Background: Proteolysis of KLF4 is involved in estrogen signaling and cell growth.
Results: Accumulation of KLF4 due to estrogen-induced inhibition of VHL facilitates estrogen-mediated mitogenic growth.
Conclusion: Proteolytic regulation of KLF4 abundance by UPS orchestrates estrogen signaling and homeostasis for breast cancer cells.
Significance: Demonstration of KLF4-VHL in facilitating estrogen signaling advances our knowledge of breast tumorigenesis, which provides value for breast cancer therapy.
Keywords: Breast Cancer, Cell Cycle, Kruppel-like Factor (KLF), Protein Degradation, Ubiquitylation
Abstract
Krüppel-like factor 4 (KLF4), a zinc finger-containing transcriptional factor, is a pivotal regulator of cellular fate. KLF4 has attracted considerable attention for its opposing effect in carcinogenesis as tumor suppressor (e.g. colorectal cancer) or oncoprotein (e.g. breast cancer), depending on tissue context, with the underlying mechanism remaining largely unknown. Here we report that KLF4 mediates estrogen signaling in breast cancer formation. Accumulation of KLF4 by inhibiting its turnover triggers estrogen-induced transactivation. We identified Von Hippel-Lindau, pVHL, as the protein that governs KLF4 turnover in breast cancer cells and demonstrated that estrogen-induced down-regulation of pVHL facilitates accumulation of KLF4. We provide mechanistic insights into KLF4 steady-state degradation as well as its elevation in the presence of estrogen and show that elevated levels of pVHL or depletion of KLF4 attenuates the estrogen-induced transactivation and cell growth. Finally, immunohistochemical staining revealed reduced concentration of pVHL and accumulation of KLF4 in breast cancer tissues. We thus propose that suppression of pVHL in response to estrogen signaling results in elevation of KLF4, which mediates estrogen-induced mitogenic effect.
Introduction
Krüppel-like factor 4 (KLF4), a zinc finger-containing transcription factor, regulates diverse cellular processes, including cell cycle progression, apoptosis, and metabolism as well as stem cell renewal (1, 2). Although our current attention has been largely drawn to its pivotal role in switching somatic cell into stem cell in the presence of Oct3/4, Sox2, and c-Myc, recent studies of carcinogenesis have surprisingly revealed its dual faced characteristics in tumor suppression and tumor promotion, depending on the tissue type and physiological context, with the underlying mechanism of its functional switch remaining largely unknown (1). The tumor-suppressive effect of KLF4 has been characterized in many types of cancer, including gastrointestinal cancer (3–8), whereas its oncogenic effect has been reported in breast and squamous cell carcinoma (9–13). Previous genetic and biochemical studies have provided a basic sketch of how KLF4 functions in regulating gene expression and how KLF4 itself is regulated during cell division or in response to extracellular signaling. Principally, the up-regulation of tumor suppressors p21/WAF1 and p27/KIP1 (11, 14, 15) and down-regulation of oncogenes cyclin B and cyclin D1 (16, 17) are thought to mediate its tumor-suppressive function. As to how KLF4 exerts its oncogenic function, it remains largely unknown. It was speculated for a time that the oncogenic function of KLF4 was probably dependent on its ability to inhibit apoptosis through the suppression of p53 and its target gene Bax, a proapoptotic factor, and to promote cell migration and invasion through the up-regulation of Notch signaling (11, 18, 19). Adding to the current understanding, our present study in estrogen receptor signal transduction and breast cancer formation provides a new view into the mystery of how KLF4 acts as a mitogenic factor to enhance cellular growth, leading to possible insight into its functional switch between tumor suppressor and tumor enhancer.
KLF4 is a highly regulated protein that responds to intrinsic and extrinsic signaling. The function of KLF4 is regulated at both transcriptional and post-transcriptional levels. Previous studies revealed that KLF4 is down-regulated by promoter hypermethylation and loss of heterozygosity for many types of cancer (3). KLF4 can be induced by a variety of stimuli, including serum starvation (20, 21), oxidative stress (22), sodium butyrate (23), selenium (24), interferon-γ (IFN-γ) (25), and cAMP (26). In addition, KLF4 expression can be elevated or repressed, depending on the extent of DNA damage (27). Stimuli-elicited KLF4 regulation is governed by multiple mechanisms, including increased transcription, decreased mRNA stability, and increased protein stability. Recent proteomic and biochemical studies have revealed that KLF4 is subjected to multiple post-translational modifications, such as acetylation (28) and SUMOylation (29); both modifications are thought to be involved in regulating the KLF4-mediated transactivation (29). A recent study has implicated the importance of the ubiquitin-proteasome system in KLF4 regulation in response to serum stimulation and during cell cycle progression (20). However, how KLF4 is regulated by the ubiquitin-proteasome system and which E3 ligase is involved in the serum-responsive KLF4 ubiquitylation remain unknown. Our present work has explored whether von Hippel-Lindau (VHL)2 could be the ubiquitin-protein ligase that governs KLF4 protein stability, where alteration of KLF4 function by mitogenic signaling, such as estrogen via regulating the VHL-mediated ubiquitin-proteasome pathway, could be a critical mechanism that orchestrates breast cell growth and transformation.
The results from the recent studies using an animal model and histological analyses have left us perplexed in regard to the dual roles of KLF4 as a tumor suppressor in gastrointestinal cancer but also as a tumor enhancer in breast cancer (1). Studies in breast cancer have uncovered that more than 70% of human breast cancer has shown increased KLF4 protein expression, which occurs as early as the stage of ductal carcinoma in situ (9). Furthermore, the nuclear localization of KLF4 is associated with a more aggressive phenotype (10). In addition, the association of elevated KLF4 with progression of breast cancer was further validated in canine mammary carcinomas, a natural animal model for the comparative study with human breast cancer (30). The aforementioned pathological observation in breast cancer studies led us to ask several important questions: 1) whether disruption of proteolytic regulation of KLF4 correlates with accumulated KLF4 protein levels in breast cancer cells; 2) whether stabilization of KLF4 is a causative element for the genesis and/or progression of breast cancer; and 3) whether any of three major signaling pathways via estrogen receptor, progesterone receptor, and epidermal growth factor receptor signaling govern the homeostasis of normal breast cells as well as breast cancer cells and if modulating the above signaling pathways would affect KLF4 turnover and in turn impede cell growth that contributes to breast carcinogenesis (31). To answer these questions, we have dissected the proteolytic regulation of KLF4 in controlling the growth of breast cancer cells using a combinatorial approach, including genetics, biochemistry, and cell biology as well as pathology. Our results suggest that KLF4 is a fast turnover protein and that its steady state level is partly governed by degradation as mediated by VHL E3 ligase. KLF4 protein stability is highly regulated in response to estrogen receptor signaling. Down-regulation of VHL E3 ligase by elevated estrogen-initiated signal results in a profound accumulation of KLF4 that in turn facilitates the estrogen-governed mitogenic growth and could be a critical mechanism for malignant growth of breast cancer cells. Information from our present work could fill the knowledge gap regarding how KLF4 proteolytic regulation is involved in breast carcinogenesis, which provides further insight for possible targets for new therapeutic treatment.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
HEK293T, MCF-7, T-47D, and MDA-MB-231 were obtained from the American Type Culture Collection (Manassas, VA). The viral packaging line Phoenix-A cells were a gift from Edward V. Prochownik (University of Pittsburgh). All cells were maintained in DMEM supplemented with 5 or 10% FBS, 1× antibiotic/antimycotic solution (100 units/ml streptomycin and 100 units/ml penicillin) (all from Invitrogen). All cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. For estrogen stimulation, MCF-7 or T-47D cells were cultured in phenol red-free DMEM supplemented with 5% charcoal-stripped FBS for 48 h. Then 10 nm 17β-estradiol (Sigma) was added to treat the cells.
Plasmids and Transfection
ERE-Luc reporter plasmid was a gift from Dr. Pamela A. Hershberger (University of Pittsburgh). pRetroSuper-KLF4 shRNA was a gift from Dr. Daniel S. Peeper, Netherlands Cancer Institute) (11). VHL and KLF4 constructs were generated by PCR amplification of the full-length or partial coding sequence of human KLF4 and VHL and subsequent subcloning into mammalian expression vectors with FLAG-HA or HA tag. KLF4 construct with a Lys-43 point mutation was engineered using the site-directed mutagenesis kit (Stratagene). The primer sequences were as follows: 5′-GGAGCTCTCCCACATGAGGCGACTTCCCC-3′ (forward primer) and 5′-GGGGAAGTCGCCTCATGTGGGAGAGCTCC-3′ (reverse primer). For transfection, cells were plated to form a 50–70% confluent culture. The HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen).
To construct pLenti6-V5-KLF4 and pLenti-V5-VHL, the coding sequence of KLF4 or VHL was amplified and cloned into pENTR/D-TOPO, an entry vector for the Gateway system. The resulting plasmids, pENTR-KLF4, pENTR-KLF4-K43R, and pENTR-VHL, were used for LR recombination reaction (recombination between attL and attR sites) together with the destination vector pLenti6/V5-Dest, leading to the generation of pLenti6-V5-KLF4, pLenti6-V5-KLF4-K43R, and pLenti6-V5-VHL.
Lentiviral and Retroviral Infection
pLenti6/V5-KLF4, pLenti6/V5-VHL, pRetroSuper-KLF4-shRNA, or pLKO.1-VHL-shRNA (1, RHS3979-9607015; 2, RHS3979-9607016; Open Biosystems) was co-transfected with pVSV-G, pRRE, and pRSV-REV into HEK293T or Phoenix-A cells. Lipofectamine 2000 was used. The packaged lentiviral or retroviral particles were collected, mixed with Polyprene, and added into MCF-7 target cells. The stable cell lines were established by culturing the cells in the medium containing antibiotic blasticidin (20 μg/ml) or puromycin (2 μg/ml).
RNA Inference
siRNAs (Sigma) specifically targeted to different E3 ubiquitin ligases were synthesized and transfected into MCF-7 cells using Lipofectamine 2000. Cells were collected at 48 h post-transfection for the immunoblotting assay. The synthesized siRNA sequences are as follows: BRCA1, 5′-GGAACCTGTCTCCACAAAG-3′; β-TrCP1/2, 5′-GTGGAATTTGTGGAACATC-3′; Skp2, 5′-ATTCAGCTGGGTGATGGTCTC-3′; VHL, 5′-CCATCTCTCAATGTTGACGGA-3′; and Cdh1 (32).
RNA Extraction, cDNA Synthesis, and Real-time PCR
Total RNA was isolated from various samples using TRIzol reagent (Invitrogen). 2 μg of RNA was primed by oligo(dT) (Promega) and reverse transcribed into cDNA with Moloney murine leukemia virus reverse transcriptase (Promega). Real-time PCR was carried out on a StepOne® Plus Real-time PCR system (Applied Biosystems) using Fast SYBR® Green master mix (Applied Biosystems). The primers for KLF4 and β-actin are as follows: KLF4, 5′-ACCTACACAAAGAGTTCCCATC-3′ (forward primer); 5′-TGTGTTTACGGTAGTGCCTG-3′ (reverse primer); β-actin, 5′-GGCGGCACCACCATGTACCCT-3′ (forward primer); 5′-AGGGGCCGGACTCGTCATACT-3′ (reverse primer).
Western Blotting and Immunoprecipitation Assay
Cells were harvested and lysed in radioimmune precipitation assay lysis buffer (Upstate Biotechnology) containing protease inhibitor mixture (Sigma). The protein concentration was determined using Bio-Rad protein assay reagent. Western blotting was performed using anti-KLF4 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), Cdh1 (Calbiochem), β-TrCP (Cell Signaling), VHL (Cell Signaling), BRCA1 (Santa Cruz Biotechnology), Skp2 (Santa Cruz), V5 (Invitrogen), FLAG (Sigma), β-actin (Sigma), and HRP-conjugated goat anti-mouse or anti-rabbit secondary antibody (Promega). Signals were detected with ECL reagents (Amersham Biosciences). Semiquantification of data was performed using NIH Image. For immunoprecipitation assay, cell lysate was incubated with anti-FLAG M2 gel (Sigma) or anti-KLF4 (Santa Cruz Biotechnology) antibody overnight at 4 °C on a rotator, followed by the addition of protein A/G plus agarose (Pierce) to the reaction containing anti-KLF4 antibody for 2 h at 4 °C. After five washes with radioimmune precipitation assay lysis buffer supplemented with protease inhibitor mixture, complexes were released from the anti-FLAG M2 gel and protein A/G plus agarose by boiling for 5 min in 2× SDS-PAGE loading buffer. Western blotting was used to detect ubiquitin conjugates with anti-Myc (Santa Cruz Biotechnology) and anti-ubiquitin antibody (BD Biosciences), respectively.
ERE-Luc Reporter Assay
Cells were plated in 24-well plates. After 24 h, cells were co-transfected with ERE-Luc reporter plasmid and Renilla luciferase control vector using Lipofectamine 2000 according to the manufacturer's instruction. Firefly and Renilla luciferase activities were measured using a dual luciferase kit (Promega). The firefly luciferase data for each sample was normalized based on transfection efficiency as determined by Renilla luciferase activity. Each experiment was performed in triplicate and repeated at least three times.
Cell Viability Assay
2 × 103 MCF-7 cells were seeded into a 96-well plate and cultured overnight. Then the cells were treated with ICI 182780 for 96 h. Cell viability was determined using the CellTiter 96 AQueous One Solution (Promega) according to the manufacturer's protocol.
Immunohistochemical Staining
Tissue sections of human breast cancer and adjacent tissues were purchased from Pantomics Inc. Tissue sections were treated as described previously (33). The sections were then incubated with primary antibody against KLF4 (1:100) and VHL (1:100) at 4 °C overnight. After PBS washes, sections were incubated with biotinylated secondary antibody at 1:200 for 30 min. After incubating with Vectastain ABC reagent (Vector Laboratories, Inc., Burlingame, CA) for 30 min, the sections were developed with DAB (3,3-Diaminobenzidine) (Sigma-Aldrich). Sections were counterstained with hematoxylin, followed by coverslip mounting. Negative controls were obtained by omitting the primary antibody.
To evaluate the expression of KLF4 and VHL, the percentage of positive tumor cells was determined semiquantitatively by assessing the entire tumor section. Each sample was assigned to one of the following categories: 0 (0–4%), 1 (5–24%), 2 (25–49%), 3 (50–74%), or 4 (75–100%). The intensity of immunostaining was determined as 0 (negative), 1+ (weak), 2+ (moderate), or 3+ (strong). A final immunoreactive score between 0 and 12 was calculated by multiplying the percentage of positive cells with the staining intensity score. All slides were blind evaluated for immunostaining without any knowledge of the clinical outcome or other clinical or pathological data. Statistical analysis was performed using SPSS statistical software (SPSS Inc., Chicago, IL). The results were presented as means ± S.D. p < 0.05 was considered statistically significant.
RESULTS
KLF4 Protein Levels Are Profoundly Up-regulated in Response to Estrogen Signaling, Which Is Mediated through Ubiquitin-Proteasome Pathway
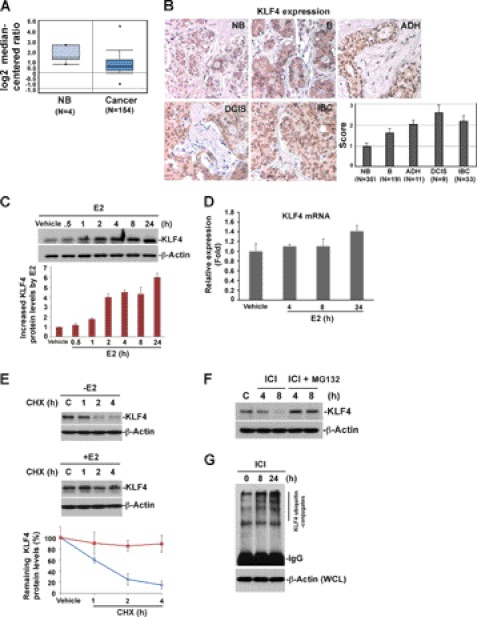
Recent studies have implicated KLF4 as a tumor enhancer in breast cancer (1, 11). Also, an increase in KLF4 protein level has been observed in both human and canine breast cancer (10, 30, 34), but the underlying mechanisms remain unknown. Unexpectedly, recent microarray studies from Oncomine demonstrated a lower expression level of KLF4 mRNA in breast cancer as compared with normal tissue (Fig. 1A) (35). Interestingly, KLF4 has been recently defined as a fast turnover protein (20), further implicating that deregulated KLF4 degradation could be a possible mechanism contributing to its accumulation in breast cancer. To confirm the hypothesis that KLF4 proteolytic regulation plays a role in breast tumorigenesis, we have initially measured KLF4 protein expression levels at various stages during the development of breast cancer by immunohistochemistry. As shown in Fig. 1B, while KLF4 protein was maintained at moderate levels in normal breast tissue, its expression was observed to be significantly higher in cancer status, including benign, atypical ductal hyperplasia (ADH), ductal carcinoma in situ (DCIS), and invasive breast cancer (IBC). Previous studies have suggested that KLF4 is a tightly regulated protein in response to a variety of environmental signals and stresses (1, 36, 37). However, to date, whether and how KLF4 protein is regulated by estrogen signaling remains unknown. We thus examined the impact of estrogen on KLF4 function, which could potentially provide insight into the observed deregulation of KLF4 in breast cancer. To our surprise, KLF4 protein levels in MCF-7 cells dramatically increased in response to 17β-estradiol (E2) (Fig. 1C), whereas alteration of its mRNA level remained insignificant (Fig. 1D). This result suggests that estrogen is a critical player governing KLF4 function and that the E2-induced KLF4 up-regulation is through post-transcriptional regulation. Further, KLF4 protein levels remained unchanged in response to E2 treatment in ER-negative cell MDA-MB-231 (supplemental Fig. 1), indicating that the regulation of KLF4 by E2 is specific to ER-positive cells.
FIGURE 1.
Proteolytic regulation of KLF4 by estrogen signaling contributes to its overexpression in breast cancer. A, KLF4 mRNA level is down-regulated in breast cancer. Oncomine was used to analyze the previously published microarray data. B, KLF4 protein level is consistently increased at different stages of breast cancer progression. The level of KLF4 protein was analyzed and scored by an immunohistochemistry assay of the tissue array. NB, normal breast tissue; B, benign; ADH, atypical ductal hyperplasia; DCIS, ductal carcinoma in situ; IBC, invasive breast cancer. C, kinetics of KLF4 protein accumulation in response to 17β-estradiol (E2). MCF-7 cells were hormone-stripped for 48 h and then treated with E2 at 10 nm for the indicated time. KLF4 protein levels were analyzed by immunoblotting. β-Actin was used as a loading control. D, real-time PCR analysis of KLF4 mRNA level in response to E2 stimulation. Total RNA was extracted from cells treated with E2 for the indicated time. A real-time PCR assay was performed using reverse transcribed cDNA. β-Actin served as internal control. E, E2 extends the half-life of KLF4 protein. Vehicle or E2-pretreated cells were treated with 20 μm protein synthesis inhibitor cycloheximide (CHX) for the indicated time. The KLF4 protein turnover rate was measured by immunoblotting. F, proteasome inhibitor MG132 attenuates the ICI (estrogen receptor antagonist)-induced drop of KLF4 protein levels. Cells were treated with ICI 182780 (1 μm) alone or together with MG132 (20 μm) for the indicated time and harvested for immunoblotting analysis of KLF4 protein level. G, KLF4 ubiquitylation is enhanced when the E2 effect is blocked by ICI. Cells were treated with ICI for the indicate time in the presence of MG132. Cells were lysed for immunoprecipitation using anti-KLF4 antibody. The immunocomplex was further probed for the level of ubiquitylated KLF4 by immunoblotting using anti-ubiquitin antibody. WCL, whole cell lysates. Error bars, S.D.
To ask whether E2-induced KLF4 protein accumulation is due to increased protein stability, we measured the rate of KLF4 protein turnover. As shown in Fig. 1E, in the absence of estrogen signaling, KLF4 rapidly degraded with a half-life of less than 1 h, whereas KLF4 degradation decreased in the presence of E2, suggesting that estrogen could stabilize KLF4 and that estrogen-induced KLF4 accumulation could be crucial for the mitogenic effect in breast cancer development. This notion is further confirmed by the observation that blockade of estrogen signaling by ICI 182780 (antagonist of estrogen receptor) resulted in a fall of KLF4 levels, whereas the ICI-induced drop of KLF4 was attenuated by the presence of MG132, a proteasomal inhibitor (Fig. 1F). In addition, ICI-caused KLF4 degradation was accompanied by increased KLF4 ubiquitylation (Fig. 1G). Taken together, the above results suggest that KLF4 is profoundly regulated in response to estrogen signaling. The estrogen-induced accumulation of KLF4 is mediated by the ubiquitin-proteasomal pathway. The loss of KLF4 control due to deregulated estrogen signaling may be linked to breast cancer formation.
KLF4 Levels Elevated by Estrogen Mediate Estrogen-induced Transactivation, Which in Turn Facilitates Estrogen-induced Mitogenic Effect
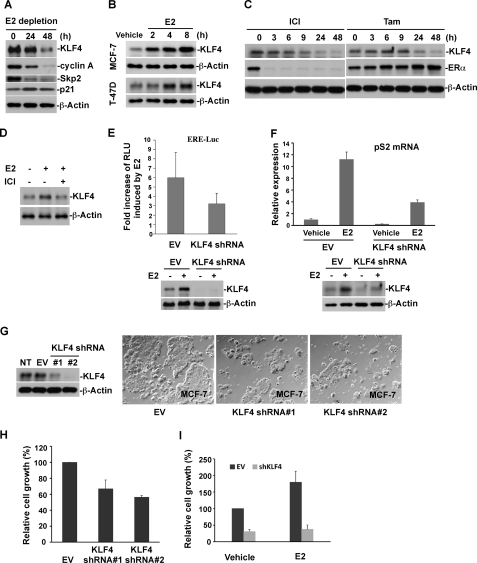
Our current observations suggest the KLF4 is tightly regulated in response to estrogen signaling with disrupted regulation of KLF4 in breast tumorigenesis (Fig. 1, A–G). The next questions include the role of KLF4 in estrogen signal transduction and the impact of estrogen-induced KLF4 accumulation on the estrogen-mediated mitogenic effect. We initially detected the effect of E2 depletion in the culture medium on KLF4 and observed that deprivation of E2 in the culture medium led to significant reduction of cellular KLF4 (Fig. 2A), which correlated with decreased cellular growth (data not shown). Similar to KLF4, the abundance of cyclin A and Skp2 significantly decreased in parallel with an increase in p21 abundance. Adding back E2 to a physiological level (10 nm) in E2-depleted culture medium significantly drove KLF4 accumulation in both MCF-7 and T-47D cells (Fig. 2B). Blockade of estrogen signaling in regular culture medium by either ICI or tamoxifen (antagonist of estrogen receptor) caused a drastic drop in KLF4 levels (Fig. 2C). Moreover, E2-induced KLF4 accumulation was muted by the addition of ICI (Fig. 2D).
FIGURE 2.
Elevation of KLF4 protein levels by E2 is required to facilitate estrogen signaling and its mitogenic effect. A, depletion of E2 in the culture medium leads to a drop of KLF4 protein levels. MCF-7 cells were hormone-stripped for the indicated time and probed for the expression of KLF4 and several cell cycle-related proteins, as indicated by immunoblotting. B, E2 promotes KLF4 protein accumulation in both MCF-7 and T-47D cells. Hormone-stripped cells were treated with E2 for the indicated time and probed for KLF4 protein expression by immunoblotting. C, the treatment of ER antagonists leads to down-regulation of KLF4 protein levels. MCF-7 cells were treated with either ICI or 4-hydroxytamoxifen (TAM) (1 μm) for the indicated time and collected for immunoblotting using various antibodies as indicated. D, E2-induced accumulation of KLF4 protein could be abolished by antagonist of estrogen receptor. Hormone-stripped cells were treated with E2 alone or E2 together with ICI. Equal amount of cell lysate was probed for KLF4 protein expression by immunoblotting. E, KLF4 knockdown (KD) results in the attenuation of E2-induced ERE-Luc activity. ERE-Luc and internal control Renilla luciferase vector (pRL-TK) were co-transfected into KLF4-KD or control cells. Cells were then cultured in hormone-stripped medium overnight followed by 24 h of E2 stimulation. Cells were harvested for the dual luciferase assay, and results are expressed as firefly luciferase activity normalized to Renilla luciferase activity (relative light units; RLU). KLF4 protein levels in KLF4-KD cells in the absence and presence of E2 signaling were analyzed by immunoblotting. F, KLF4 knockdown attenuates the E2-induced pS2 mRNA levels. KLF4-KD or control cells were hormone-stripped for 48 h and then treated with E2 for 24 h. Total RNA was isolated from treated cells followed by real-time PCR analysis of pS2 mRNA level. G and H, depletion of KLF4 by shRNA significantly suppresses cellular growth. MCF-7 cells with stable expression of two different KLF4 shRNAs were used for immunoblotting and the cell proliferation assay. I, KLF4 knockdown significantly abolishes E2-induced cell growth. KLF4-KD or control cells were cultured in hormone-stripped medium supplemented with E2 or vehicle for 5 days, and cell growth was assayed by MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium). Error bars, S.D.
In the current paradigm of estrogen action, binding of E2 to the estrogen receptor (ERα) causes homodimerization of ERα. The ligand-bound ERα in turn translocates to bind to cognate DNA sequence, estrogen response elements (EREs), and cooperates with recruited co-regulators to activate target gene transcription (38). Given that KLF4 is a transcriptional factor, a possible theory is that elevation of KLF4 by estrogen is necessary for E2-activated transcription. To test this hypothesis, the alteration of KLF4 by RNA interference on estrogen-mediated transactivation was assessed by a dual luciferase assay. As shown in Fig. 2E, while a 6-fold increase of ERE-Luc activity by E2 stimulation is observed, depletion of KLF4 significantly attenuates E2-induced transcriptional activation. This observation is further supported by the effect of altering KLF4 on pS2 mRNA (a well known E2-inducible gene) with E2-induced increase of pS2 largely absent in KLF4 knockdown cells (Fig. 2F). To further examine the impact of KLF4 regulation by estrogen signaling, we estimated the effect of alteration of KLF4 by RNA interference on estrogen-mediated mitogenic growth by an MTS assay. As shown in Fig. 2, G–I, depletion of KLF4 significantly suppresses E2-induced cell proliferation. The present results suggest that up-regulation of KLF4 by estrogen is necessary for estrogen-induced transactivation and mitogenic effect. Suppression of estrogen-mediated mitogenic growth by KLF4 depletion further points to the tumor-enhancing role for KLF4 in breast cancer cell proliferation.
pVHL Acts as Ubiquitin-Protein Ligase Governing KLF4 Steady-state Turnover in Breast Cancer Cells
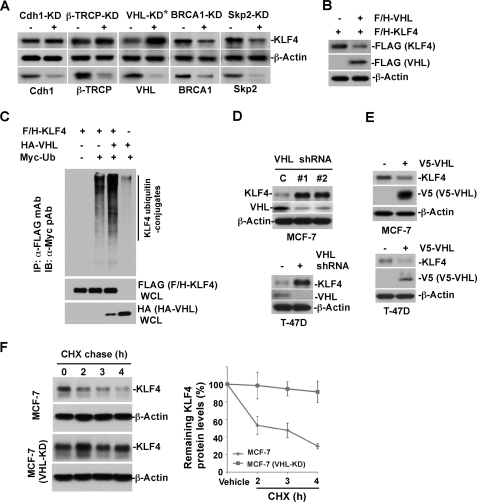
Our present results revealed that KLF4 abundance is tightly orchestrated by a ubiquitin-proteasomal pathway and further implicated that down-regulation of the protein degradation machinery by estrogen signaling could be the mechanism contributing to the estrogen-induced KLF4 accumulation. To identify a potential E3 ubiquitin-protein ligase that governs KLF4 degradation in order to maintain homeostasis in breast cancer cells, we examined a cluster of possible E3 ligases that have been previously connected to cell cycle control or signal transduction in breast cancer cells, including Cdh1 (33), β-TrCP (39), VHL (40), BRCA1, and Skp2 (41). The role for each candidate E3 ligase in KLF4 turnover was evaluated by utilizing knockdown-based transfection of duplex siRNA in MCF-7 cells. To our surprise, although no obvious KLF4 alteration was detected in response to silencing of Cdh1, β-TrCP, BRCA1, and Skp2, we observed an acute increase of KLF4 protein abundance when VHL was depleted by RNA interference in breast cancer cells (Fig. 3A). We further noticed that elevated VHL expression resulted in significant down-regulation of KLF4 levels in cultured HEK293T cells (Fig. 3B). Moreover, KLF4 ubiquitylation was largely enhanced by elevation of VHL expression in vivo (Fig. 3C). To confirm the observed connection between KLF4 turnover and VHL, we conducted similar experiments in both MCF-7 and T-47D cells. As shown in Fig. 3, D and E, depletion of VHL led to significant accumulation of KLF4, whereas overexpression of VHL correlated with down-regulation of KLF4 levels. We also observed the KLF4 protein half-life is significantly extended in VHL knockdown MCF-7 cells (Fig. 3F). In addition, the endogenous KLF4 ubiquitylation in MCF-7 cells was elevated by increased expression of VHL (Fig. 3G). All together, our results suggest that VHL is an ubiquitin-protein ligase that governs KLF4 turnover in breast cancer cells.
FIGURE 3.
VHL E3 ligase plays critical role governing KLF4 steady-state turnover in breast cancer cells. A, evaluation of candidate E3 ubiquitin ligases involving in KLF4 protein turnover. MCF-7 cells were transfected with a series of siRNAs against various candidate E3 ubiquitin ligases as indicated. An equal amount of cell lysate was probed for the expression of various proteins, as indicated by immunoblotting. B, KLF4 is down-regulated by elevated expression of VHL. FLAG/HA-tagged KLF4 was co-transfected with VHL construct or empty vector into HEK293T cells. Transfected cells were probed for the expression of the indicated proteins by immunoblotting. C, VHL catalyzes KLF4 ubiquitylation. HEK293T cells were transfected with the indicated plasmids and subjected to immunoprecipitation by anti-FLAG M2 beads. The immunocomplex was probed for the KLF4 ubiquitin conjugates by immunoblotting using anti-Myc antibody. D, VHL knockdown leads to KLF4 accumulation. Both MCF-7 and T-47D cells were infected by VHL-shRNAs as described under “Experimental Procedures” and probed for KLF4 and VHL expression by immunoblotting. E, elevated VHL down-regulates KLF4 expression. Both MCF-7 and T-47D cells were infected with V5-tagged VHL and probed for KLF4 and VHL expression by immunoblotting. F, VHL knockdown extends the half-life of KLF4 protein. VHL-KD or control cells were treated with CHX for the indicated time and probed for KLF4 expression by immunoblotting. CHX, cycloheximide; Error bars, S.D.
Down-regulation of pVHL Results in Accumulation of KLF4 Protein Levels Ensuring Estrogen-induced Transactivation and Mitogenic Effect
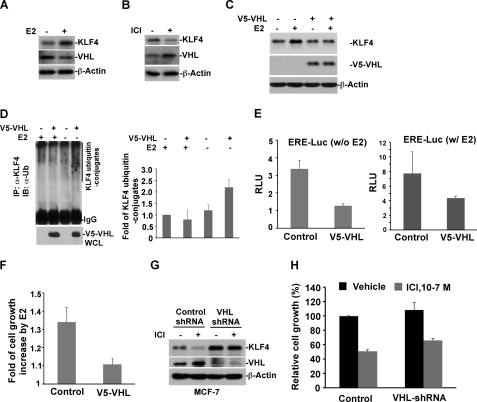
The above results have sketched a possible role for VHL in regulating KLF4 steady-state degradation, which also supports the hypothesis that suppression of VHL function by estrogen could be the mechanism leading to KLF4 accumulation. To test this hypothesis, we have examined the alteration of VHL in the presence and absence of estrogen signaling and have further evaluated the impact of VHL in estrogen-mediated transactivation and cellular proliferation. Interestingly, we observed that VHL protein levels were down-regulated in response to estrogen signaling (Fig. 4A). We further observed that blockade of estrogen signaling by the addition of antagonist of estrogen receptor led to elevated VHL and decreased KLF4 levels (Fig. 4B). Overexpression of VHL abolished the estrogen-induced KLF4 accumulation (Fig. 4C). Moreover, stimulation of cells with estrogen muted the VHL-catalyzed formation of KLF4-ubiquitin conjugates (Fig. 4D). To further corroborate the regulatory function of VHL on KLF4 stability in the context of estrogen signaling, we have assessed the impact of VHL alteration on estrogen-mediated transactivation and mitogenic proliferation. As shown in Fig. 4E, whereas stimulation of cells with E2 led to ∼6-fold increase of ERE-Luc activity, overexpression of VHL caused a drop of ERE-Luc activity. Furthermore, elevated VHL significantly abolished the estrogen-induced mitogenic growth (Fig. 4F). Moreover, the ICI-induced up-regulation of VHL and down-regulation of KLF4 was eliminated by depletion of VHL (Fig. 4G). In addition, ICI-inhibited MCF-7 cell growth was attenuated in VHL knockdown cells (Fig. 4H). Taken together, the above findings suggest that VHL is an E3 ligase that governs KLF4 turnover in MCF-7 cells. Down-regulation of KLF4 steady-state turnover rate by suppressing VHL could be a mechanism that ensures estrogen-induced KLF4 accumulation.
FIGURE 4.
The proteolytic regulation of KLF4 by VHL is involved in estrogen signaling and its mitogenic effect. A, stimulation with estrogen results in down-regulation of VHL, which in turn leads to accumulated KLF4. Hormone-stripped MCF-7 cells were treated with E2 or vehicle. Whole cell lysates were prepared, and immunoblots were probed with the indicated antibodies. B, blockade of estrogen receptor signaling by ICI leads to elevated VHL, which in turn abolishes KLF4 protein degradation. C, elevated expression of VHL attenuates E2-induced KLF4 accumulation. V5-VHL-transfected or control cells were hormone-stripped, followed by E2 treatment. Whole cell lysates were prepared for immunoblotting. D, estrogen abolishes the VHL-catalyzed formation of KLF4 ubiquitin conjugates. E, elevated VHL suppresses ERE-Luc activity. ERE-Luc and internal control Renilla luciferase vector were co-transfected into V5-VHL or control cells. After 24 h of hormone stripping, cells were treated with E2 or vehicle and harvested for the dual luciferase assay. F, elevated VHL attenuates E2-induced cell growth. V5-VHL or control cells were cultured in hormone-stripped medium supplemented with E2 or vehicle for 4 days. Cell viability was measured by MTS. G, VHL knockdown abolishes ICI-induced KLF4 degradation. VHL-shRNA or control cells were treated with ICI and probed for the expression of VHL and KLF4 by immunoblotting. H, VHL knockdown attenuates ER antagonist-induced cell growth inhibition. VHL-shRNA or control cells were treated with ICI for 4 days. Cell growth was analyzed by an MTS assay. IP, immunoprecipitation. Error bars, S.D.
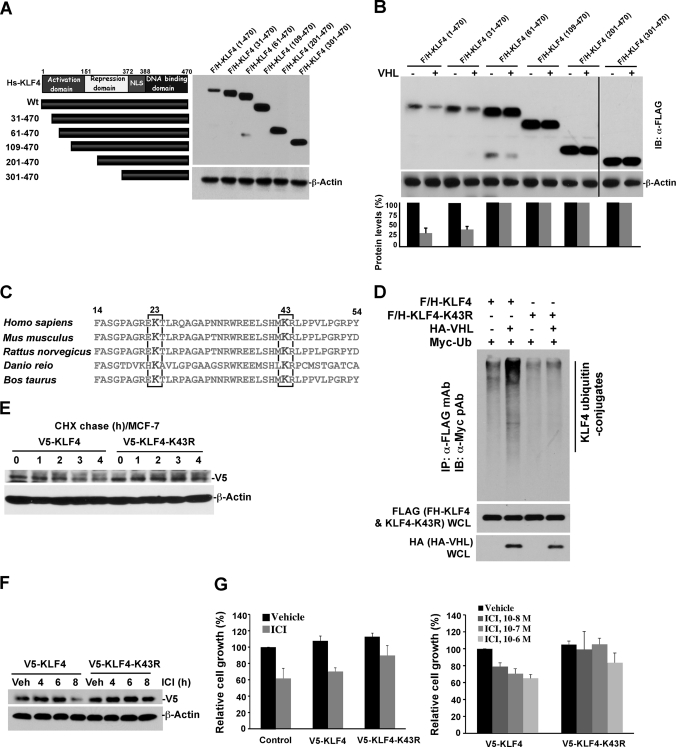
Mechanistic Role of Lysine in KLF4 Amino Terminus in Regulation by pVHL
Our demonstration of VHL as an E3 ligase that regulates KLF4 turnover in breast cancer cell led us to the mechanism by which KLF4 is catalyzed by VHL for ubiquitylation and then degradation. KLF4 is a member of the zinc finger transcription factor family, which bears activation and repression domains at the amino terminus and nuclear localization sequence as well as a DNA binding domain at the carboxyl terminus (Fig. 5A) (36, 42). To identify the molecular region of KLF4 responsible for its ubiquitylation by VHL, we generated a series of FLAG/HA-tagged deletion mutants (Fig. 5A). All of the deletion mutants retain nuclear localization sequence and DNA binding domain to avoid any potential change of subcellular localization and DNA binding properties. Deletion mutants were co-transfected with VHL into HEK293T cells. Degradation of wild-type KLF4 or various KLF4 mutants by VHL was evaluated by immunoblotting. As shown in Fig. 5B, the 60 residues at the amino terminus play a pivotal role in mediating KLF4 degradation by VHL. To further identify detailed motifs on the amino-terminal 60 residues that facilitate the KLF4 ubiquitylation by VHL, we have searched all possible information on the 60 residues and found two conserved lysine residues at positions 23 and 43 (Fig. 5C). Deletion of the first 31 residues at the amino terminus did not affect KLF4 protein stability, suggesting that lysine 23 is not the critical residue involved in KLF4 ubiquitylation and degradation (Fig. 5B). To test this hypothesis, we engineered a lysine 43 mutant KLF4 by replacing lysine with arginine (K43R) (Fig. 5C). To examine the impact of lysine 43 on KLF4 on facilitating KLF4 ubiquitylation by VHL, K43R mutant and VHL were co-transfected into HEK293 cells. As shown in Fig. 5D, whereas the wild-type KLF4 is significantly ubiquitylated by the co-transfected VHL, mutation of lysine 43 on KLF4 significantly attenuates KLF4 ubiquitylation, suggesting that lysine 43 is the critical molecular element mediating the ubiquitylation of KLF4 by VHL. To further confirm the role of lysine 43 in the proteolytic regulation of KLF4, we examined the turnover rate of wild type and mutant KLF4. MCF-7 cells were infected with lentivirus expressing V5-tagged KLF4 wild type or K43R mutant. Cycloheximide was used to block the protein synthesis, and the protein turnover rate was measured by immunoblotting using anti-V5 antibody. As shown in Fig. 5E, the half-life of wild type KLF4 was about 3 h, whereas the half-life for K43R mutant was greatly extended, further verifying the critical role of lysine 43 on KLF4 stability. As demonstrated in Fig. 4, G and H, KLF4 protein stability is affected by ICI, an antagonist of estrogen receptor. Thus, it is anticipated that, like VHL knockdown, K43R mutation could abolish ICI-induced KLF4 degradation. As expected, at 8 h of ICI treatment, only a minor amount of V5-tagged KLF4 remained. By contrast, no effect by ICI was observed for K43R mutant (Fig. 5F). In addition, an MTS assay was performed to evaluate the effect of KLF4 stabilization on the cell growth inhibition by ICI. MCF-7 cells stably expressing V5-tagged wild type or mutant KLF4 were treated with 100 nm ICI for 96 h, with cell viability examined by MTS reagent. As shown in Fig. 5G, expression of K43R mutant attenuated ICI-mediated growth inhibition (Fig. 5G, left). All together, the present results suggest that lysine 43 on KLF4 is critical to facilitate KLF4 ubiquitylation by VHL for degradation. Stabilization of KLF4 significantly mimics the estrogen-induced mitogenic effect and recapitulates the cellular growth that is suppressed by blockade of estrogen signaling by ICI. This result further indicates that molecular management of KLF4 protein stability is a central point that orchestrates the effects of estrogen signal, which has translational value for endocrine therapy in breast cancer treatment (43).
FIGURE 5.
Mapping the critical regions on KLF4 that mediate its degradation by VHL. A, generation of a series of KLF4 deletion mutants. B, evaluation of protein stability for a set of designed KLF4 mutants. KLF4 wild-type or deletion mutant was co-transfected with the VHL construct into HEK293T cells. Whole cell lysates were prepared, and immunoblots were probed with anti-FLAG for ectopic KLF4 protein. Although the loss of the amino-terminal 30 residues has no effect on KLF4 degradation, deletion of the amino-terminal 60 residues results in stabilization of KLF4, suggesting that the region from residue 30 to 60 is crucial to facilitate KLF4 degradation. C, identification of two conserved lysine residues on the amino terminus of KLF4 through an informatics search that are highlighted in red. D, KLF4-K43R is resistant to ubiquitylation catalyzed by VHL. KLF4 wild-type or KLF4-K43R was co-transfected with HA-tagged VHL and Myc-tagged ubiquitin constructs into HEK293T cells. Whole cell lysates were prepared for immunoprecipitation by anti-FLAG M2-agarose. The immunocomplex was probed for ubiquitin conjugates by immunoblotting using anti-Myc. E, KLF4-K43R has a lower turnover rate as compared with its wild-type counterpart. MCF-7 cells were infected with pLenti6-V5-KLF4 or V5-KLF4-K43R and treated with CHX. At the indicated time points, cells were harvested for an immunoblotting assay of ectopic KLF4 expression using anti-V5. F, KLF4-K43R is resistant to the antagonist of estrogen receptor-induced degradation. MCF-7 cells, infected with pLenti6-V5-KLF4 or pLenti6-V5-KLF4-K43R, were treated with ICI for the indicated time. Whole cell lysates were prepared, and immunoblots were probed by anti-V5 for the expression of ectopic KLF4. G, KLF4-K43R attenuates the antagonist of estrogen receptor-induced cell growth inhibition. MCF-7 cells stably expressing V5-KLF4 or V5-KLF4-K43R were treated with the indicated dose of ICI for 4 days. Cell viability was measured by MTS. Error bars, S.D.
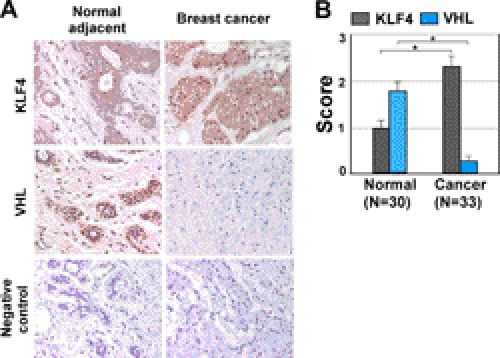
Increased KLF4 in Breast Cancer Correlates with Reduced pVHL Levels
Previous studies have demonstrated the tumor suppressor function for VHL in various types of cancer (44). Our result based on a cultured cell model suggests that, in breast cancer cells, VHL could antagonize the estrogen-induced mitogenic effect, suggesting the mechanism of its growth suppression in breast carcinogenesis. To validate this conclusion obtained by molecular and biochemical analyses, we have performed immunohistochemistry to examine the expression of both VHL and KLF4 in breast cancer and normal adjacent tissue samples. The immunohistochemistry was done on sections from paraffin-embedded specimens with a representative stain shown in Fig. 6A. In agreement with previous studies (40), VHL protein level decreased in breast cancer tissue in comparison with adjacent normal tissue (Fig. 6, A and B). In the same cohort, KLF4 protein level was significantly up-regulated in comparison with adjacent normal tissue, which shows a reversible pattern compared with VHL expression. The complementary staining pattern further corroborates the notion that KLF4 protein is accumulated via VHL down-regulation to achieve its oncogenic function during breast carcinogenesis. Our measurement of KLF4 expression at different stages of breast cancer formation showed that elevated KLF4 expression crossed from benign to invasion stage (Fig. 1A). We speculate that deregulated VHL could be a reason for accumulated KLF4 in breast cancer tissue. We predict that disruption in the regulatory circuitry of VHL-KLF4 and estrogen signal transduction could be an etiological factor during early onset of breast tumorigenesis.
FIGURE 6.
Regulation of KLF4 by VHL is validated in human breast cancer tissue. A and B, immunohistochemistry analysis of KLF4 and VHL protein expression in human breast cancer. Sections from breast cancer and adjacent normal tissues were analyzed by immunostaining using antibodies against KLF4 and VHL. Staining without primary antibody served as negative control. A quantification of tissue staining, as described under “Experimental Procedures,” is shown in B. *, p < 0.001. Error bars, S.D.
DISCUSSION
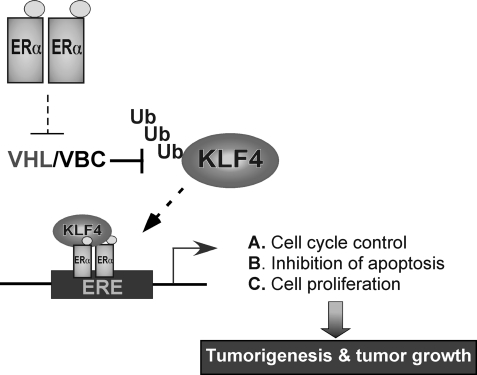
KLF4 has been well known for its mysterious role in the switch of the cellular program from somatic to stem cell, with its putative role in tumorigenesis attracting considerable interest given its ambiguous nature from tumor suppressor to oncogene. Results from the recent combinatorial dissection of KLF4 have revealed its role in carcinogenesis through regulating cell cycle, apoptosis, genomic integrity, and signal transduction (1, 36, 45). The present work explores the function of KLF4 in breast carcinogenesis, especially for its role in orchestrating estrogen signal transduction. We have demonstrated for the first time that the proteolytic regulation of KLF4 is crucial to facilitate estrogen-induced transactivation and mitogenic effect. We identified VHL as a ubiquitin-protein ligase that governs KLF4 degradation and demonstrated that accumulation of KLF4 via suppression of VHL via estrogen signaling is necessary for physiological response of estrogen signaling. These results indicate that KLF4, as an oncogenic factor, might play an important role in the genesis of breast cancer, and the VHL-KLF4 molecular axis could be a new target in endocrine therapy for breast cancer treatment (Fig. 7).
FIGURE 7.
Model for the regulation of KLF4 by VHL in the estrogen signal transduction and concomitant mitogenic effect.
New Insight into Role of VHL-KLF4 Axis in Estrogen Signal Transduction
One of the major features of KLF4 in tumorigenesis is its tissue-dependent nature for its function from tumor suppression to oncogenesis (1). Previous studies defined the role for KLF4 as a tumor suppressor in gastrointestinal cancer through up-regulating transcription of several key cell cycle inhibitors, such as p21, p27, and p57 (11, 14, 15), and down-regulating cell cycle progression factors, such as cyclin B and cyclin D1 (16, 17). Intriguingly, recent studies in breast cancer revealed that, unlike its role in gastrointestinal tissue, KLF4 predominantly promotes tumor genesis and progression during breast cancer formation (11, 19). However, how KLF4 exerts its oncogenic functions remains unknown. Our present study has uncovered a mechanism by which KLF4 acts as an oncogenic factor in breast tumorigenesis via mediating estrogen signal transduction. Particularly, we demonstrated that ubiquitin-dependent proteolysis of KLF4 plays a critical role in regulating estrogen-activated mitogenic effect via orchestrating turnover of KLF4. Given the critical impact of estrogen signaling in mammary gland development and breast tumor initiation, our work provides a new insight into the VHL-KLF4 molecular axis in estrogen signal transduction and breast tumorigenesis (38).
The classical model of estrogen signaling involves ligand-receptor binding, which induces receptor dimerization and subsequent binding to specific DNA sequence. The ligand-bound ER recruits and complexes with co-activators, such as SRC1 and -3, and co-regulators, such as histone acetyltransferases, and activates target gene transcription. ER also regulates gene transcription by interacting with other transcription factors, such as activating protein 1 (AP1) and specificity protein 1 (SP1) (46). Although a known factor in orchestrating transcription, the detailed mechanism by which KLF4 is involved in estrogen-mediated transactivation still remains under investigation. Previous studies have shown that KLF4 is a gene-specific activator or repressor (36), where the transactivation domain on its amino terminus governs its essential activity (47). Mechanistically, the function of the amino-terminal domain depends on its interaction with co-activators, such as p300/CBP (28, 47, 48) and Tip 60 (49). Both p300/CBP and Tip60 have histone acetyltransferase activity, which catalyzes the acetylation of localized histone at the promoter. The acetylated histone in turn recruits other transcription factors as well as the basal transcriptional machinery to activate transcription. Intriguingly, both p300/CBP and Tip60 also function as co-activators for several nuclear hormone receptors, including ERα (50, 51). Thus, it could be possible to speculate that KLF4 complexes with ERα and certain co-activators to exert its promoting function in estrogen-activated transcription.
One surprise conclusion based on recent studies on estrogen signaling is its innovative aspect in regulating cell cycle checkpoint and DNA repair (52, 53). E2/ER inhibited ATR activation elicited by DNA damage-inducing stimuli in breast cancer and normal mammary epithelial cells (53). As a result, the downstream signaling of ATR was blocked, including G2 checkpoint and p53/p21-mediated G1/S checkpoint. The inhibition of DNA damage-induced cell cycle checkpoints might allow accumulation of unrepaired mutations, which further promotes the onset of breast carcinogenesis. Indeed, estrogen was reported to induce DNA double strand breaks in breast cancer cells (52). Other than its function in cell cycle progression and proliferation, the role of KLF4 in DNA damage response has been well documented as well (16, 18, 27, 54, 55). It is noteworthy that KLF4 could inhibit p53 expression to achieve its anti-apoptotic effect when cells were challenged with severe DNA damage (27). In addition, depletion of KLF4 from breast cancer cells was shown to cause p53-dependent apoptosis (11). Collectively, these observations could support a possible hypothesis that estrogen-induced KLF4 accumulation might have a role in regulating the cell cycle checkpoint whose deregulation may contribute to genome instability and tumorigenesis.
Proteolytic Regulation of KLF4 and Its Impact in Breast Carcinogenesis
As a critical component that governs stem cell pluripotency, cell cycle progression, genomic integrity, and signal transduction, it is not surprising that KLF4 must be tightly regulated at different levels, including transcriptional and post-translational regulation. Indeed, previous studies have uncovered a regulatory circuitry for KLF4, including promoter methylation, loss of heterozygosity, phosphorylation, acetylation, and sumoylation (3, 36). In addition to the current KLF4 regulatory paradigm, recent studies have demonstrated that KLF4 is a fast-turnover protein that fluctuates during the cell cycle progression, suggesting proteolytic regulation of KLF4, although the ubiquitin-protein ligase was unknown (20).
Intriguingly, increased KLF4 expression was observed in up to 70% of primary human breast cancers (9). KLF4 overexpression at the stage of ductal carcinoma in situ suggests that this represents an early event in breast carcinogenesis. Our studies showed that elevation of KLF4 level is evident even in benign breast and is greatly enhanced in atypical ductal hyperplasia. Indeed, recent studies from other groups have consistently reported up-regulation of KLF4 in breast cancer, further corroborating the notion that KLF4 is an important player in breast carcinogenesis, and accumulated KLF4 protein levels are well correlated with the prognosis of breast cancer (30, 34). Similar to the novel observation of the oncogenic function of KLF4 in breast cancer, if the accumulation of KLF4 is disrupted and that is the causative factor for breast carcinogenesis, then the question becomes the mechanism involved in accumulating KLF4. The comprehensive studies in the present work, including microarray data sets from Oncomine, our results from pulse-chase, treatment with estrogen receptor antagonist, MG132 blockade, and ubiquitylation assay, support a hypothesis that KLF4 protein half-life needs to be accurately controlled by the ubiquitin-proteasome system for cellular homeostasis. Disruption of the proteolytic regulation of KLF4 would lead to accumulation that in turn would enhance oncogenesis and proliferation in mammary gland and breast cancer cells. These results explain our hypothesis that estrogen, a mitogenic stimulus, drives accumulation of KLF4 and that estrogen-induced KLF4 elevation facilitates the estrogen-mediated mitogenic effect and breast tumorigenesis. Indeed, rapid protein turnover is a typical feature of many important proteins (e.g. p53, p27, and Myc). Selective degradation of these proteins is essential for normal cell growth and differentiation, whereas abnormal accumulation or hyperactive degradation of these regulatory proteins is associated with carcinogenesis (56, 57).
Regulation of KLF4 Degradation by VHL in Breast Cancer Development
An increasing number of abnormalities in the ubiquitin-proteasome pathway have been identified in breast cancer. Several critical ubiquitin-protein ligases, including BRCA1, Skp2, and APC/Cdh1, have been connected to breast tumorigenesis (33, 39–41). Our finding for the first time demonstrates that VHL is the E3 ubiquitin ligase that governs KLF4 degradation in order to maintain cell homeostasis, with the disruption of the VHL-KLF4 axis contributing to KLF4 overexpression in breast cancer. The evidence at multiple levels supports the finding that VHL governs KLF4 turnover in breast cancer cells, including the following. 1) Depletion and elevated expression of VHL correlates well with up-regulation as well as down-regulation of KLF4. 2) VHL directly targets KLF4 for ubiquitylation. 3) An inverse relationship in the expression of VHL and KLF4 protein was observed in normal and malignant breast tissue. The best characterized function of VHL is as a substrate-specific adaptor of VCB-Cul2 E3 ubiquitin ligase complex that targets HIFα for proteolytic degradation. The current dogma for recognition of HIF1 and HIF2 by pVHL is contingent on post-translational modification of conserved HIF proline residues located in LXXLAP motifs by specific prolyl-hydroxylases, a process involving molecular oxygen (58). Under normoxic or atmospheric oxygen levels, HIFα subunits are efficiently modified by prolyl-hydroxylases, ubiquitylated by CBCVHL, and rapidly degraded by the 26S proteasome. We indeed found LXXLAP motifs on the amino terminus of KLF4 and mutated LXXLAP motifs. To our surprise, mutation of LXXLAP motifs on KLF4 did not affect the VHL-catalyzed KLF4 degradation, suggesting the presence of an additional substrate recognition mechanism for KLF4 ubiquitylation by VHL, which is our next mission (data not shown). Importantly, with our endeavor to molecularly map the degron, we successfully uncovered the mechanism that governs KLF4 ubiquitylation by VHL and identified that lysine 43 on KLF4 is the pivotal amino residue mediating the ubiquitin-conjugating chain on KLF4 for its ubiquitylation and further degradation. The finding allows us to engineer KLF4 stable mutants that further allow us to dissect the impact of KLF4 stabilization in the estrogen-mediated mitogenic effect.
VHL functions as a tumor suppressor whose mutations or loss of function predisposes to the formation of tumors (58). Most studies have linked VHL tumor suppressor function to its ability to degrade HIFα. However, recent studies suggest that HIF-independent function of VHL also plays a role in tumorigenesis. For example, certain VHL mutations predispose affected individuals to develop certain type of tumors without impairing its ability to degrade HIFα (59). Moreover, overexpression of constitutively active HIF failed to cause hemangioblastomas or renal carcinomas (60). Therefore, the mechanisms of tumor suppression by VHL are far from simple. No mutations on VHL have been reported thus far in breast cancer (61). Studies from us and other groups (40) have shown that VHL was down-regulated in breast cancer in comparison with normal tissues, although the underlying mechanisms remain unknown. It is noteworthy that overexpression of HIFα resulting from VHL knockout was not sufficient for breast carcinogenesis (62), further corroborating the role of HIF-independent function of VHL in tumorigenesis. The VHL-KLF4 circuit, as revealed by our current studies, could at least represent one important mechanism for breast cancer development.
In addition to the oncogenic function, estrogen signaling is also essential for normal mammary gland development. Most recent studies suggest that estrogen is involved in the control of mammary stem cell function, including number and regenerative activity (63, 64). Intriguingly, KLF4 is one of the four well known factors that could reprogram somatic cells into pluripotent cells. This role was confirmed in a recent study that revealed a critical role of KLF4 in the maintenance of breast cancer stem cells (19). It would be of great worth to test whether the E2-KLF4 cascade, as revealed by our current study, plays a role in mammary stem cell biology. Surprisingly, VHL deletion has been shown to impair the differentiation of progenitor cells into alveolar epithelium, implicating that VHL has a role in stem cell renewal (62). Importantly, co-deletion of HIF1α could not rescue the vhl−/−-dependent phenotype, suggesting that additional VHL-regulated genes besides HIF1A function to regulate the regenerative potential of the breast epithelium. Thus, whether the VHL-KLF4 circuit is involved in mammary stem cell biology warrants further investigation.
Implication of VHL-KLF4 Circuitry in Anti-breast Cancer Endocrine Therapy
Estrogen/ER signaling plays a critical role in breast carcinogenesis. Strategies of interfering with estrogen action have demonstrated great success in breast cancer therapy. Adjuvant therapy with tamoxifen has made significant contributions to decreasing breast cancer mortality in the past 2 decades. However, a significant percentage of breast cancer patients will eventually develop endocrine resistance, which represents a severe challenge in the field of breast cancer therapy. Deregulation of various aspects of estrogen signaling has been thought to be a common strategy for resistance. The critical role of KLF4 regulation by VHL in estrogen signaling, as revealed in our current studies, provides novel insight into the genesis of breast malignancy and suggests that VHL-KLF4 axis could be a potential target for breast cancer therapy. Our data suggest the presence of a protein turnover cascade that ensures the transmission of signal upon occupancy of receptor by estrogen, whereas down-regulation of VHL in response to the stimulation with estrogen results in the accumulation of KLF4. Our pilot study suggests that estrogen-induced down-regulation of VHL is via a ubiquitin-dependent event, although the involved E3 ligase needed to be identified. Our results now shed new light on a previously unknown avenue to manage the estrogen signaling at two novel targeting steps: estrogen-induced VHL degradation and estrogen-induced elevation of KLF4. Further efforts to develop small molecules that could interrupt the biochemical catalysis of estrogen-induced VHL degradation or KLF4 accumulation could provide a new approach to enhance the current anti-breast cancer endocrine therapy.
Supplementary Material
Acknowledgments
We are grateful to Drs. Daniel S. Peeper, Vincent W. Yang, Pamela A. Hershberger, and Edward V. Prochownik for kindly providing plasmids and cell lines. We are grateful for technical support from Drs. Armin Gamper, Richard A. Steinman, and Hyun Kim. We thank Drs. Steffi Oesterreich and Adrian Lee for constructive discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant CA115943. This work was also supported by the Pittsburgh Woman Cancer Fund.

This article contains supplemental Fig. 1.
- VHL
- Von Hippel-Lindau
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- ER
- estrogen receptor
- ERE
- estrogen response element
- KD
- knockdown.
REFERENCES
- 1. Rowland B. D., Peeper D. S. (2006) KLF4, p21, and context-dependent opposing forces in cancer. Nat. Rev. Cancer 6, 11–23 [DOI] [PubMed] [Google Scholar]
- 2. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 3. Wei D., Kanai M., Huang S., Xie K. (2006) Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis 27, 23–31 [DOI] [PubMed] [Google Scholar]
- 4. Wang N., Liu Z. H., Ding F., Wang X. Q., Zhou C. N., Wu M. (2002) Down-regulation of gut-enriched Kruppel-like factor expression in esophageal cancer. World J. Gastroenterol. 8, 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tetreault M. P., Yang Y., Travis J., Yu Q. C., Klein-Szanto A., Tobias J. W., Katz J. P. (2010) Esophageal squamous cell dysplasia and delayed differentiation with deletion of Krüppel-like factor 4 in murine esophagus. Gastroenterology 139, 171–181.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ohnishi S., Ohnami S., Laub F., Aoki K., Suzuki K., Kanai Y., Haga K., Asaka M., Ramirez F., Yoshida T. (2003) Down-regulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem. Biophys. Res. Commun. 308, 251–256 [DOI] [PubMed] [Google Scholar]
- 7. Hu W., Hofstetter W. L., Li H., Zhou Y., He Y., Pataer A., Wang L., Xie K., Swisher S. G., Fang B. (2009) Putative tumor-suppressive function of Kruppel-like factor 4 in primary lung carcinoma. Clin. Cancer Res. 15, 5688–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guan H., Xie L., Leithäuser F., Flossbach L., Möller P., Wirth T., Ushmorov A. (2010) KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood 116, 1469–1478 [DOI] [PubMed] [Google Scholar]
- 9. Foster K. W., Frost A. R., McKie-Bell P., Lin C. Y., Engler J. A., Grizzle W. E., Ruppert J. M. (2000) Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 60, 6488–6495 [PubMed] [Google Scholar]
- 10. Pandya A. Y., Talley L. I., Frost A. R., Fitzgerald T. J., Trivedi V., Chakravarthy M., Chhieng D. C., Grizzle W. E., Engler J. A., Krontiras H., Bland K. I., LoBuglio A. F., Lobo-Ruppert S. M., Ruppert J. M. (2004) Nuclear localization of KLF4 is associated with an aggressive phenotype in early stage breast cancer. Clin. Cancer Res. 10, 2709–2719 [DOI] [PubMed] [Google Scholar]
- 11. Rowland B. D., Bernards R., Peeper D. S. (2005) The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 7, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 12. Foster K. W., Ren S., Louro I. D., Lobo-Ruppert S. M., McKie-Bell P., Grizzle W., Hayes M. R., Broker T. R., Chow L. T., Ruppert J. M. (1999) Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells. Transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 10, 423–434 [PubMed] [Google Scholar]
- 13. Foster K. W., Liu Z., Nail C. D., Li X., Fitzgerald T. J., Bailey S. K., Frost A. R., Louro I. D., Townes T. M., Paterson A. J., Kudlow J. E., Lobo-Ruppert S. M., Ruppert J. M. (2005) Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene 24, 1491–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang W., Geiman D. E., Shields J. M., Dang D. T., Mahatan C. S., Kaestner K. H., Biggs J. R., Kraft A. S., Yang V. W. (2000) The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J. Biol. Chem. 275, 18391–18398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wei D., Kanai M., Jia Z., Le X., Xie K. (2008) Kruppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res. 68, 4631–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoon H. S., Yang V. W. (2004) Requirement of Krüppel-like factor 4 in preventing entry into mitosis following DNA damage. J. Biol. Chem. 279, 5035–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shie J. L., Chen Z. Y., Fu M., Pestell R. G., Tseng C. C. (2000) Gut-enriched Krüppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 28, 2969–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghaleb A. M., Katz J. P., Kaestner K. H., Du J. X., Yang V. W. (2007) Krüppel-like factor 4 exhibits antiapoptotic activity following γ-radiation-induced DNA damage. Oncogene 26, 2365–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu F., Li J., Chen H., Fu J., Ray S., Huang S., Zheng H., Ai W. (2011) Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 30, 2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Z. Y., Wang X., Zhou Y., Offner G., Tseng C. C. (2005) Destabilization of Krüppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 65, 10394–10400 [DOI] [PubMed] [Google Scholar]
- 21. Shields J. M., Christy R. J., Yang V. W. (1996) Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J. Biol. Chem. 271, 20009–20017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cullingford T. E., Butler M. J., Marshall A. K., Tham el L., Sugden P. H., Clerk A. (2008) Differential regulation of Krüppel-like factor family transcription factor expression in neonatal rat cardiac myocytes. Effects of endothelin-1, oxidative stress, and cytokines. Biochim. Biophys. Acta 1783, 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shie J. L., Chen Z. Y., O'Brien M. J., Pestell R. G., Lee M. E., Tseng C. C. (2000) Role of gut-enriched Krüppel-like factor in colonic cell growth and differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 279, G806–G814 [DOI] [PubMed] [Google Scholar]
- 24. Liu S., Zhang H., Zhu L., Zhao L., Dong Y. (2008) Kruppel-like factor 4 is a novel mediator of selenium in growth inhibition. Mol. Cancer Res. 6, 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Z. Y., Shie J., Tseng C. (2000) Up-regulation of gut-enriched Krüppel-like factor by interferon-γ in human colon carcinoma cells. FEBS Lett. 477, 67–72 [DOI] [PubMed] [Google Scholar]
- 26. Birsoy K., Chen Z., Friedman J. (2008) Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 7, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Q., Hong Y., Zhan Q., Shen Y., Liu Z. (2009) Role for Kruppel-like factor 4 in determining the outcome of p53 response to DNA damage. Cancer Res. 69, 8284–8292 [DOI] [PubMed] [Google Scholar]
- 28. Evans P. M., Zhang W., Chen X., Yang J., Bhakat K. K., Liu C. (2007) Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J. Biol. Chem. 282, 33994–34002 [DOI] [PubMed] [Google Scholar]
- 29. Du J. X., McConnell B. B., Yang V. W. (2010) A small ubiquitin-related modifier-interacting motif functions as the transcriptional activation domain of Krüppel-like factor 4. J. Biol. Chem. 285, 28298–28308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chu P. Y., Hsu N. C., Liao A. T., Yeh K. T., Hou M. F., Liu C. H. (2011) Elevated Krüppel-like factor 4 transcription factor in canine mammary carcinoma. BMC Vet. Res. 7, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tyson J. J., Baumann W. T., Chen C., Verdugo A., Tavassoly I., Wang Y., Weiner L. M., Clarke R. (2011) Dynamic modeling of oestrogen signaling and cell fate in breast cancer cells. Nat. Rev. Cancer 11, 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu W., Wu G., Li W., Lobur D., Wan Y. (2007) Cdh1 anaphase-promoting complex targets Skp2 for destruction in transforming growth factor β-induced growth inhibition. Mol. Cell. Biol. 27, 2967–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fujita T., Liu W., Doihara H., Date H., Wan Y. (2008) Dissection of the APCCdh1-Skp2 cascade in breast cancer. Clin. Cancer Res. 14, 1966–1975 [DOI] [PubMed] [Google Scholar]
- 34. Chen C. J., Lin S. E., Lin Y. M., Lin S. H., Chen D. R., Chen C. L. (2011) Association of expression of Kruppel-like factor 4 and Kruppel-like factor 5 with the clinical manifestations of breast cancer. Pathol. Oncol. Res., in press [DOI] [PubMed] [Google Scholar]
- 35. Gluck S., Ross J. S., Royce M., McKenna E. F., Jr., Perou C. M., Avisar E., Wu L. (2011) TP53 genomics predict higher clinical and pathologic tumor response in operable early stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res. Treat., in press [DOI] [PubMed] [Google Scholar]
- 36. Evans P. M., Liu C. (2008) Roles of Krüpel-like factor 4 in normal homeostasis, cancer, and stem cells. Acta Biochim. Biophys. Sin. 40, 554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McConnell B. B., Ghaleb A. M., Nandan M. O., Yang V. W. (2007) The diverse functions of Krüppel-like factors 4 and 5 in epithelial biology and pathobiology. BioEssays 29, 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Musgrove E. A., Sutherland R. L. (2009) Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer 9, 631–643 [DOI] [PubMed] [Google Scholar]
- 39. Kudo Y., Guardavaccaro D., Santamaria P. G., Koyama-Nasu R., Latres E., Bronson R., Yamasaki L., Pagano M. (2004) Role of F-box protein βTrcp1 in mammary gland development and tumorigenesis. Mol. Cell. Biol. 24, 8184–8194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zia M. K., Rmali K. A., Watkins G., Mansel R. E., Jiang W. G. (2007) The expression of the von Hippel-Lindau gene product and its impact on invasiveness of human breast cancer cells. Int. J. Mol. Med. 20, 605–611 [PubMed] [Google Scholar]
- 41. Signoretti S., Di Marcotullio L., Richardson A., Ramaswamy S., Isaac B., Rue M., Monti F., Loda M., Pagano M. (2002) Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J. Clin. Invest. 110, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Schuetz A., Nana D., Rose C., Zocher G., Milanovic M., Koenigsmann J., Blasig R., Heinemann U., Carstanjen D. (2011) The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell. Mol. Life Sci. 68, 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brown R. J., Davidson N. E. (2006) Adjuvant hormonal therapy for premenopausal women with breast cancer. Semin. Oncol. 33, 657–663 [DOI] [PubMed] [Google Scholar]
- 44. Kaelin W. G. (2007) Von Hippel-Lindau disease. Annu. Rev. Pathol. 2, 145–173 [DOI] [PubMed] [Google Scholar]
- 45. Hu D., Wan Y. (2011) Regulation of Krüppel-like factor 4 by the anaphase-promoting complex pathway is involved in TGF-β signaling. J. Biol. Chem. 286, 6890–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas C., Gustafsson J. Å. (2011) The different roles of ER subtypes in cancer biology and therapy. Nat. Rev. Cancer 11, 597–608 [DOI] [PubMed] [Google Scholar]
- 47. Geiman D. E., Ton-That H., Johnson J. M., Yang V. W. (2000) Transactivation and growth suppression by the gut-enriched Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res. 28, 1106–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Feinberg M. W., Cao Z., Wara A. K., Lebedeva M. A., Senbanerjee S., Jain M. K. (2005) Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J. Biol. Chem. 280, 38247–38258 [DOI] [PubMed] [Google Scholar]
- 49. Ai W., Zheng H., Yang X., Liu Y., Wang T. C. (2007) Tip60 functions as a potential corepressor of KLF4 in regulation of HDC promoter activity. Nucleic Acids Res. 35, 6137–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanstein B., Eckner R., DiRenzo J., Halachmi S., Liu H., Searcy B., Kurokawa R., Brown M. (1996) p300 is a component of an estrogen receptor coactivator complex. Proc. Natl. Acad. Sci. U.S.A. 93, 11540–11545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gaughan L., Brady M. E., Cook S., Neal D. E., Robson C. N. (2001) Tip60 is a co-activator specific for class I nuclear hormone receptors. J. Biol. Chem. 276, 46841–46848 [DOI] [PubMed] [Google Scholar]
- 52. Williamson L. M., Lees-Miller S. P. (2011) Estrogen receptor α-mediated transcription induces cell cycle-dependent DNA double strand breaks. Carcinogenesis 32, 279–285 [DOI] [PubMed] [Google Scholar]
- 53. Pedram A., Razandi M., Evinger A. J., Lee E., Levin E. R. (2009) Estrogen inhibits ATR signaling to cell cycle checkpoints and DNA repair. Mol. Biol. Cell 20, 3374–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoon H. S., Chen X., Yang V. W. (2003) Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J. Biol. Chem. 278, 2101–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoon H. S., Ghaleb A. M., Nandan M. O., Hisamuddin I. M., Dalton W. B., Yang V. W. (2005) Krüppel-like factor 4 prevents centrosome amplification following γ-irradiation-induced DNA damage. Oncogene 24, 4017–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hoeller D., Dikic I. (2009) Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 [DOI] [PubMed] [Google Scholar]
- 57. Sun Y. (2006) E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia 8, 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kaelin W. G., Jr. (2008) The von Hippel-Lindau tumor suppressor protein. O2 sensing and cancer. Nat. Rev. Cancer 8, 865–873 [DOI] [PubMed] [Google Scholar]
- 59. Hoffman M. A., Ohh M., Yang H., Klco J. M., Ivan M., Kaelin W. G., Jr. (2001) von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to down-regulate HIF. Hum. Mol. Genet. 10, 1019–1027 [DOI] [PubMed] [Google Scholar]
- 60. Elson D. A., Thurston G., Huang L. E., Ginzinger D. G., McDonald D. M., Johnson R. S., Arbeit J. M. (2001) Induction of hypervascularity without leakage or inflammation in transgenic mice overexpressing hypoxia-inducible factor-1α. Genes Dev. 15, 2520–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sourvinos G., Miyakis S., Liloglou T. L., Field J. K., Spandidos D. A. (2001) Von Hippel-Lindau tumour suppressor gene is not involved in sporadic human breast cancer. Tumour Biol. 22, 131–136 [DOI] [PubMed] [Google Scholar]
- 62. Seagroves T. N., Peacock D. L., Liao D., Schwab L. P., Krueger R., Handorf C. R., Haase V. H., Johnson R. S. (2010) VHL deletion impairs mammary alveologenesis but is not sufficient for mammary tumorigenesis. Am. J. Pathol. 176, 2269–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Asselin-Labat M. L., Vaillant F., Sheridan J. M., Pal B., Wu D., Simpson E. R., Yasuda H., Smyth G. K., Martin T. J., Lindeman G. J., Visvader J. E. (2010) Control of mammary stem cell function by steroid hormone signaling. Nature 465, 798–802 [DOI] [PubMed] [Google Scholar]
- 64. Joshi P. A., Jackson H. W., Beristain A. G., Di Grappa M. A., Mote P. A., Clarke C. L., Stingl J., Waterhouse P. D., Khokha R. (2010) Progesterone induces adult mammary stem cell expansion. Nature 465, 803–807 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.