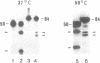Abstract
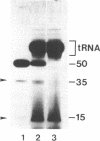
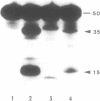
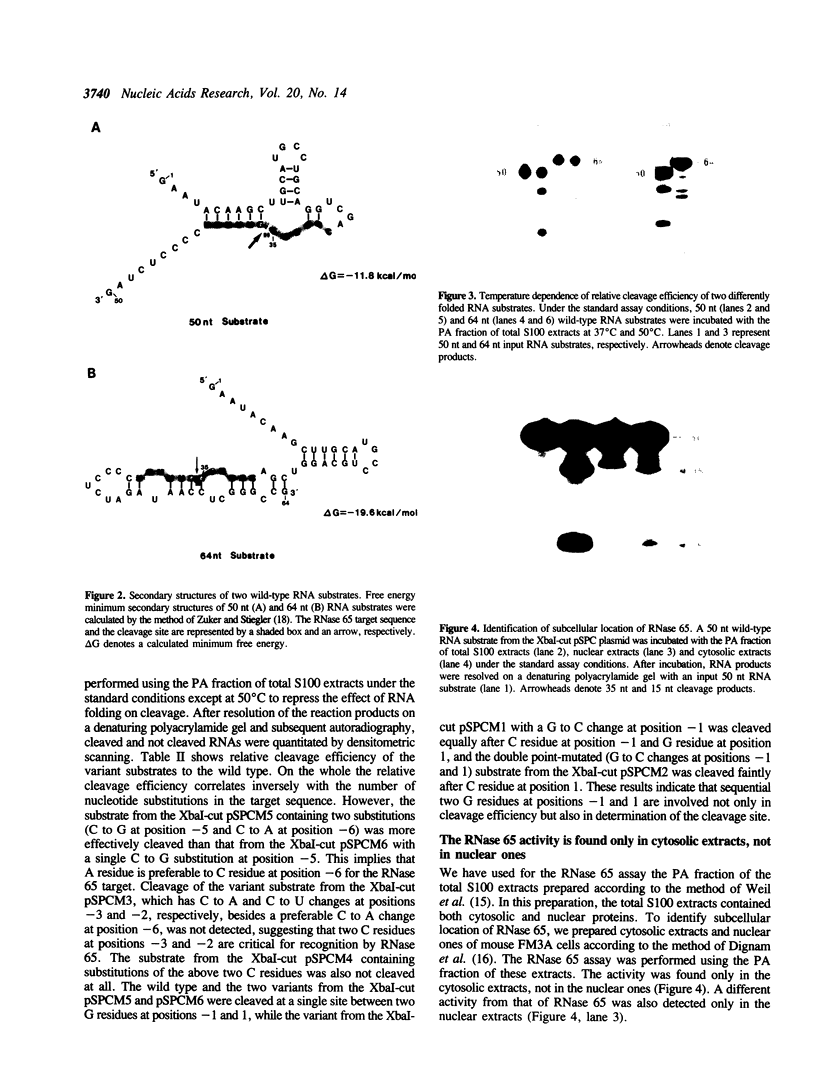
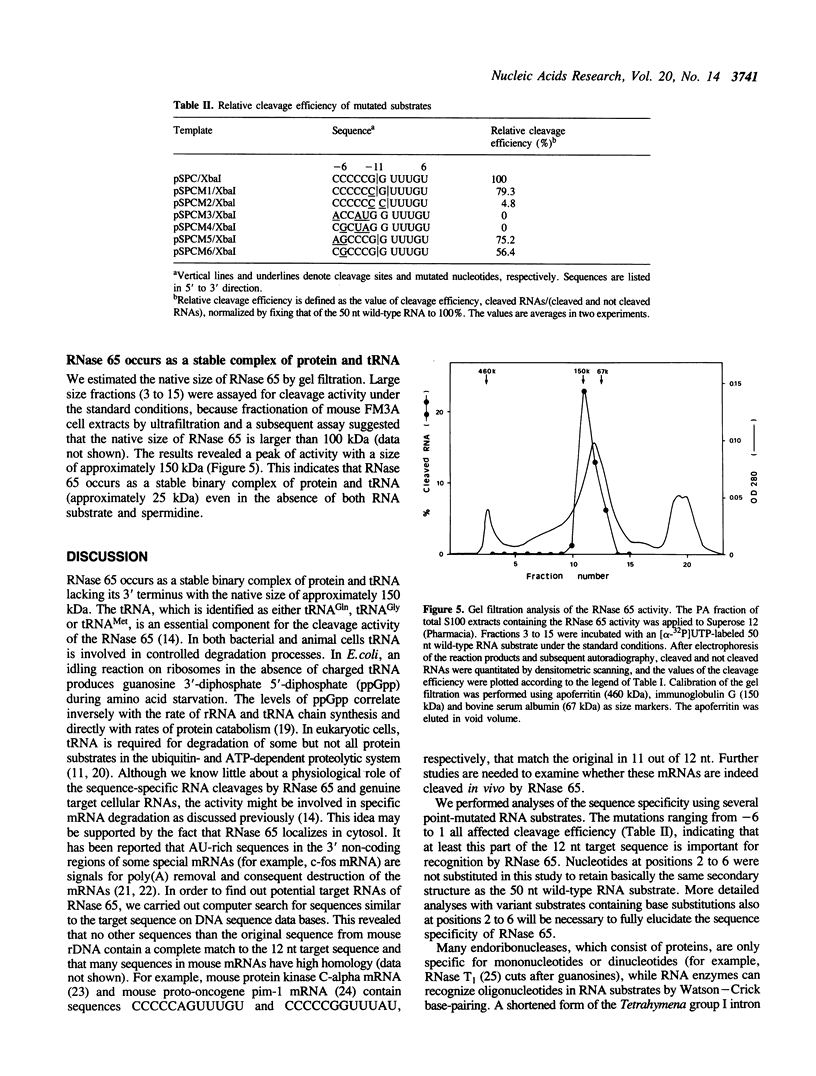
The spermidine-dependent, sequence-specific endoribonuclease (RNase 65) in mouse FM3A cells consists of protein and transfer RNA lacking its 3' terminus. In vitro properties of this enzyme were characterized using partially purified enzyme. The RNase 65 activity requires spermidine, which is not replaceable with spermine or Mg++. The enzyme cleaves an RNA substrate on the 3' side of the phosphodiester bond. The cleavage reaction has a temperature optimum around 50 degrees C and a pH optimum around 7.0. The optimum KCl concentration for the activity is around 10 mM. Relative cleavage efficiency of two differently folded RNA substrates with the common target sequence was analyzed at 37 degrees C and 50 degrees C. The results of this analysis suggest that unfolding of the target sequence is critical for recognition by RNase 65. Furthermore, in experiments using several point-mutated RNA substrates designed to form basically the same secondary structure as the wild type, one to three nucleotide substitutions in the target sequence all reduced cleavage efficiency. The RNase 65 activity is found only in cytosolic extracts, not in nuclear ones. Gel filtration analysis suggests that the native size of the endoribonuclease is approximately 150 kDa.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cashel M. Regulation of bacterial ppGpp and pppGpp. Annu Rev Microbiol. 1975;29:301–318. doi: 10.1146/annurev.mi.29.100175.001505. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987 Mar 6;235(4793):1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Wolin S. L., Steitz J. A., Lodish H. F. Transfer RNA is an essential component of the ubiquitin- and ATP-dependent proteolytic system. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1341–1345. doi: 10.1073/pnas.82.5.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egami F., Oshima T., Uchida T. Specific interaction of base-specific nucleases with nucleosides and nucleotides. Mol Biol Biochem Biophys. 1980;32:250–277. doi: 10.1007/978-3-642-81503-4_21. [DOI] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Ferber S., Ciechanover A. Transfer RNA is required for conjugation of ubiquitin to selective substrates of the ubiquitin- and ATP-dependent proteolytic system. J Biol Chem. 1986 Mar 5;261(7):3128–3134. [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Harada F., Sawyer R. C., Dahlberg J. E. A primer ribonucleic acid for initiation of in vitro Rous sarcarcoma virus deoxyribonucleic acid synthesis. J Biol Chem. 1975 May 10;250(9):3487–3497. [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Kass S., Tyc K., Steitz J. A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990 Mar 23;60(6):897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Nashimoto M., Kominami R., Nishi S., Mishima Y. A novel spermidine-dependent endoribonuclease activity caused by RNA-protein complex in mouse FM3A cell extracts. Biochem Biophys Res Commun. 1991 May 15;176(3):1163–1169. doi: 10.1016/0006-291x(91)90407-x. [DOI] [PubMed] [Google Scholar]
- Nashimoto M., Ogata K., Mishima Y. In vitro sequence-specific cleavage in transcribed spacer of mouse precursor ribosomal RNA. J Biochem. 1988 Jun;103(6):992–997. doi: 10.1093/oxfordjournals.jbchem.a122399. [DOI] [PubMed] [Google Scholar]
- Nashimoto M., Sakai M., Nishi S. Transfer RNA lacking its 3' terminus is required for spermidine-dependent ribonuclease 65 activity in mouse FM3A cell extracts. Biochem Biophys Res Commun. 1991 Aug 15;178(3):1247–1252. doi: 10.1016/0006-291x(91)91027-a. [DOI] [PubMed] [Google Scholar]
- Rose-John S., Dietrich A., Marks F. Molecular cloning of mouse protein kinase C (PKC) cDNA from Swiss 3T3 fibroblasts. Gene. 1988 Dec 30;74(2):465–471. doi: 10.1016/0378-1119(88)90179-5. [DOI] [PubMed] [Google Scholar]
- Schön A., Krupp G., Gough S., Berry-Lowe S., Kannangara C. G., Söll D. The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature. 1986 Jul 17;322(6076):281–284. doi: 10.1038/322281a0. [DOI] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Boelens W., Robanus-Maandag E., Verbeek J., Domen J., van Beveren C., Berns A. The primary structure of the putative oncogene pim-1 shows extensive homology with protein kinases. Cell. 1986 Aug 15;46(4):603–611. doi: 10.1016/0092-8674(86)90886-x. [DOI] [PubMed] [Google Scholar]
- Waters L. C., Mullin B. C., Ho T., Yang W. K. Ability of tryptophan tRNA to hybridize with 35S RNA of avian myeloblastosis virus and to prime reverse transcription in vitro. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2155–2159. doi: 10.1073/pnas.72.6.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]
- Wilson T., Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3' AU-rich sequences. Nature. 1988 Nov 24;336(6197):396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]