Background: PpsR controls synthesis of heme and bacteriochlorophyll in purple photosynthetic bacteria.
Results: PpsR binds heme as a co-factor and changes its DNA binding pattern.
Conclusion: Heme affects the ability of PpsR to regulate tetrapyrrole gene expression.
Significance: A cysteine axial ligand to heme is in the helix-turn-helix domain providing a way for heme to affect DNA binding properties of PpsR.
Keywords: Bacterial Genetics, DNA Transcription, Gene Regulation, Heme, Photosynthesis, Rhodobacter sphaeroides, Tetrapyrrole Biosynthesis
Abstract
Heme-mediated regulation, presented in many biological processes, is achieved in part with proteins containing heme regulatory motif. In this study, we demonstrate that FLAG-tagged PpsR isolated from Rhodobacter sphaeroides cells contains bound heme. In vitro heme binding studies with tagless apo-PpsR show that PpsR binds heme at a near one-to-one ratio with a micromolar binding constant. Mutational and spectral assays suggest that both the second Per-Arnt-Sim (PAS) and DNA binding domains of PpsR are involved in the heme binding. Furthermore, we show that heme changes the DNA binding patterns of PpsR and induces different responses of photosystem genes expression. Thus, PpsR functions as both a redox and heme sensor to coordinate the amount of heme, bacteriochlorophyll, and photosystem apoprotein synthesis thereby providing fine tune control to avoid excess free tetrapyrrole accumulation.
Introduction
Heme-binding proteins are involved in a wide range of biological functions including electron transfer (1), oxygen metabolism (1), iron metabolism (2), nitric oxide synthesis (3) and neuron-degenerated disease (4, 5). As a co-factor, heme can modulate transcription (6), translation (7), protein targeting (8), and degradation (2, 9, 10) as well as functioning as a sensor and carrier of a variety of gas molecules (11–15). Among photosynthetic organisms, the heme and bacteriochlorophyll biosynthetic pathways share common intermediates from δ-aminolevulinic acid to protoporphyrin IX. Unbound heme and bacteriochlorophyll are both toxic; thus, the vast majority of these tetrapyrroles are sequestered within proteins. However, it is not clear how cells coordinate the synthesis of defined amounts of tetrapyrrole end products that utilize common intermediates with the synthesis of apoproteins that bind tetrapyrroles. In recent years, several heme-sensing regulatory proteins have been identified in prokaryotic (2) and eukaryotic organisms (9, 16) that are able to sense free heme. To date, identified heme sensors contain one or more heme regulatory motifs (HRMs)2 comprising a conserved -Cys-Pro- (8, 17).
In Rhodobacter sphaeroides, heme and bacteriochlorophyll biosynthesis are co-regulated by the redox and light controlled PpsR-AppA system (18). PpsR is a DNA-binding transcription factor that recognizes conserved palindromes present in a number of tetrapyrrole and photosynthesis promoters (19, 20). Under aerobic conditions, a pair of redox active Cys in PpsR undergo oxidation to stimulate binding of PpsR to target promoters to block transcription (18). PpsR is also regulated by the flavin-containing antirepressor AppA that responds to changes in both redox and light intensity. AppA inactivates PpsR by forming an inactive PpsR2-AppA complex under anaerobic dark conditions (18). Recently, it was reported that AppA also binds heme as a co-factor (21, 22), although the role of heme bound to AppA remains ambiguous.
In this study, we present evidence that PpsR is a heme-binding protein and that the redox active Cys424 present in the DNA binding domain of PpsR is critical for heme interaction. The binding of heme to PpsR was found to change its DNA binding pattern and to induce increased transcription of several PpsR-regulated genes. These results are consistent with the hypothesis that excess heme can quickly change the state of photosynthetic gene expression from inhibition to activation, thus providing a mechanism for bacteria to react to the toxic unbound tetrapyrrole products. It is also worth noting that PpsR does not contain a typical HRM, suggesting that heme-sensing regulators may be more widespread than was previously understood.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
R. sphaeroides strain HRIF (18) was used as the parent strain. PpsR knock-out strain (PPSR1) was made by replacing the PpsR gene with a kanamycin-resistant cassette. PpsR-FLAG strain was made by adding a FLAG tag to the end of ppsR gene with a single cross-event. Strain BL21 (DE3) (Novagen) was used for protein overexpression in E. coli. Luria broth (LB) medium was used for agar-solidified plates and liquid cultures for E. coli and R. sphaeroides. Kanamycin was used at 50 μg/ml for agar-solidified plates and at 25 μg/ml for liquid cultures as needed.
Construction of Expression Vectors
All proteins were expressed in E. coli using a modified SUMO-I (LifeSensors) overexpression system. For full-length PpsR, the coding region was cloned from pETPpsR (18) into the NdeI-SacI restriction sites of pSUMO, resulting in the recombinant plasmid pSUMO-PpsR. Plasmid pSUMO-PpsR was subjected to site-directed mutagenesis (Herculase II; Stratagene) to construct stop codons to form truncated PpsR variants as well as PpsR point mutations.
Purification of PpsR from R. sphaeroides
PpsR-FLAG strain culture was collected and resuspended in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mg/ml lysozyme, and 20 μl/per sample of protease inhibitor mixture (Sigma)). Cells were kept on ice, disrupted by sonication of 10 s with 10 s interval, for 10 min in total. Cracked cells were centrifuged at 15,000 × g for 30 min, and the supernatant was incubated with 50 μl of EZviewTM red anti-FLAG M2 affinity gel (Sigma) at 4 °C overnight. The anti-FLAG gel was then spun down and washed with TBS buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl). PpsR-FLAG was eluted with 300 μl of 3× FLAG peptide (Sigma).
Purification of PpsR from E. coli
SUMO-tagged PpsR was overexpressed by induction of T7 polymerase with 1 mm isopropyl-β-d-thiogalactopyranoside at 16 °C overnight in LB. Cells were disrupted by three passages through a cell cracker and clarified by centrifugation at 15,000 × g for 30 min. PpsR-SUMO was purified by binding to HisTrapTM HP columns (GE Healthcare), washed with ∼10 column volumes of 50 mm imidazole, and then eluted with a 50–500 mm linear gradient of imidazole in a buffer comprising 20 mm Tris-HCl, pH 8.0, 500 mm NaCl, and 5% glycerol. Eluted PpsR-SUMO was then cleaved from the SUMO tag with SUMO-protease-I (LifeSensors) for 1 h at room temperature. Following cleavage, tagless PpsR was then isolated by gel filtration chromatography using a Sephacryl S-200 HP resin (GE Healthcare) equilibrated in 20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 5% glycerol.
Protein–Co-factor Interactions
Heme stock solutions were freshly dissolved in 0.01 m NaOH. The final concentration was determined spectrophotometrically using a coefficient of 58.4 mm−1 cm−1 (23). To quantify heme binding, purified protein was incubated with a 2-fold excess of heme at room temperature for >20 min. The unbound heme was then removed by passing through MidiTrapTM G-25 column (GE Healthcare) or HiTrapTM Q HP column (GE Healthcare). Heme bound to protein was determined by using the pyridine hemochromagen assay as described previously (24). All the protein-heme interactions, including the reactions used for later analysis, were carried out under oxidizing conditions unless noted otherwise.
Spectroscopy
Electronic absorption UV-visible spectra were recorded using a Beckman DU 640 spectrophotometer. Purified protein was incubated with various concentrations of heme at room temperature for more than 20 min before the measurements were taken. To measure the reduced spectrum of protein-heme complex, 1 m sodium dithionite stock was made freshly with the buffer of 2 m Tris-HCl, pH 8.0, with a final concentration of 5 mm sodium dithionite added to the sample. Carbon monoxide was added by briefly bubbling CO into the protein sample. The deoxy-PpsR-heme spectrum was obtained using the same method for reduced spectrum, except that all buffer and samples were completely degassed, and the whole procedure was done in an anaerobic hood and with flow-stop cuvette.
Tryptophan Fluorescence Quenching Assay
Fluorescence measurements were performed at room temperature using a LS50B luminescence spectrometer (PerkinElmer Life Sciences) in a 2-ml sample volume with protein in 20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 5% glycerol. Heme was added into the sample to final concentration of 0.1–2.5 μm. After incubation for 2 min, the sample was excited at 272 nm. Three emissions from 290 to 400 nm were then recorded and averaged. Data were processed using nonlinear curve fitting with one-to-one binding model using Origin software (OriginLab). The experiments were carried out in the PpsR-heme concentration range that exhibits linear relationship to protein fluorescence (supplemental Fig. 1).
Gel Mobility Shift Analysis
Probes for gel mobility shift assays were prepared by colony PCR amplification of the puc (262 bp) and the hemE (344 bp) promoter regions (supplemental Table 1). 10 nm DNA probes were incubated with different concentrations of purified protein for 30 min at room temperature, in 40 mm Tris-HCl, pH 8.0, 250 mm NaCl, 50 mm KCl, 5 mm MgSO4, 5% glycerol, 1 mm EDTA, and 20 μg/ml heparin. The mixtures were then subjected to 6% PAGE at 4 °C in a buffer containing 50 mm Tris-HCl, pH 8.0, 380 mm glycine, and 2 mm EDTA. After the electrophoresis, the gel was stained with SYBR Green dye (Invitrogen) as described by the manufacturer and scanned with a Typhoon 9210 variable mode imager (GE Healthcare). When reducing condition was required, fresh dithionite of 5 μm final concentration was added into all of the buffers and the reactions.
DNase I Footprinting
Promoter region of puc (262 bp) was amplified with a 6-FAM-labeled primer on the forward strand and 5-HEX-labeled primer on the reverse strand. A DNA binding reaction similar to that described for the gel mobility shift assays was set up in a 20- μl volume to which 5 μl of DNase I was added for 30 min at room temperature to digest DNA. DNA digestion was then stopped by the addition of 20 μl of 0.5 m EDTA. DNA digestion products were recovered using a Min Elute PCR purification kit (Qiagen), with sample eluted with 15 μl of elution buffer containing 0.75 μl of 500 LIZTM Size Standard (Applied Biosystems). The samples were separated and detected with a 3730 DNA Analyzer (Applied Biosystems) and analyzed with Peak Scanner Software v1.0 (Applied Biosystems).
Quantitative PCR (qPCR) Analysis of Gene Expression
Different R. sphaeroides strains were grown in LB medium under aerobic conditions until the absorbance at 660 nm reached 0.2–0.3. Cells were then harvested by centrifugation, washed, and resuspended with Sistrom's minimal medium, then continued growth under aerobic conditions. After 20 min of starvation, freshly made heme (in 50% ethanol, 0.05 m NaHCO3) was added into the cultures to a final concentration of 25 μm, and 1-ml samples were then extracted and stored on ice at different time points. The RNA was purified with ISOLATE RNA mini kit (Bioline) and Turbo DNA-free (Ambion). Reverse transcription and real-time PCR were performed with Brilliant II SYBR Green QPCR Master Mix kit (Agilent Technologies) on either Quantitative PCR Stratagene MX3000P (Agilent Technologies) or StepOnePlus Real-Time PCR system (Applied Biosystems). The transcription level of rpoZ gene (encoding the RNA polymerase omega subunit) was used to normalize the levels of all other genes. At least three biological replications were measured on each condition, and qPCR was performed for each sample at least twice. For each biological replication, a mixture of all RNA samples was used to determine the efficiency of the primers with serial dilution methods. Primer sequences for qPCR analysis are shown in supplemental Table 2.
RESULTS
PpsR Binds Heme in Vivo and in Vitro
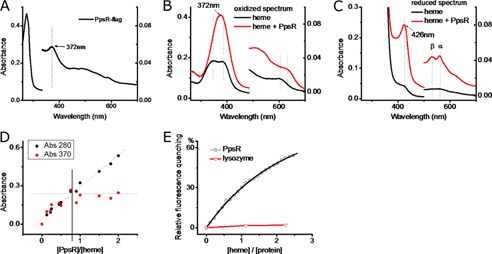
We constructed a chromosomally encoded carboxyl-terminal FLAG-tagged PpsR construct in R. sphaeroides at its native chromosomal location by recombination. Affinity purification of FLAG-tagged PpsR yielded a colored preparation that was judged to be >95% pure based on SDS-PAGE chromatography. An absorption spectrum of the purified FLAG-tagged PpsR showed the presence of a distinct heme peak at 372 nm as well as additional broad peaks in the 440–600-nm region (Fig. 1A and supplemental Fig. 2A). We subsequently tested the ability of tagless apo-PpsR, purified from an E. coli overexpression system, to bind to heme-agarose using a pulldown assay. The results demonstrate that tagless PpsR is indeed capable of binding to heme-agarose in vitro (supplemental Fig. 2B).
FIGURE 1.
PpsR binds heme. A, UV-visible spectrum of purified PpsR-FLAG from R. sphaeroides. B, UV-visible spectrum of heme and PpsR-heme (the spectrum of PpsR is subtracted) under oxidizing conditions. 5 μm heme was incubated with 10 μm PpsR for at least 20 min before the spectrum was taken. C, UV-visible spectrum of heme and PpsR-heme under reducing conditions. D, titration of PpsR in to heme. The change of the absorbance at 280 nm indicates the change of protein concentration whereas the change of Soret peaks at 370 nm indicating the formation of PpsR-heme complex. E, binding constant of PpsR-heme interaction. Heme was titrated into 1 μm PpsR, with at least 5 min of incubation time between each step. The data were fitted with one-to-one binding model. The same settings were applied to 1 μm lysozyme.
We further investigated the ability of tagless PpsR to bind heme by monitoring changes in the absorption spectrum of heme upon addition of purified tagless apo-PpsR under oxidizing conditions (oxidized spectrum). The spectrum in Fig. 1B shows that upon the addition of apo-PpsR, the broad doublet Soret peak of free heme intensified and was converted to a single peak at 372 nm concomitant with peaks in the 500–700-nm region of the spectrum that is virtually indistinguishable from absorbance characteristics of FLAG-tagged PpsR that was isolated from R. sphaeroides cells (Fig. 1A). Reduction of the heme-bound iron from Fe3+ to Fe2+ by the addition of dithionate resulted in a red shift of Soret peak from 372 nm to 426 nm as well as the appearance of α and β peaks in the 500–600-nm region (Fig. 1C, reduced spectrum). Analysis under same reducing conditions but with no oxygen present (deoxy-PpsR-heme spectrum) shows that the 426-nm Soret peak and the α and β peaks in the 500–600-nm region, all disappear (supplemental Fig. 2C). Adding carbon monoxide to reduced PpsR-heme species also resulted in a characteristic carbon monoxy heme spectrum with a Soret peak at 418 nm (supplemental Fig. 2D), typical for ferrous-heme-CO complex. Interestingly, myoglobin under low pH conditions demonstrates a UV-visible spectrum almost identical to that observed with PpsR-heme (25). A well studied heme-binding protein, myoglobin, under low pH conditions is pentacoordinated under oxidizing conditions and hexacoordinated when reduced (25). The similarity of the UV-visible spectrum suggests that PpsR-heme binding could utilize the same mechanism.
The stoichiometry of heme binding to PpsR was addressed by reconstituting PpsR with heme in vitro and then removing the PpsR-heme complex from free heme either by size exclusion chromatography or by ion exchange chromatography. With these approaches, we could obtain PpsR-heme complex with a molar ratio of 0.85 ± 0.08 PpsR to 1 heme. To confirm spectrally the chromatography results we titrated heme into PpsR and measured the absorbance change that occurs at 370 nm upon binding. The plot of the absorbance change versus the PpsR:heme concentration ratio indicates that a PpsR-heme complex is formed with a stoichiometry of 0.84 ± 0.05 [PpsR]:[heme] (Fig. 1D). Tryptophan quenching data of heme titrated into PpsR can also be fitted with a one-to-one binding model with a Kd for heme binding of 1.9 μm (Fig. 1E). This micromolar binding affinity is similar to a dissociation constant that has been reported for heme binding by R-transferase (9).
His275 and Cys424 Are Critical for PpsR-Heme Interaction
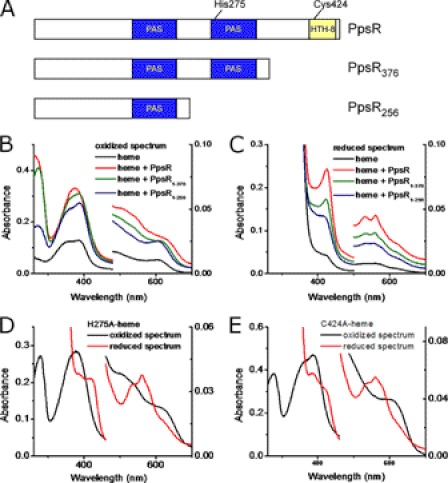
Prior inspection of the PpsR sequence indicates the presence of two PAS domains followed by a carboxyl-terminal helix-turn-helix (HTH) DNA binding domain (Fig. 2A) (26). It is well known that several PAS domains use heme as a co-factor to sense environmental signals (11, 13), so several deletion constructs were made to test their involvement. Unlike full-length PpsR, spectral analysis of truncated constructs PpsR256 and PpsR376 did not show the formation of a single Soret peak upon addition of heme under oxidizing conditions (Fig. 2B). This type of spectrum change, the loss of the single 370-nm Soret peak and the appearance of two overlapping peaks, has been observed for regulatory proteins that have a HRM and assigned to the lost Cys-heme coordination (17). Under reducing conditions the larger construct that contained both PAS domains but no HTH domain (PpsR376 in Fig. 2A) showed spectroscopic changes similar to those shown by wild-type PpsR with the exception that the intensity was not as increased (Fig. 2C). Thus, both the carboxyl terminus HTH domain and the second PAS domain appear to be involved in the interaction with heme.
FIGURE 2.
Characterization of the heme-binding domain in PpsR. A, domains structure of PpsR, PpsR1–376, and PpsR1–256. B, UV-visible spectrum of heme and different PpsR constructs under oxidizing conditions. 4.6 μm heme was incubated with 9.4 μm different constructs for at least 20 min before the spectrum were taken. C, UV-visible spectrum of heme and different PpsR constructs under reducing conditions. The same samples used for oxidized spectrum were reduced with 5 mm dithionite for the reduced spectrum. D, UV-visible spectrum of PpsRH275A-heme. 4.1 μm heme was incubated with 8.2 μm PpsRH275A for at least 20 min before the spectrum were taken. E, UV-visible spectrum of PpsRC424A-heme. 4.1 μm heme was incubated with 8.2 m μm PpsRC424A for at least 20 min before the spectrum was taken.
The spectrum of the PpsR-heme complex is similar to that of several heme sensors that characteristically have a single Soret peak at ∼370 nm under oxidizing conditions that also red shifts to ∼420 nm under reducing conditions (2, 9). For example, the heme-sensing protein Irr has a Soret peak at 377 nm under oxidizing conditions that red shifts to 413 nm under reduced conditions (2, 27). The appearance of single Soret peak has been attributed to the coordination between axial residue and the iron in heme (17). In the case of Irr, His and Cys are utilized as axial ligands to iron of heme which are two of the most common axial residues found in heme-binding proteins (2, 9, 17, 28, 29). Inspection of the second PAS domain and carboxyl-terminus region indicates that there is only one His present at position 275 (His275), and only one Cys (Cys424) located within the HTH DNA binding domain. Consequently, we used site-directed mutagenesis to convert His275 and Cys424 to Ala (H275A and C424A, respectively) and then assayed what effect these mutations had on heme interaction. When PpsRH275A was assayed under oxidizing conditions the spectrum was similar to that observed with oxidized wild-type PpsR (Fig. 2, D and B, respectively). This was contrasted by spectrally assaying heme binding by PpsRH275A under reducing conditions where ferrous heme did not show a sharp and distinguish Soret peak at 426 nm and also exhibited a reduced β peak at 530 nm (Fig. 2D). Analysis of heme binding with PpsRC424A under reducing conditions shows a similar spectrum as reduced PpsRH275A (Fig. 2E). However, under oxidizing conditions, the single Soret peak of heme observed with wild-type PpsR is converted to a double peak with PpsRC424A (Fig. 2E), very similar to the spectrum observed with PpsR376 (Fig. 2B). In addition, the reconstitution of PpsRC424A with heme only achieved a molar ratio of ∼0.38 ([heme]:[PpsRC424A]), which is considerably less than that observed with wild-type PpsR, further confirming that Cys424 is critical for PpsR-heme interaction. These observations, combined with the results presented earlier, can be explained with a model in which Cys424 forms an axial ligand under oxidizing conditions where heme is pentacoordinated, whereas both Cys424 and His275 form axial ligands under reducing conditions where heme is hexacoordinated.
Heme Affects DNA Binding Activity of PpsR
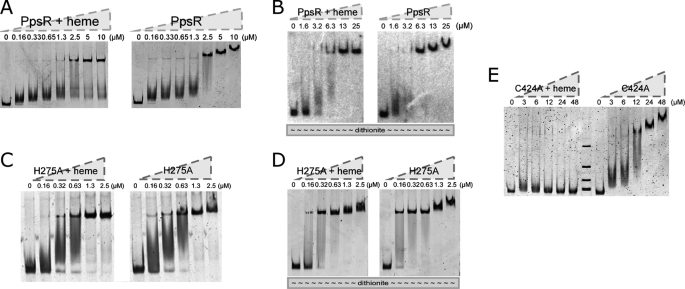
Previous mutational and biochemical analyses demonstrated that the redox state of Cys424 affects the DNA binding activity of PpsR (18, 30). One could envision, therefore, that binding of heme to Cys424 may also affect the DNA binding activity of PpsR. To explore this hypothesis, we performed gel mobility shift assays to the puc promoter that contains two PpsR binding sites located 8 bp apart with and without heme bound to PpsR. Incubation of PpsR with a 262-bp puc promoter probe resulted in a distinct shift as reported previously (18, 20). The PpsR-DNA complex exhibited progressively slower electrophoretic mobility (supershift) as the concentration of PpsR increased (Fig. 3A, right panel). Similar analysis with PpsR-heme shows the presence of the PpsR-DNA complexes. However, unlike PpsR alone, PpsR-heme does not form a supershift as protein concentration increased (Fig. 3A, left panel). Analysis of the mobility of permuted DNA segments indicates no significant alteration in PpsR-mediated DNA bending with or without heme present. Thus, heme appears to inhibit the ability of PpsR to form a higher ordered PpsR-DNA complex (supplemental Fig. 3).
FIGURE 3.
Heme regulates PpsR-DNA binding. PpsR or its mutant was incubated with 2-fold of heme for 20 min before adding DNA when needed. 10 nm probe was used to bind PpsR unless noted otherwise. A, effect of heme on wild-type PpsR-puc interaction under oxidizing conditions. B, effect of heme on wild-type PpsR-puc interaction under reducing conditions. C, effect of heme on the PpsRH275A-puc interaction under oxidizing conditions. D, effect of heme on the PpsRH275A-puc interaction under reducing conditions. E, effect of heme on the PpsRC424A-puc interaction under oxidizing conditions.
We also addressed the effect of increasing heme concentration to PpsR-puc complex. A ratio of [PpsR]:[heme] from 1:1 up to ∼1:2 results in the disappearance of PpsR-DNA supershift, whereas a higher heme level completely releases PpsR from the puc DNA probe (supplemental Fig. 4A). Similar analysis with a hemE promoter probe, which contains two PpsR binding sites separated by 252 bp, showed a similar pattern except that the binding of PpsR was more sensitive to disruption by addition of heme than it was with the puc probe (supplemental Fig. 4B). Additional control assays with hemE also showed that neither protoporphyrin IX, zinc porphyrin, nor ferric ion affects PpsR-DNA complex mobility (supplemental Fig. 4C).
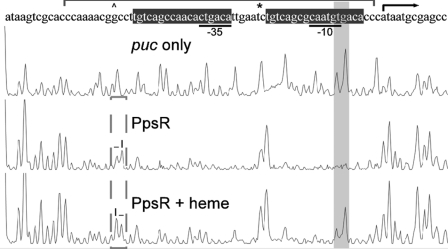
Changes in the mobility shift patterns of the PpsR-DNA complex observed with and without heme suggest that PpsR may interact differently with target promoter upon binding heme. To test this possibility, we performed DNase I footprint analysis of PpsR binding to puc promoter (Fig. 4). The protection pattern exhibited by PpsR alone showed complete protection of the two PpsR binding sites (TGT-N11-ACA) that overlaps the −35 and −10 promoter recognition sequences, as well as two nucleic acids remaining DNase I-accessible (marked with * in Fig. 4) between the two binding sites. In the presence of heme, two previously protected nucleic acids were now DNase I-accessible (shaded peaks, Fig. 4). Another small but consistent change was also observed where the DNase I sensitivity of two adjacent nucleic acids is switched from “low-high” to “high-low” (marked with a dashed box in Fig. 4). In summary, the footprint assay confirms the different conformations of the PpsR-puc complexes with and without bound heme.
FIGURE 4.
DNase I footprinting of PpsR and puc promoter. 10 μm PpsR was incubated with puc probe before being applied to DNase I digestion. PpsR binding sites are represented with brackets. Changes in binding are indicated with shaded and dashed boxes.
His275 and Cys424 Are Critical for Heme Regulation of PpsR-DNA Interaction
Because His275 and Cys424 appear to be critical for the interaction with heme, we examined the ability of isolated PpsRH275A and PpsRC424A to bind to a puc promoter segment containing a PpsR binding site. Under oxidizing conditions without heme, wild-type PpsR and PpsRH275A both form a similar supershifted complex, as protein is increased (Fig. 3, A and C, respectively). Similar results occur under reducing conditions without heme where a supershifted complex is observed with both wild-type PpsR and PpsRH275A as protein concentration is increased (Fig. 3, B and D, respectively). However, very different results are observed in the presence of heme. Under oxidizing conditions with heme we observed that wild-type PpsR and PpsRH275A are both incapable of forming the supershifted complex as protein is increased (Fig. 3A and 3C, respectively). This is contrasted by reducing conditions with heme where wild-type PpsR is incapable of forming the supershifted complex (Fig. 3B, left panel), whereas the PpsRH275A mutant is still capable of forming the supershift complex (Fig. 3D, left panel).
Under oxidizing conditions, larger amounts of PpsRC424A were required to bind to the puc promoter compared with wild-type PpsR, which is in agreement with that of previous studies (18). Specifically, whereas 10 nm puc probe was completely shifted by the addition of 2.5 μm wild-type PpsR (Fig. 3A), more than 20 μm PpsRC424A was needed to bind to the same amount of puc probe (Fig. 3E). Furthermore, as observed with wild-type PpsR, a supershift with the puc probe was formed as the concentration of PpsRC424A was increased (Fig. 3E). When the same experiments were performed in the presence of heme, PpsRC424A was directly released from the puc promoter probe instead of exhibiting the relaxation of supershifted PpsR-puc complex as observed with wild-type PpsR (Fig. 3E). Similar DNA binding studies with PpsRC424A under reducing conditions were not feasible as this mutant is not stable under such conditions in vitro. In summary, these observations are consistent with the model we proposed earlier, where Cys424 is critical for PpsR-heme interaction under oxidizing conditions, and both Cys424 and His275 are critical for PpsR-heme interaction under reducing conditions.
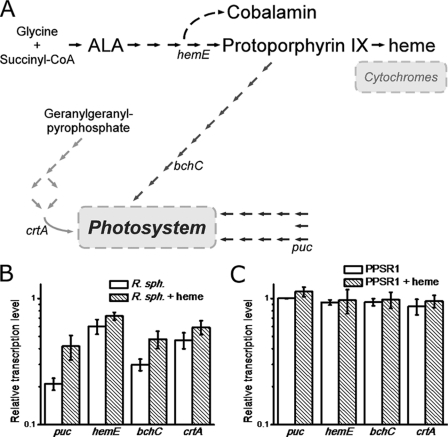
Heme Induces in Vivo Increase of Transcription of Subset of PpsR-controlled Genes
To explore how PpsR responds to the presence of heme in vivo, we used qPCR to measure the level of several genes known to be regulated by PpsR. Specifically, we tested expression of light harvesting represented by puc, heme/bacteriochlorophyll common trunk represented by hemE, bacteriochlorophyll branch represented by bchC, and carotenoid biosynthesis represented by crtA (Fig. 5A and supplemental Fig. 5). Interestingly, addition of heme to growing cultures of R. sphaeroides at 25 μm induced different responses on the tested promoters (supplemental Fig. 6). After 20-min incubation with heme, the level of puc and bchC expression increased whereas the level of hemE and crtA did not show significant changes (Fig. 5B). A control experiment with the PpsR knock-out strain PPSR1 showed essentially the same level of all four genes tested, with and without heme (Fig. 5C), indicating that the change in the original strain observed by addition of heme was mediated through PpsR.
FIGURE 5.
Heme effect on the transcription levels of PpsR-regulated genes. A, PpsR-regulated photosynthesis and tetrapyrrole biosynthesis genes. The detailed diagram is described in supplemental Fig. 5. B, changes in relative transcription level of various PpsR-regulated genes in wild-type R. sphaeroides culture after addition of heme to the culture. After 20 min of starvation, 25 μm heme was added into the culture, and the samples were taken after another 20 min of incubation and normalized (supplemental Fig. 6). Data shown are mean ± S.E. (error bars). C, similar to B with the exception that mRNA levels were analyzed in the ppsR deletion strain, PPSR1.
DISCUSSION
This study demonstrates that PpsR binds heme with an oxidized spectrum that is similar to several heme-sensing proteins such as Irr (2), HAP1 (17), and R-transferase (9). Indeed, the binding constant of PpsR-heme is close to that of the regulator R-transferase (9), but significantly weaker than hemoproteins that tightly bind heme to perform functions such as electron transport or transporting/sensing gas molecules (29). This is consistent with the fact that only a small fraction of bound heme is present when PpsR is harvested from R. sphaeroides (Fig. 1A). The oxidized PpsR-heme spectrum is almost identical to that of oxidized myoglobin (met-myoglobin) at low pH conditions, which is in a pentacoordinated state (25). Under reducing conditions, the PpsR-heme spectrum shares several characteristics with reduced myoglobin with CO (myoglobin-CO) at low pH conditions including the α and β peaks which is typical for hexacoordinated heme (25). Thus, we propose that PpsR-heme has coordination states similar to those of myoglobin under low pH conditions. Myoglobin is in a U state under such conditions, representing a stable intermediate on the myoglobin unfolding pathway (25, 31). This mechanism and the relatively low binding constant would suggest a flexible heme binding pocket, consistent with the proposed PpsR role as a heme-sensing protein. A flexible heme binding pocket has also been observed in IsdC (32) which is part of the heme uptake system in Staphylococcus aureus.
His275 and Cys424 are clearly important for PpsR to interact with heme. Our mutational analysis of His275 shows a spectral alteration only under reducing conditions. This indicates that His275 likely constitutes an axial ligand only to Fe2+ and not when the iron is oxidized to Fe3+. This result is contrasted by an Ala substitution of Cys424 that leads to a loss of the Soret peak under both oxidizing and reducing conditions. Because this type of spectrum change has been observed for regulatory proteins that have a HRM (17) and assigned to a Cys-heme coordination, we deduce that Cys424 likely forms an axial ligand under both oxidizing and reducing conditions. This model is also supported by our gel mobility shift assays with the PpsR mutants. Further studies would be needed to examine this model on the structural level in the future.
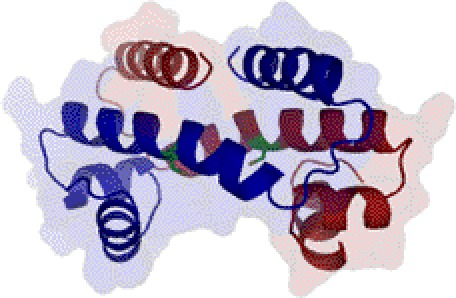
Heme clearly affects the DNA binding activity of PpsR as demonstrated by the ability of heme to inhibit the formation of a higher ordered PpsR-puc supercomplex. At even higher concentrations of heme, PpsR is completely released from the puc promoter. Interestingly, Cys424 is located within a well conserved HTH DNA binding domain (33). As show in previous study, oxidation of Cys424 in the absence of heme stimulates the DNA binding activity of PpsR (18). The same effect of redox regulation was observed while heme was present (Fig. 3, A and B). Thus, it appears that Cys424 could be a versatile target for different types of modification, such as forming/breaking disulfide bridge (18), coordinating to heme as presented in this study, and potentially being oxidized into different oxidation states much like the case of OxyR (34). A three-dimensional model of the PpsR HTH domain based on homology to the Fis HTH domain structure (33) indicates that heme could potentially be accommodated by a cleft near Cys424 (Fig. 6). If this is indeed the location of heme binding then inclusion of heme could affect the pitch of the protein helix resulting in the altered footprint pattern that was observed. Other types of modification of Cys424 could also change the PpsR-DNA interaction correspondingly.
FIGURE 6.
Model of PpsR-heme interaction. Location of Cys424 in the HTH domain is based on structure modeled after the HTH domain of Fis. The two chains of PpsR DNA binding domain are in red and blue with Cys424 shown in green. According to a theoretical model of Fis, bent DNA would interact with PpsR at the bottom of this molecule (33).
One common feature of heme-sensing proteins is that they use Cys as a heme axial ligand (29). One notable difference with PpsR is that Cys424 is followed by an Ile whereas other heme sensors have a -Cys-Pro- as the HRM. The discovery of PpsR as a heme sensor with Cys-Ile instead of the Cys-Pro suggests that heme-sensing-based regulation may be more widespread. Nonetheless, PpsR utilizes cysteine as the critical axial residue as is the case for other HRM-containing heme sensors. Thus, it is feasible that Cys coordinated to Fe2+/3+ in heme is a conserved strategy for heme sensing.
Heme has a more obvious affect on puc expression than on hemE expression via PpsR (Fig. 5). However, in vitro gel shift assays show that heme affects PpsR binding to both puc and hemE promoters (supplemental Fig. 4). This suggests that different promoters respond to the presence of heme with different sensitivities. As shown in supplemental Fig. 4C, a PpsR:heme ratio of 1:1 caused complete release of PpsR from hemE promoter, whereas a ratio of 1:2 PpsR:heme can still produces a stable PpsR-puc complex (Fig. 3A). Meanwhile, puc and bchC genes which exhibit greater effects upon the addition of heme in vivo (Fig. 5) also have two PpsR recognition sites close to each other (7 and 8 bp apart, respectively) (supplemental Fig. 5). This is contrasted by crtA and hemE genes that have two PpsR recognition sites relatively far from each other (55 and 252 bp apart, respectively) that have no obvious effect of heme on expression in vivo (supplemental Fig. 5). This difference in the type of PpsR binding sites may be a reason for the observed differing heme sensitivity at different PpsR-regulated genes.
Why would PpsR have evolved as a heme-sensing transcription factor? In purple bacteria, all three branches of the tetrapyrrole biosynthetic pathway are essential for photosynthesis with bacteriochlorophyll needed for light absorption and electron donation, heme needed for electron transfer, and cobalamin (vitamin B12) needed for synthesis of bacteriochlorophyll (35). The heme and bacteriochlorophyll branches share common intermediates from δ-aminolevulinic acid to protoporphyrin IX with both branches regulated by PpsR (19, 20, 36) (Fig. 5A and supplemental Fig. 5). In the case of the puc and bchC promoters, our in vivo results suggest that an excess of free heme can induce an increased synthesis of light harvesting II peptides and bacteriochlorophyll to which it binds. Interestingly, there is no significant increase in the level of hemE expression when heme is in excess. Thus, the interaction of PpsR with heme could potentially divert tetrapyrrole biosynthesis away from the production of excess toxic heme toward bacteriochlorophyll. This could be an advantage for photosynthetic bacteria that must coordinate the synthesis of heme with that of bacteriochlorophyll as well as with various tetrapyrrole-binding apoproteins. A similar strategy has been found in animal cells, which use heme as feedback signals between the circadian clock network and clock-controlled metabolic pathways to maintain a balance homeostasis (37). Finally, it has been shown that AppA, an antirepressor of PpsR, is also a heme-binding protein. Evidence suggests that heme bound to AppA can increase its affinity to PpsR (21), further underscoring the important role that heme has in directly and indirectly regulating the DNA binding activity of PpsR.
Supplementary Material
Acknowledgments
We thank Dr. Shinji Masuda for providing PPSR1 and PpsR-FLAG strain and Dr. Sébastien Zappa at Indiana University for valuable advice on gel shift, DNase I footprinting, and qPCR methodologies. We also thank Prof. Kara Bren at University of Rochester for the discussion of interpreting the UV-visible spectrum.
This work was supported, in whole or in part, by National Institutes of Health Grant R37 GM040941 (to C. E. B.).

This article contains supplemental Figs. 1–6 and Tables 1 and 2.
- HMR
- heme regulatory motif
- HTH
- helix-turn-helix
- Irr
- iron response regulator
- qPCR
- quantitative PCR
- PAS
- Per-Arnt-Sim.
REFERENCES
- 1. Marks G. S. (1969) Heme and Chlorophyll, pp. 6–25, D. Van Nostrand Company, Princeton, NJ [Google Scholar]
- 2. Qi Z., Hamza I., O'Brian M. R. (1999) Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. U.S.A. 96, 13056–13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boon E. M., Huang S. H., Marletta M. A. (2005) A molecular basis for NO selectivity in soluble guanylate cyclase. Nat. Chem. Biol. 1, 53–59 [DOI] [PubMed] [Google Scholar]
- 4. Atamna H., Killilea D. W., Killilea A. N., Ames B. N. (2002) Heme deficiency may be a factor in the mitochondrial and neuronal decay of aging. Proc. Natl. Acad. Sci. U.S.A. 99, 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atamna H., Frey W. H., 2nd (2004) A role for heme in Alzheimer's disease: heme binds amyloid β and has altered metabolism. Proc. Natl. Acad. Sci. U.S.A. 101, 11153–11158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pfeifer K., Kim K. S., Kogan S., Guarente L. (1989) Functional dissection and sequence of yeast HAP1 activator. Cell 56, 291–301 [DOI] [PubMed] [Google Scholar]
- 7. Méndez R., Moreno A., de Haro C. (1992) Regulation of heme-controlled eukaryotic polypeptide chain initiation factor 2 α-subunit kinase of reticulocyte lysates. J. Biol. Chem. 267, 11500–11507 [PubMed] [Google Scholar]
- 8. Lathrop J. T., Timko M. P. (1993) Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science 259, 522–525 [DOI] [PubMed] [Google Scholar]
- 9. Hu R. G., Wang H., Xia Z., Varshavsky A. (2008) The N-end rule pathway is a sensor of heme. Proc. Natl. Acad. Sci. U.S.A. 105, 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang J., Kim K. D., Lucas A., Drahos K. E., Santos C. S., Mury S. P., Capelluto D. G., Finkielstein C. V. (2008) A novel heme-regulatory motif mediates heme-dependent degradation of the circadian factor period 2. Mol. Cell. Biol. 28, 4697–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gilles-Gonzalez M. A., Gonzalez G., Perutz M. F., Kiger L., Marden M. C., Poyart C. (1994) Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autooxidation. Biochemistry 33, 8067–8073 [DOI] [PubMed] [Google Scholar]
- 12. Shelver D., Kerby R. L., He Y., Roberts G. P. (1997) CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc. Natl. Acad. Sci. U.S.A. 94, 11216–11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delgado-Nixon V. M., Gonzalez G., Gilles-Gonzalez M. A. (2000) Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39, 2685–2691 [DOI] [PubMed] [Google Scholar]
- 14. Huang L. E., Willmore W. G., Gu J., Goldberg M. A., Bunn H. F. (1999) Inhibition of hypoxia-inducible factor 1 activation by carbon monoxide and nitric oxide: implications for oxygen sensing and signaling. J. Biol. Chem. 274, 9038–9044 [DOI] [PubMed] [Google Scholar]
- 15. Gilles-Gonzalez M. A., Ditta G. S., Helinski D. R. (1991) A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature 350, 170–172 [DOI] [PubMed] [Google Scholar]
- 16. Ogawa K., Sun J., Taketani S., Nakajima O., Nishitani C., Sassa S., Hayashi N., Yamamoto M., Shibahara S., Fujita H., Igarashi K. (2001) Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20, 2835–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L., Guarente L. (1995) Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 14, 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Masuda S., Bauer C. E. (2002) AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110, 613–623 [DOI] [PubMed] [Google Scholar]
- 19. Elsen S., Ponnampalam S. N., Bauer C. E. (1998) CrtJ bound to distant binding sites interacts cooperatively to aerobically repress photopigment biosynthesis and light harvesting II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 273, 30762–30769 [DOI] [PubMed] [Google Scholar]
- 20. Ponnampalam S. N., Bauer C. E. (1997) DNA binding characteristics of CrtJ: a redox-responding repressor of bacteriochlorophyll, carotenoid, and light harvesting-II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 272, 18391–18396 [DOI] [PubMed] [Google Scholar]
- 21. Han Y., Meyer M. H., Keusgen M., Klug G. (2007) A haem cofactor is required for redox and light signalling by the AppA protein of Rhodobacter sphaeroides. Mol. Microbiol. 64, 1090–1104 [DOI] [PubMed] [Google Scholar]
- 22. Moskvin O. V., Kaplan S., Gilles-Gonzalez M. A., Gomelsky M. (2007) Novel heme-based oxygen sensor with a revealing evolutionary history. J. Biol. Chem. 282, 28740–28748 [DOI] [PubMed] [Google Scholar]
- 23. Dawson R. M., Elliott D. C., Elliot W. H., Jones K. M. (1969) Data for Biochemical Research, 2nd Ed., p. 316, Oxford University Press, Oxford [Google Scholar]
- 24. Berry E. A., Trumpower B. L. (1987) Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 161, 1–15 [DOI] [PubMed] [Google Scholar]
- 25. Sage J. T., Morikis D., Champion P. M. (1991) Spectroscopic studies of myoglobin at low pH: heme structure and ligation. Biochemistry 30, 1227–1237 [DOI] [PubMed] [Google Scholar]
- 26. Nickens D. G., Bauer C. E. (1998) Analysis of the puc operon promoter from Rhodobacter capsulatus. J. Bacteriol. 180, 4270–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang J., Ishimori K., O'Brian M. R. (2005) Two heme binding sites are involved in the regulated degradation of the bacterial iron response regulator (Irr) protein. J. Biol. Chem. 280, 7671–7676 [DOI] [PubMed] [Google Scholar]
- 28. Reedy C. J., Elvekrog M. M., Gibney B. R. (2008) Development of a heme protein structure-electrochemical function database. Nucleic Acids Res. 36, D307–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Igarashi J., Kitanishi K., Martinkova M., Murase M., Iizuka A., Shimizu T. (2008) The roles of thiolate-heme proteins, other than the P450 cytochromes, in the regulation of heme-sensor proteins. Acta Chim. Slov. 55, 67–74 [Google Scholar]
- 30. Masuda S., Dong C., Swem D., Setterdahl A. T., Knaff D. B., Bauer C. E. (2002) Repression of photosynthesis gene expression by formation of a disulfide bond in CrtJ. Proc. Natl. Acad. Sci. U.S.A. 99, 7078–7083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Irace G., Bismuto E., Savy F., Colonna G. (1986) Unfolding pathway of myoglobin: molecular properties of intermediate forms. Arch. Biochem. Biophys. 244, 459–469 [DOI] [PubMed] [Google Scholar]
- 32. Villareal V. A., Pilpa R. M., Robson S. A., Fadeev E. A., Clubb R. T. (2008) The IsdC protein from Staphylococcus aureus uses a flexible binding pocket to capture heme. J. Biol. Chem. 283, 31591–31600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yuan H. S., Finkel S. E., Feng J. A., Kaczor-Grzeskowiak M., Johnson R. C., Dickerson R. E. (1991) The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding. Proc. Natl. Acad. Sci. U.S.A. 88, 9558–9562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim S. O., Merchant K., Nudelman R., Beyer W. F., Jr., Keng T., DeAngelo J., Hausladen A., Stamler J. S. (2002) OxyR: a molecular code for redox-related signaling. Cell 109, 383–396 [DOI] [PubMed] [Google Scholar]
- 35. Gough S. P., Petersen B. O., Duus J. O. (2000) Anaerobic chlorophyll isocyclic ring formation in Rhodobacter capsulatus requires a cobalamin cofactor. Proc. Natl. Acad. Sci. U.S.A. 97, 6908–6913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moskvin O. V., Gomelsky L., Gomelsky M. (2005) Transcriptome analysis of the Rhodobacter sphaeroides PpsR regulon: PpsR as a master regulator of photosystem development. J. Bacteriol. 187, 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. (2007) Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.