entire neurons (28). It is described so far as migration of microglia to the site of lesion and an axon outgrowth that is probably cytoskeleton-driven. We asked whether the leech macins can also affect non-leech neuronal cells. Interestingly, both neuromacin and theromacin not only induce nerve repair (i.e. the axonal regrowth in leech) but they also increased the number of viable murine neuroblastoma cells. This observation might extend the role of leech macins in nerve repair as they are able to modulate cytoskeletal functions in leech and enhance the proliferation of neuroblastoma cells. Therefore, the nerve repair process in leech might also include the proliferation of neurons de novo. The mechanism of proliferation induction remains to be determined. The fast uptake and initial inhomogeneous distribution of theromacin might be a hint that endocytosis is involved. As proliferation of neuroblastoma cells was also induced by hydramacin-1, which in contrast does not exert a nerve repair activity in leech, both processes might be induced by separate mechanisms, i.e. due to different structural properties of the three molecules. However, as Hydra also possesses a primitive nervous system (29), an impact of hydramacin-1 on its nerve cells cannot be excluded, in particular as Hydra is well known for its regenerative capacity (30).
With respect to the nerve repair activity in leech, neuromacin appears to be a chimera of theromacin and hydramacin-1. On the one hand, it follows the barnacle model like hydramacin-1, and on the other hand, it possesses a nerve repair capacity like theromacin. Structurally, neuromacin also appears to be a chimera of theromacin and hydramacin-1. Comparing their primary structures, the three molecules share differently conserved amino acid residues. Some of them are only conserved between neuromacin and theromacin, others are only conserved between neuromacin and hydramacin-1. It is tempting to assume that at least with respect to the nerve repair character the six separately conserved amino acid residues alone might be the structural basis of that functional characteristic. By contrast, the induction of the proliferation must originate in a part all three macins share. Again, this may be further corroborated by a rational chimeric protein design as performed with the human β-defensins 2 and 3 (3).
The development of antimicrobial pharmaceuticals based on AMPs represents a promising alternative approach against bacterial infections. However, the rational design of novel antimicrobial agents necessitates a significantly improved understanding of the structure-activity relationships of AMPs (31, 32). Evidently, this remains valid for instrumentalization of the potential of their secondary functions. Obviously, the macin AMPs do not reflect perfect candidates for drug development because in their current form the antimicrobial activities are already diminished by low ionic strength. However, the macins or AMPs in general should not be limited to their antimicrobial activity. Besides their potential as templates for the development of alternative antibiotics, the various “secondary” activities of AMPs might be beneficial for the development of pharmaceuticals suitable in other medical fields. In particular, the nerve repair activity of the leech macins or the proliferation effect exerted by all macins, respectively, work at physiological salt concentrations, suggesting a mechanism different from the antimicrobial activity. Further investigations of the nerve repair activity might eventually lead to findings that support the development of pharmaceuticals effective against neural diseases, e.g. paraplegia, which is of course purely speculative.
The results of the present study clearly expand our knowledge about this interesting family of macin proteins sharing antimicrobial and nerve repair activities in particular. In general, the macins also corroborate once more that AMPs are more than just antimicrobials. The presented data form a valuable basis for a comprehensive structure-activity study to elaborate the molecular mechanisms of killing and regeneration exerted by the macins. It may well be warranted in the future to dissect the antimicrobial and nerve-repairing activity of macins by analyzing artificially designed chimeras of structurally defined regions of the three macins.
Supplementary Material
Acknowledgments
We thank Silvia Voss (Institute for Infection Medicine) and Heidrun Liessegang (Institute of Zoology, Zoophysiology) for excellent technical assistance.
This work was supported by the excellence cluster “Inflammation at Interfaces,” the Ministere de l'Enseignement de la Recherche et des Technologies, and the CNRS.

This article contains supplemental Fig. SF1 and Table 1.
The atomic coordinates and structure factors (code 2ln8) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Hydramacin-1 was generated as described (10).
C.-W. Hung, S. Jung, J. Grötzinger, and A. Tholey, manuscript in preparation.
- AMP
- antimicrobial peptide/protein
- TM
- theromacin
- NM
- neuromacin
- FP
- fusion protein
- TOCSY
- total correlation spectroscopy
- MBC
- minimal bactericidal concentration
- HBD
- human β-defensin.
REFERENCES
- 1. Jenssen H., Hamill P., Hancock R. E. (2006) Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zasloff M. (2002) Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 3. Jung S., Mysliwy J., Spudy B., Lorenzen I., Reiss K., Gelhaus C., Podschun R., Leippe M., Grötzinger J. (2011) Human β-defensin 2 and β-defensin 3 chimeric peptides reveal the structural basis of the pathogen specificity of their parent molecules. Antimicrob. Agents Chemother. 55, 954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwab U., Gilligan P., Jaynes J., Henke D. (1999) In vitro activities of designed antimicrobial peptides against multidrug-resistant cystic fibrosis pathogens. Antimicrob. Agents Chemother. 43, 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Otero-González A. J., Magalhães B. S., Garcia-Villarino M., López-Abarrategui C., Sousa D. A., Dias S. C., Franco O. L. (2010) Antimicrobial peptides from marine invertebrates as a new frontier for microbial infection control. FASEB J. 24, 1320–1334 [DOI] [PubMed] [Google Scholar]
- 6. Neonakis I. K., Spandidos D. A., Petinaki E. (2011) Confronting multidrug-resistant Acinetobacter baumannii: a review. Int. J. Antimicrob. Agents 37, 102–109 [DOI] [PubMed] [Google Scholar]
- 7. Wilmes M., Cammue B. P., Sahl H. G., Thevissen K. (2011) Antibiotic activities of host defense peptides: more to it than lipid bilayer perturbation. Nat. Prod. Rep. 28, 1350–1358 [DOI] [PubMed] [Google Scholar]
- 8. Schikorski D., Cuvillier-Hot V., Leippe M., Boidin-Wichlacz C., Slomianny C., Macagno E., Salzet M., Tasiemski A. (2008) Microbial challenge promotes the regenerative process of the injured central nervous system of the medicinal leech by inducing the synthesis of antimicrobial peptides in neurons and microglia. J. Immunol. 181, 1083–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang G., Li X., Wang Z. (2009) APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 37, D933–D937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jung S., Dingley A. J., Augustin R., Anton-Erxleben F., Stanisak M., Gelhaus C., Gutsmann T., Hammer M. U., Podschun R., Bonvin A. M., Leippe M., Bosch T. C., Grötzinger J. (2009) Hydramacin-1, structure and antibacterial activity of a protein from the basal metazoan Hydra. J. Biol. Chem. 284, 1896–1905 [DOI] [PubMed] [Google Scholar]
- 11. Tasiemski A., Vandenbulcke F., Mitta G., Lemoine J., Lefebvre C., Sautire P. E., Salzet M. (2004) Molecular characterization of two novel antibacterial peptides inducible upon bacterial challenge in an annelid, the leech Theromyzon tessulatum. J. Biol. Chem. 279, 30973–30982 [DOI] [PubMed] [Google Scholar]
- 12. Dingley A. J., Lorenzen I., Grötzinger J. (2008) NMR analysis of viral protein structures. Methods Mol. Biol. 451, 441–462 [DOI] [PubMed] [Google Scholar]
- 13. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 14. Johnson B. A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 15. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 16. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 18. Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8, 52–56, 29 [DOI] [PubMed] [Google Scholar]
- 19. van Gunsteren W. F., Billeter S. R., Eising A. A., Hünenberger P. H., Krüger P., Mark A. E., Scott W. R. P., Tironi I. G. (1996) Biomolecular Simulation: the GROMOS96 Manual and User Guide, vdf Hochschulverlag; AG, Zürich [Google Scholar]
- 20. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 21. Petrey D., Honig B. (2003) GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 374, 492–509 [DOI] [PubMed] [Google Scholar]
- 22. Sahly H., Schubert S., Harder J., Rautenberg P., Ullmann U., Schröder J., Podschun R. (2003) Burkholderia is highly resistant to human β-defensin 3. Antimicrob. Agents Chemother. 47, 1739–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herbst R., Ott C., Jacobs T., Marti T., Marciano-Cabral F., Leippe M. (2002) Pore-forming polypeptides of the pathogenic protozoon Naegleria fowleri. J. Biol. Chem. 277, 22353–22360 [DOI] [PubMed] [Google Scholar]
- 24. Leippe M., Ebel S., Schoenberger O. L., Horstmann R. D., Müller-Eberhard H. J. (1991) Pore-forming peptide of pathogenic Entamoeba histolytica. Proc. Natl. Acad. Sci. U.S.A. 88, 7659–7663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pick U. (1981) Liposomes with a large trapping capacity prepared by freezing and thawing of sonicated phospholipid mixtures. Arch. Biochem. Biophys. 212, 186–194 [DOI] [PubMed] [Google Scholar]
- 26. Tasiemski A., Salzet M. (2010) Leech immunity: from brain to peripheral responses. Adv. Exp. Med. Biol. 708, 80–104 [DOI] [PubMed] [Google Scholar]
- 27. Boidin-Wichlacz C., Vergote D., Slomianny C., Jouy N., Salzet M., Tasiemski A. (2012) Morphological and functional characterization of leech circulating blood cells: role in immunity and neural repair. Cell. Mol. Life Sci., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duan Y., Panoff J., Burrell B. D., Sahley C. L., Muller K. J. (2005) Repair and regeneration of functional synaptic connections: cellular and molecular interactions in the leech. Cell. Mol. Neurobiol. 25, 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schaller H. C., Hermans-Borgmeyer I., Hoffmeister S. A. (1996) Neuronal control of development in Hydra. Int. J. Dev. Biol. 40, 339–344 [PubMed] [Google Scholar]
- 30. Bosch T. C. (2007) Why polyps regenerate and we don't: towards a cellular and molecular framework for Hydra regeneration. Dev. Biol. 303, 421–433 [DOI] [PubMed] [Google Scholar]
- 31. Zucca M., Scutera S., Savoia D. (2011) New antimicrobial frontiers. Mini Rev. Med. Chem. 11, 888–900 [DOI] [PubMed] [Google Scholar]
- 32. Nguyen L. T., Haney E. F., Vogel H. J. (2011) The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 29, 464–472 [DOI] [PubMed] [Google Scholar]
- 33. Yang Y. S., Mitta G., Chavanieu A., Calas B., Sanchez J. F., Roch P., Aumelas A. (2000) Solution structure and activity of the synthetic four-disulfide bond Mediterranean mussel defensin (MGD-1). Biochemistry 39, 14436–14447 [DOI] [PubMed] [Google Scholar]
- 34. Lay F. T., Schirra H. J., Scanlon M. J., Anderson M. A., Craik D. J. (2003) The three-dimensional solution structure of NaD1, a new floral defensin from Nicotiana alata and its application to a homology model of the crop defense protein alfAFP. J. Mol. Biol. 325, 175–188 [DOI] [PubMed] [Google Scholar]
- 35. Blanc E., Romi-Lebrun R., Bornet O., Nakajima T., Darbon H. (1998) Solution structure of two new toxins from the venom of the Chinese scorpion Buthus martensi Karsch blockers of potassium channels. Biochemistry 37, 12412–12418 [DOI] [PubMed] [Google Scholar]
- 36. Lamberty M., Caille A., Landon C., Tassin-Moindrot S., Hetru C., Bulet P., Vovelle F. (2001) Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities. Biochemistry 40, 11995–12003 [DOI] [PubMed] [Google Scholar]
- 37. Cook W. J., Zell A., Watt D. D., Ealick S. E. (2002) Structure of variant 2 scorpion toxin from Centruroides sculpturatus Ewing. Protein Sci. 11, 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.