Background: Cardiomyopathy-causing mutations in the N-terminal domain of cTnT disrupt thin filament function.
Results: Amino acid substitutions at cTnT residue 92 alter motility, whereas computational analysis reveals significant changes in structural forces.
Conclusion: The observed changes in thin filament function can be correlated to changes in bending forces.
Significance: Coupling computation to in vitro measurements extends our understanding of disease mechanisms.
Keywords: Biophysics, Cardiac Hypertrophy, Cardiac Muscle, Contractile Protein, Molecular Dynamics, Cardiac Troponin T, Thin Filament
Abstract
Nearly 70% of all of the known cTnT mutations that cause familial hypertrophic cardiomyopathy fall within the TNT1 region that is critical to cTn-Tm binding. The high resolution structure of this domain has not been determined, and this lack of information has hindered structure-function analysis. In the current study, a coupled computational experimental approach was employed to correlate changes in cTnT dynamics to basic function using the regulated in vitro motility assay (R-IVM). An in silico approach to calculate forces in terms of a bending coordinate was used to precisely identify decreases in bending forces at residues 105 and 106 within the proposed cTnT “hinge” region. Significant functional changes were observed in multiple functional properties, including a decrease in the cooperativity of calcium activation, the calcium sensitivity of sliding speed, and maximum sliding speed. Correlation of the computational and experimental findings revealed an association between TNT1 flexibility and the cooperativity of thin filament calcium activation where an increase in flexibility led to a decrease in cooperativity. Further analysis of the primary sequence of the TNT1 region revealed a unique pattern of conserved charged TNT1 residues altered by the R92W and R92L mutations and may represent the underlying “structure” modulating this central functional domain. These data provide a framework for further integrated in silico/in vitro approaches that may be extended into a high-throughput predictive screen to overcome the current structural limitations in linking molecular phenotype to genotype in thin filament cardiomyopathies.
Introduction
Cardiac troponin (cTn)3 is a key modulator of cardiac contraction via its regulation of calcium activation of the thin filament. The thin filament consists primarily of actin, Tm, and cTn. Filamentous F-actin provides the backbone for the thin filament. Tm exists as a coiled-coil dimer spiraling along actin. As strands of Tm spiral around actin, each head-to-tail overlap of adjacent N and C termini of Tm binds to cTn via cTnT. cTn consists of three subunits, cTnC (the calcium-binding subunit), cTnI (inhibitory unit), and cTnT (Tm-binding subunit), with each interacting with the other. TNT1 is an N-terminal tail domain of cTnT that interacts directly with overlapping Tm as shown in Fig. 1. The juxtaposition of TNT1 and overlapping Tm supports the role of troponin as a key modulator of myofilament function and assembly (1, 2).
FIGURE 1.
Orientation to TNT1 with respect to the thin filament. Yellow, cTnT with TNT1 thickened; blue, cTnI; red, cTnC; cyan, calcium; light/dark gray, actin; green, two overlapping Tm associated with top cTn; orange, two overlapping Tm associated with bottom cTn.
We have shown previously that changes in protein flexibility caused by single amino acid substitutions at the same residue of cTnT (Arg-92), implicated in the pathogenesis of FHC, correlate with both in vitro changes in myocellular properties and in vivo physiology in transgenic mice (3). Our work has extended biochemical and biophysical studies that have shown that this tail domain is flexible and thus critical in modulating the protein-protein interactions between the thin filament and myosin during contraction (4, 5). The domain of the troponin complex that we studied has not been crystallized and diffracted, and we determined that a likely cause of this is the large inherent flexibility in the region as originally surmised by Takeda et al. (3). We further demonstrated that a single amino acid mutation in the TNT1 domain is sufficient to cause distinct alterations in TnT structure and dynamics (6). We therefore have been engaging in integrated computational and experimental studies of known human N-terminal tail domain FHC mutations to both determine the general applicability of this methodology in understanding the primary effects of cTnT mutations on protein dynamics and to eventually characterize the downstream effects of altered cTnT flexibility on troponin complex function.
From our earlier work, we observed that the effected flexible hinge region identified from the molecular dynamics (MD) simulations of cTnT FHC mutations is ∼18 angstroms from the point mutation at Arg-92 and lies between residues 104–110 (3), an unexpected result. Understanding how a single amino acid substitution in cTnT causes such a distant effect was addressed by characterizing the dynamics of WT and mutant cTnT segments using MD. Our results demonstrated that the removal of a charged residue in R92L and R92W mutants is the central modulator of dynamics in the region of residues 104–110 of cTnT (6). These findings provided valuable clues to the link between cTnT structure and function in the context of the TNT1 N-terminal tail domain of cTnT (4, 7). TNT1 is a modulator in the formation of a TnT-Tm-Tm complex involving the N and C terminus overlap region of Tm (1, 7–9).
A concern with the application of the previous concepts is that flexibility is not in and of itself a rigorously defined mathematical quantity, which can be used to assess and predict biological function. To directly address this limitation, we now calculate forces acting on a bending coordinate within a model of cTnT residues 70–170 and correlate these data with results generated in R-IVM functional studies. We have found that the in silico parameter for flexibility and the in vitro parameter for cooperativity of calcium activation were correlated statistically, whereas others were not. A primary sequence analysis was performed and showed the highest degree of conservation of charged residues is found in TNT1, providing further support for the hypothesis that this region is crucial in the normal function of the thin filament and a hot spot for FHC-related mutations. Thus, we have now demonstrated that a strong correlation between molecular flexibility as quantified by a force constant can be linked to clinically relevant changes in thin filament function, a finding that sets the stage for eventual approaches to predict pathogenicity of novel TNT1 mutations in this complex disorder.
EXPERIMENTAL PROCEDURES
Development of in Silico Model
MD simulations were conducted as described previously (3, 6). α-Helical models of residues 70–170 of murine cTnT sequences for the WT protein, R92L, R92W, and R92K were made using the commercial software INSIGHTII (Accelrys, Inc., San Diego, CA). To simulate the protein environment, charged residues were assigned partial charges corresponding to physiological conditions. Minimization, equilibration, and production runs of MD were performed using CHARMM29 macromolecular simulation package with a classical potential for all atoms (10, 11). The nonbonded interaction cutoff was set to 12 angstroms and smoothed to zero between 12 and 14 angstroms. Starting structures were created by minimizing each cTnT segment using the steepest descent method with a step size of 200 fs for 20,000 cycles or until the gradient of the energy converged (11). For the remaining phases of the simulation, we used the SHAKE algorithm to constrain all bonds involving hydrogen atoms and an integration time step of 1 fs. We slowly increased the temperature to 300 K over 10 ps, at which point, we allowed the systems to equilibrate for 50 ps prior to proceeding with the production run. Initial velocities were assigned from a Boltzmann distribution at a low temperature and rescaled every 100 steps during the heating phase. During the production run, the dynamics were simulated for an additional 300 ps. We monitored the coordinates and system properties every five fs for the WT, R92L, R92W, and R92K models, using Visual Molecular Dynamics (VMD) to create movies of the production run dynamics (30).
Our earlier work demonstrated that this model maintained thermodynamic and structural equilibrium for 100 ps (3). Thus, 300 ps is more than adequate to explore changes in average structure and dynamics. The advantage to the study of fragments of large protein assemblages is not that you can run them for long periods of time but rather that short periods of time suffice to answer questions. Furthermore, we performed four separate simulations for each variant with different initial conditions to ensure that the phenomena we observed were not merely due to inadequate sampling of conformation space. It is well known that multiple short simulations are far superior to single long ones for exploring conformation space. The results we obtained were qualitatively invariant to initial conditions.
As was done previously, a means of characterizing structural change in the helix was by identifying changes in distance between α-carbons four amino acids apart (or simply n − (n + 4) distance, where n is the α carbon residue number) along the helices throughout the MD production run. We designate a shortening in n − (n + 4) distance as a compaction; and a lengthening in this distance is designated as an expansion, or helical opening.
To determine the bending force throughout the hinge region, we defined an intrinsic bending coordinate, β. β defines an internal coordinate system along the backbone of a protein based on relative positions of three contiguous α-carbons as shown in Fig. 2. From the chain rule, we can introduce the coordinate β.
 |
The −∂U/∂r⃗2 term is solved by CHARMM in terms of Cartesian coordinates. Therefore, the ∂r⃗2/∂β term also must be evaluated in terms of Cartesian coordinates. This yields an expression of the force on α-carbon two in terms of a coordinate system defined by three adjoining α-carbons and an angle β, which can be described explicitly in terms of Cartesian coordinates, allowing us to use the output of CHARMM to express this force. The needed derivative was calculated analytically (shown in the supplemental data), and the CHARMM source code was modified to evaluate the force at each time step of the simulation following the minimization. For instance, to calculate the forces acting on a residue 92 for WT, a coordinate transformation was performed using the positions of residues 91, 92, and 93. This ability to calculate bending forces gives us a quantitative property that can be compared with and correlated with experimental observation augmenting the previously reported structural results.
FIGURE 2.
Characterization of a helix. On the left is the traditional description of structural change in the helix, where m is the number of amino acids per helical turn, and p is the distance the helix rises along its axis per turn. For a standard α-helix, m = 3.6 amino acids and p = 5.4 Angstroms (29). We adapted this method to track α-carbons four amino acids apart, which we referred to as n − (n + 4) distance in our earlier work (6). The characterization of helical change depicted on the right uses three contiguous α-carbons to define an intrinsic coordinate, β, by which changes in local force can be measured.
mcTnT Site-directed Mutagenesis
cDNA of adult mouse cardiac TnT (mcTnT) previously cloned into a pSBETa vector (12) (provided by Dr. M. Sumandea) was cloned into pET3D vector (Novagen catalog no. 69421-3) and mutated selectively using the Quik-Change site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Sense and antisense primers (from Invitrogen) were used for the respective mutations. Only the sense primers are shown below, with the mutated residues underlined: cTnT R92L, 5′-ggactttgatgacatccacctaaagcgcgtggagaaggacc-3′; cTnT R92W, 5′-ctttgatgacatccactggaagcgcgtggagaa-3′; cTnT R92K, 5′-ctttgatgacatccacaagaagcgcgtggagaagg-3′. The identity of each mutant construct was verified by direct DNA sequencing.
Protein Expression and Purification
The cDNA containing only the coding sequence for mouse cardiac TnI (mcTnI) cloned into pET3D expression vector (Novagen) was provided by Dr. M. Sumandea and cDNA for human cardiac TnC cloned into pET3D vector was provided by Dr. J. D. Potter. Both constructs were transformed into BL21 or DH5 α competent cells and transformed into Rosetta II (DE3) competent cells (Novagen) for protein expression. Only cTnT constructs were transformed into Rosetta cells because these cells express rare codons, which cTnT has a higher amount of than cTnC and cTnI. To express cTnI, cTnC, and/or cTnT protein, Luria Broth-ampicillin agar plates were streaked with glycerol stock and incubated at 37 °C. Four to six liters of Overnight Express TB medium (Novagen) with ampicillin were inoculated with Luria Broth-ampicillin culture and grown at 37 °C overnight with shaking. Bacteria were centrifuged in a JLA-8.1 rotor at 6,000 rpm for 5 min at 4 °C. For mcTnT, bacterial pellets were resuspended in 200 ml/6 liter culture of S-Sepharose buffer (6 m urea, 50 mm Tris, pH 7.0, 2 mm EDTA, 1 mm DTT; pH 7.0). Thawed, resuspended pellet was sonicated on ice for 10 min in 30-s bursts with 2-min pauses using an Ultrasonic Cell Disruptor, Microson XL (Misonix). Sonicated sample was then centrifuged at 20,000 rpm in an SS-34 Sorvall rotor for 45 min at 4 °C to pellet bacterial debris and completely clear supernatant. The remaining steps were done at 4 °C. Supernatant was loaded at 1.3 ml/min flow rate on an SP-Sepharose column (Sigma; packed in a Bio-Rad EconoColumn with a 100-ml bed volume) pre-equilibrated with S-Sepharose buffer. The loaded column was washed with ∼1–1.5 liters of S-Sepharose buffer. mcTnT protein was eluted with a linear gradient of 0–0.6 m KCl, and fractions were collected. Pure mcTnT fractions were determined via absorbance at 280 nm and Coomassie-stained SDS-PAGE gel. Pooled fractions were dialyzed at 4 °C against 2 × 2 liters of Q-Sepharose buffer (6 m urea, 20 mm Tris, pH 7.8, 1 mm EDTA, 0.3 mm DTT; pH 7.8). Dialyzed protein loaded on a pre-equilibrated Q-Sepharose column (Sigma; packed in a Bio-Rad EconoColumn with a 100-ml bed volume), washed, and eluted with 0–0.5 m NaCl. Pure mcTnT fractions were determined as stated above. Pooled pure fractions were dialyzed at 4 °C against 2 × 4 liters of cold 0.05% trifluoroacetic acid. Dialyzed protein was flash frozen, lyophilized, and stored at −80 °C.
Human cTnC was purified on a Q-Sepharose column and eluted with a linear gradient of 0 to 0.6 m KCl. Pooled pure fractions were dialyzed at 4 °C against 4 × 2 liters of phenyl Sepharose buffer A (50 mm Tris-HCl, 1 mm CaCl2 , 50 mm NaCl, 1 mm DTT; pH 7.5). Dialyzed protein was loaded at room temperature onto a pre-equilibrated phenyl Sepharose column (Sigma; packed in a Bio-Rad EconoColumn with a 100-ml bed volume) with phenyl Sepharose buffer A + 0.5 m ammonium sulfate. The loaded column was washed with phenyl Sepharose buffer A + 0.5 m ammonium sulfate. Protein was eluted with phenyl Sepharose buffer C (50 mm Tris-HCl, 1 mm EDTA, 1 mm DTT; pH 7.5) + 0.5 m ammonium sulfate. Pure fractions were dialyzed against 4 × 4 liters of 5 mm ammonium bicarbonate solution before lyophilization and storage at −80 °C. mcTnI was purified on an SP-Sepharose column and eluted with a linear gradient of 0 to 0.6 m KCl. Pure fractions were dialyzed at 4 °C against 2 × 2 liters of Q-Sepharose buffer. The second column for mcTnI purification was a TnC affinity column prepared by manufacturer's protocol for conjugating proteins to a cyanogen bromide-activated Sepharose 4B gel (Sigma) and packed in an EconoColumn (Bio-Rad). Dialyzed mcTnI protein was loaded at 1.3 ml/min on the TnC affinity column pre-equilibrated with TnC affinity buffer (50 mm Tris, pH 7.5, 2 mm CaCl2 , 1 m NaCl, 1 mm DTT). The column was washed with 2–3 column volumes of TnC affinity buffer. Protein was eluted with a linear gradient of 0 mm EDTA, 0 m urea to 3 mm EDTA, 6 m urea. Pure fractions were dialyzed at 4 °C against 2 × 2 liters of double distilled H2O before lyophilization and storage at −80 °C.
Recombinant cTn Complex Reconstitution, Myosin and Actin Preparation, and Labeling
The recombinant heterotrimeric cTn complex was reconstituted by mixing equimolar amounts of cTnT, cTnI, and cTnC in a solubilization buffer containing 6 m urea, 50 mm Tris, pH 8.0, 1 m KCl, 5 mm MgCl2, 1 mm CaCl2, and 1 mm DTT (13). The solution was subjected to sequential dialysis using 0.7 m KCl, 0.4 m KCl, and finally 0.2 m KCl in the above buffer minus urea. Next, the cTn complex was further separated from the monomeric troponins by anion exchange on a RESOURCE-Q column (Amersham Biosciences) connected to a FPLC system. The cTn complex identity and purity were verified by 12% SDS-PAGE and then concentrated to ∼500 μl using a Centriprep 10K (Millipore). Next, the cTn complex was quantitated for concentration, aliquoted, and stored at −80 °C until it was used for motility experiments. Myosin isolation and purification from rabbit skeletal muscle are described in the supplemental data (Myosin Isolation and Purification).
Regulated in Vitro Motility Assay
Motility assay procedures and measurement were performed as described previously (14). In brief, a nitrocellulose-coated coverslip was attached to a glass microscope slide using double-faced tape, forming a flow cell chamber. Heavy meromyosin of muscle myosin II of 300 μg/ml was injected into the flow cell and allowed to incubate for 1 min. Heavy meromyosin solution was replaced by 35 μl of 1 mg/ml bovine serum albumin in assay buffer (25 mm KCl, 2 mm MgCl2, 2 mm EGTA, 25 mm MOPS, 50 mm DTT). One minute later, the flow chamber was washed with two 35-μl aliquots of assay buffer. 35 μl of 1 μm solution of sheared, unlabeled actin filaments (produced by rapid flow of actin solution through a syringe needle) was introduced into the chamber for 1 min, followed by two 35-μl aliquots of assay buffer containing 1 mm ATP. This acts to block any “dead” S-1 myosin heads. Next, the chamber was washed twice with assay buffer to clear the chamber of any ATP. Rhodamine-phalloidin actin was added in a concentration of 6 nm, washed off with assay buffer, and replaced with a buffer containing 100 nm of regulatory proteins (Tn and rabbit skeletal Tm provided by Dr. E. Homsher). After a 5-min incubation, the flow cell was washed with motility solution containing 40 mm MOPS, 1 mm MgCl2 , 3 mm MgATP, 2 mm K-EGTA (pCa 9) or 2 mm Ca-EGTA (pCa 5), 50 mm DTT, 100 nm Tn, 100 nm Tm, and 20 nm rhodamine-phalloidin. To achieve free Ca concentrations between pCa 9 and 5, the high and low Ca motility solutions were mixed accordingly as described previously (15). Thus, R-IVM was performed for WT and each mutation at pCa 9, 7.5, 7, 6.5, 6, 5.5, and 5. Photobleaching protective agents, 14 mm glucose, 240 units of glucose oxidase/ml, and 9 × 103 units of catalase/ml, were also added. The motility chamber was mounted on a temperature-controlled objective and microscope stage such that experiments can be run at 25 °C. The conditions were repeated as above for each experiment with the exception of the recombinant troponin used, which was changed accordingly for WT, R92L, R92W, and R92K variants.
Motility was viewed using an Olympus IX-71 inverted microscope under fluorescence illumination and a 100× objective. Thin filament movement was recorded using a Hamamatsu ORCA-ER cooled CCD. For each motility chamber, 10–15 sites on a surface were observed and recorded for 10–20 s each, during which photobleaching was insignificant. Using the MetaMorph 7 system, we tracked the movement of each filament in the field of view using frame-grabbing rates of 5–10 frames/sec, from which sliding speed was calculated. The lengths of moving filaments varied from 0.4 to 20 μm, and each filament was analyzed for at least 10–20 frames (1–2 s). The quality of the filament movement was also determined as described previously (14): filaments not moving have an average speed of <0.8 μm/sec; filaments moving at uniform speed had a frame-to-frame S.D. of <0.5 of the mean speed of the filament; the frame-to-frame S.D. of the erratically moving filament was >0.5 of its mean speed (15).
Correlation Analysis
Our MD results allowed us to parameterize bending force in TNT1, whereas our R-IVM results allowed us to parameterize cooperativity (n, Hill coefficient), calcium sensitivity (EC50 or pCa 50), and maximum rates of force-producing cross-bridge cycling (maximum sliding speed). With respect to R-IVM, maximum sliding speeds at highest calcium concentrations (pCa 5), are associated with actomyosin kinetics, whereas at submaximal levels of activation quantities such as cooperativity and EC50 are measured. Our MD model does not include actin or myosin, so we cannot compare MD results with maximum sliding speed results from R-IVM. The TNT1 region of cTnT is in an influential position to affect the flexibility and dynamics of Tm because TNT1 directly binds to overlapping Tm, where Tm displays important affinity for actin and shows its greatest flexibility (16). Therefore, we confined our attempt to correlate bending force from MD with cooperativity and calcium sensitivity from R-IVM. A Pearson correlation between MD and R-IVM results was performed with the statistical software package R.2.11.1 for Windows.
Statistical Analysis
Values in this study are expressed as mean ± S.E. Statistical analyses were performed using Prism5 (Graph Pad, San Diego, CA). Analysis of variance and Newman Keuls post hoc analysis was used for statistical comparisons between transgenic lines and corresponding controls for each parameter. A level of p < 0.05 was accepted as statistically significant.
RESULTS
This study was performed with the goal of associating changes in computationally derived physical parameters of TNT1 with changes in functional parameters of the thin filament due to mutations in TNT1 using experimental means. Therefore, particular care was taken to choose analogous theoretical and experimental models. We chose to compare an in silico 101-residue, helical model of murine TNT1 with an in vitro model of the thin filament consisting of recombinant cTn proteins reconstituted into a ternary complex and then added to Tm and actin. We believe these two models present analogous systems in that they are both unloaded representations of sarcomeric function. Also, these models both represent the same physiologic scale, a single cardiac thin filament. The major difference between the two models is that the experimental model contains a significant portion of the thin filament that is missing from our computational model. Our labs have shown previously that this same 101-residue, α-helical in silico model of TNT1 is capable of capturing structural changes necessary to explain observed biochemical and biophysical changes resulting from mutation (3).
We chose to analyze four variants of cTnT for this study: WT, R92L, R92W, and R92K. R92L and R92W are well described FHC-related mutations (1, 3, 6). R92K is a positive test case to test our hypothesis that size and charge of residues at residue 92 cTnT are key features with respect to the observed mutational effects.
Theoretical Results
We reproduced previous results that showed that point mutations R92L and R92W decrease bending forces at residues 105 and 106 in the hinge site of cTnT. This is the physical reason underlying the changes in flexibility of TNT1 caused by these FHC mutations (data not shown).
Data from force analyses are presented in Fig. 3 using box plots for each variant. Force analyses of the regions of interest, the mutational site at residue 92 and the hinge site at residues 105–106, show not only a decrease of median force in mutants at the hinge site but also a decrease in the variance of the distribution of mutant forces at the hinge site, as shown in Fig. 3 (right graph).
FIGURE 3.
Force analyses of mutation and hinge sites. Left, mutational site, residue 92. Middle, box plot legend. Right, hinge site, residues 105–106. Box plots of force calculated at each time step (1 fs) over 300 ps. The median is denoted with a horizontal line. Interquartile ranges (25th and 75th percentiles) are denoted using colored boxes. The whiskers extend to the 10th and 90th percentiles of the data. These data suggest that the physical change responsible for the increased bending motion of mutant segments is a decrease in local force at residues 105 and 106 of cTnT.
As would be expected, there is an inverse relationship between the median force at the hinge site for mutants in Fig. 3 (right graph) and the degree of observed bending from our previous work, which showed the flexibility of variants to be R92L > R92W > WT. Our positive test case involving substitution with lysine, R92K, resulted in virtually identical dynamics as those of WT with no change in observed forces at the site of mutation of the hinge site. This fully matches our observations that no bend is seen at the mutational site during simulations for R92K.
Experimental Results
From our R-IVM results detailed in Fig. 4 and Table 1, we observe that FHC-related point mutations at R92 cTnT are not only capable of changing the structure and dynamics of TNT1 but also the function of the thin filament as a whole. We hypothesized in our earlier work that this was due to a removal of a charged residue at hot spot Arg-92; however our R-IVM data suggest that TNT1 is sensitive to mutation beyond charge change. We note that even minor structural changes at residue 92 results in major functional changes, such as R92K, which is a substitution that is identical in charge but slightly different in structure. Of note, without Tm present, as in our MD simulations, the structure and function of R92K TNT1 appear similar to WT. This suggests that the residue 92 cTnT is indeed a key residue in cTnT-Tm interactions as proposed previously (1). Using the “knob and hole” model of helix-helix interactions (17), Arg-92 is likely a knob that precisely interacts with a hole in a binding pocket within a Tm helix. This motif of TnT-Tm interactions has been found in a recent crystal structure (8).
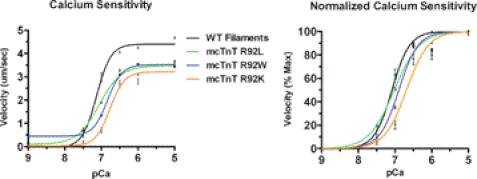
FIGURE 4.
Regulated in vitro motility assays: velocity-pCa relationships. Regulated in vitro motility assays of mcTnT WT, R92L, R92W, and R92K substitutions at varying calcium concentrations showing calcium sensitivity (left) and normalized calcium sensitivity (right). Arg-92 substitutions show slightly decreased maximal sliding speed and calcium sensitivity compared with WT, demonstrating functional impairment of the thin filament resulting from the slightest substitution at Arg-92.
TABLE 1.
Parameters of velocity-pCa relationship for cTnT substitutions
| WT | R92L | R92W | R92K | |
|---|---|---|---|---|
| Max vf (μm/s) | 4.68 ± 0.3 | 3.66 ± 0.6 | 3.44 ± 0.5 | 3.53 ± 0.4 |
| EC50 | 7.08 ± 0.02 | 7.02 ± 0.01 | 6.90 ± 0.01* | 6.69 ± 0.03* |
| n (Hill coefficient) | 1.8 ± 0.1 | 1.2 ± 0.04* | 1.5 ± 0.05 | 1.6 ± 0.05 |
*, p < 0.05.
Of note, R-IVM shows a decrease in the cooperativity of calcium activation (n, Hill coefficient) for R92L and R92W in comparison with WT, whereas R92K showed no significant alteration, as shown in Fig. 4 and Table 1. It has been suggested that thin filament FHC mutant proteins (such as the Arg-92 hot spots) markedly alter the nature of the interaction between actin and myosin (16). Consequently, the changes in cooperativity observed in the Arg-92 hot spots may be indicative of alterations in the allosteric regulation of Tm by Tn, whereby the cooperative interactions of Tn-Tm may produce structural changes allowing for strong actin-myosin binding (the transition to “open” state in the three-state model of myofilament activation) (14).
Additionally, the Arg-92 substitutions and their respective flexibility/force effects on the hinge region are manifest in altered maximum sliding speed and the percent of filaments moving uniformly and erratically in comparison with WT (shown in Fig. 5), likely indicative of the impaired interaction of cTnT with Tm and the resultant inability to fully regulate function.
FIGURE 5.
Regulated in vitro motility assays percent motile data. Regulated in vitro motility assays of mcTnT WT, R92L, R92W, R92Q, R92K, and R94L substitutions at varying calcium concentrations showing percent of filaments moving uniformly (left) and moving erratically (right). The substitutions in general show a decrease in uniform motion and an increase in erratic motion compared with WT. This is likely an indication of altered cTnT-Tm interaction, impairing proper regulation of thin filament function.
Upon completion of the in silico and in vitro experiments, we analyzed the data to correlate results. Analysis was expected to show which, if any, of these parameters are associated with bending force. Fig. 6 shows that changes in bending forces of the TNT1 hinge region directly correlate with cooperativity of the thin filament function as parameterized by the Hill coefficient. This implies that the alterations in bending force we observe in our MD simulations are associated with the functional changes we observe in our R-IVM experiments. This, in turn, suggests that changes in TNT1 dynamics alter dynamics within the cTn core as well as cTnT-Tm interactions.
FIGURE 6.
MD versus R-IVM correlation analysis. Left, correlation of bending force (in silico parameter) with cooperativity (in vitro parameter). Right, there is no apparent correlation of bending force with calcium sensitivity.
In light of our observations, we asked what characteristics of TNT1 make it sensitive to point mutations, resulting in a hinge region removed from the site of mutation and causing alterations in function? We propose that the highly conserved, charged nature of TNT1 yields an intrinsic curvature that appears necessary for proper assembly and function of the thin filament. To address this hypothesis, we performed a sequence analysis using previously published alignments for various isoforms of TnT, including human, mouse, and chicken sequences of cardiac, fast skeletal, and slow skeletal troponin T (18). Sequences were analyzed for conservation of charged residues: arginine, lysine, glutamate, and aspartate. The percentage of conserved residues was calculated for the portion of TnT represented by our model and compared with the remainder of TnT.
Fig. 7 shows the alignment of published TnT isoforms (18) with respect to conservation of charged residues. In our 101-amino acid model, ∼97% (98/101) of the residues are conserved with respect to charge. In the preceding N terminus and trailing C terminus regions of TnT, 45% (34/76) and 88% (118/134) of the residues are conserved with respect to charge, respectively.
FIGURE 7.
Sequence alignment of TnT isoforms emphasizing the conservation of charged residues in TNT1.
Our analysis suggests that the region of charged residues in TNT1 is the most highly conserved region of TnT. In addition, the sequence and intrinsic bend of this region would appear critical to normal assembly and function, and alterations in charge or structure at Arg-92 will affect the dynamics of TNT1 as well as thin filament function via cTnT-Tm interactions.
DISCUSSION
The decreases in forces experienced by residues in the hinge region of mutant segments identify a potential physical mechanism for the observed distant effects of FHC-related point mutations R92L and R92W in cTnT. Our MD calculations on murine cTnT peptides extend our earlier findings and provide a potential mechanism for the impaired troponin tail-Tm binding observed previously for FHC mutations (1, 7). These Arg-92 mutations, which have been shown to affect the head-to-tail association of Tm and the ability of Tm to bind tightly to actin, may be expected to negatively affect myofibrillar assembly, the cooperativity and extent of force development, and calcium dependence of contraction (1, 4, 7). Our in vitro motility results are supportive and provide experimental support of this hypothesis and demonstrate that local changes in structure impose significant functional alterations.
Despite nearly identical bending forces at and dynamics about the hinge region for R92K when compared with WT, there is a significant change in its function when measured using R-IVM. On the one hand, this points to the dangers of correlating fragment computations to regulated filament function but also implies that even the slightest alteration in charge or structure at Arg-92 cTnT affects cTnT-Tm interactions. This conclusion is supported by previous studies on the 70–170-cTnT peptide (human instead of murine) and the whole cTnT (bovine), which have experimentally shown that FHC-related mutations at Arg-92 displayed decreased affinity for Tm (1, 7). One of these studies also suggested that hydrophobic mutations such as R92L and R92W would disrupt cTnT Arg-92-Tm Gln-263 hydrogen bonding, and the mutational effects on helical stabilization would affect side chain packing at the hydrophobic interface so as to weaken complex formation (1).
Our R-IVM studies in Arg-92 substitutions using murine cTnT all showed slightly decreased maximal sliding velocity compared with WT (hence, a decreased heavy meromyosin ATPase activity) and decreased calcium sensitivity of thin filament sliding speed (indicative of an altered number of attached and cycling cross-bridges). A significant increase in the calcium sensitivity of force development/thin filament activation at the cardiac skinned fiber level has been one of the most consistent in vitro findings in thin filament models of FHC (19–21). Although this may seem discordant with our R-IVM findings, it is important to note that the motility assay is an unloaded system unlike the skinned fiber experiment; hence, making velocity-pCa and force-pCa measurements difficult to compare (16, 22). Previous studies have shown that when comparable plots of the force-pCa curve were made using skinned muscle fibers contracting under the same conditions as in vitro motility assays, the EC50 for the force-pCa curves were 0.4–0.8 pCa units smaller than the velocity-pCa curves (16, 23). Additionally, our current R-IVM utilizes a heterogeneous combination of skeletal Tm and cTn. Our initial R-IVM studies establishing this assay as our functional test utilized rabbit skeletal Tm (αα- or αβ-Tm isoform dimers). Thus, for comparison between substitutions and WT, and reproducibility of data, we maintained the use of this skeletal Tm isoform. A study comparing the differential effects of cardiac versus skeletal muscle regulatory proteins on R-IVM showed that the skeletal Tm/cTn combination resulted in a decreased EC50 in comparison to cardiac Tm/cTn (24). Thus, these caveats may potentially explain the differences between the EC50 values reported for our R-IVM substitutions and the increase in calcium sensitivity previously observed in fiber studies for these mutations. Future R-IVM experiments utilizing rodent cardiac Tm (predominantly αα-Tm homodimers, as well as the use of rodent cardiac heavy meromyosin, with our murine cardiac Tn will also be established for our assay, potentially addressing these differences. Nonetheless, the Arg-92 velocity-pCa sensitivity from our current R-IVM exhibited comparable change in comparison with WT as the force-pCa sensitivity results of Harada et al. (21).
The characterization of in silico structural changes in FHC-related cTnT mutations and their functional correlate is a novel approach, which has revealed important observations regarding thin filament biology. Because our initial study characterizing the structural and functional effects of R92L and R92W FHC mutations via computational and physiological methodologies (3), other groups also have begun to utilize an integrative approach to better understand the structural mechanisms at work in their respective systems (25, 26).
A central aspect of biochemistry and biophysics regarding the study of single molecules is that structure implies function. This concept with respect to protein complexes and alterations that cause disease has become an important area of research utilizing integrative biophysical, biochemical, and physiological approaches. The results of this study provide insight into the structure-function relationship within cardiac troponin and how FHC mutations may alter it. Our previous and current studies demonstrate that there is likely to be a conserved network of residues lying with TNT1 that have distant effects on the cTn core (27, 28). An integrated approach as described here proves necessary to link mechanistic and functional changes at the molecular level. This integrative approach provides a potentially predictive screen for specific mutations to be further studied experimentally at the in vitro and in vivo levels.
Supplementary Material
This work has been supported in part by National Institutes of Health Grant HL075619 (to J. C. T.), and Predoctoral Training Grants 1F31HL085915-01 (to P. J. G.) and American Heart Association 10PRE3610021 (to E. P. M.), and Medical Scientist Training Grant GM007288.

This article contains supplemental data.
- cTn
- cardiac troponin
- cTnT
- cardiac troponin T
- cTnI
- cardiac troponin I
- cTnC
- cardiac troponin C
- mcTnT
- mouse cardiac troponin T
- mcTnI
- mouse cardiac troponin I
- TnT
- troponin T
- Tn
- troponin
- Tm
- tropomyosin
- FHC
- familial hypertrophic cardiomyopathy
- R-IVM
- regulated in vitro assay
- MD
- molecular dynamics.
REFERENCES
- 1. Palm T., Graboski S., Hitchcock-DeGregori S. E., Greenfield N. J. (2001) Disease-causing mutations in cardiac troponin T: Identification of a critical tropomyosin-binding region. Biophys. J. 81, 2827–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tobacman L. (1996) Thin filament-mediated regulation of cardiac contraction. Annu. Rev. Physiol. 58, 447–481 [DOI] [PubMed] [Google Scholar]
- 3. Ertz-Berger B. R., He H., Dowell C., Factor S. M., Haim T. E., Nunez S., Schwartz S. D., Ingwall J. S., Tardiff J. C. (2005) Changes in the chemical and dynamic properties of cardiac troponin T cause discrete cardiomyopathies in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 102, 18219–18224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hinkle A., Goranson A., Butters C. A., Tobacman L. S. (1999) Roles for the troponin tail domain in thin filament assembly and regulation. A deletional study of cardiac troponin T. J. Biol. Chem. 274, 7157–7164 [DOI] [PubMed] [Google Scholar]
- 5. Tobacman L. S., Nihli M., Butters C., Heller M., Hatch V., Craig R., Lehman W., Homsher E. (2002) The troponin tail domain promotes a conformational state of the thin filament that suppresses myosin activity. J. Biol. Chem. 277, 27636–27642 [DOI] [PubMed] [Google Scholar]
- 6. Guinto P., Manning E., Schwartz S., Tardiff J. (2007) Computational characterization of mutations in cardio troponin T known to cause familial hypertrophic cardiomyopathy. J. Theor. Comp. Chem. 6, 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hinkle A., Tobacman L., (2003) Folding and function of the troponin tail domain. Effects of cardiomyopathic troponin T mutations. J. Biol. Chem. 278, 506–513 [DOI] [PubMed] [Google Scholar]
- 8. Murakami K., Stewart M., Nozawa K., Tomii K., Kudou N., Igarashi N., Shirakihara Y., Wakatsuki S., Yasunaga T., Wakabayashi T. (2008) Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin T. Proc. Natl. Acad. Sci. U.S.A. 105, 7200–7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takeda S., Yamashita A., Maeda K., Maéda Y. (2003) Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature 424, 35–41 [DOI] [PubMed] [Google Scholar]
- 10. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–196 [Google Scholar]
- 12. Chandra M., Montgomery D. E., Kim J. J., Solaro R. J. (1999) The N-terminal region of troponin T is essential for the maximal activation of rat cardiac myofilaments. J. Mol. Cell Cardiol. 31, 867–880 [DOI] [PubMed] [Google Scholar]
- 13. Sumandea M. P., Pyle W. G., Kobayashi T., de Tombe P. P., Solaro R. J. (2003) Identification of a functionally critical protein kinase C phosphorylation residue of cardiac troponin T. J. Biol. Chem. 278, 35135–35144 [DOI] [PubMed] [Google Scholar]
- 14. Homsher E., Nili M., Chen I. Y., Tobacman L. S. (2003) Regulatory proteins alter nucleotide binding to acto-myosin of sliding filaments in motility assays. Biophys. J. 85, 1046–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Homsher E., Kim B., Bobkova A., Tobacman L. (1996) Calcium regulation of thin filament movement in an in vitro motility assay. Biophys. J. 70, 1881–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gordon A. M., Homsher E., Regnier M. (2000) Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924 [DOI] [PubMed] [Google Scholar]
- 17. Crick F. (1953) The packing of α-helices: Simple coiled coils. Acta Crystallogr. 6, 689–698 [Google Scholar]
- 18. Jin J. P., Zhang Z., Bautista J. A. (2008) Isoform diversity, regulation, and functional adaptation of troponin and calponin. Crit. Rev. Eukaryot. Gene Expr. 18, 93–124 [DOI] [PubMed] [Google Scholar]
- 19. Chandra M., Rundell V. L., Tardiff J. C., Leinwand L. A., De Tombe P. P., Solaro R. J. (2001) Ca2+ activation of myofilaments from transgenic mouse hearts expressing R92Q mutant cardiac troponin T. Am. J. Physiol. Heart Circ. Physiol. 280, H705–713 [DOI] [PubMed] [Google Scholar]
- 20. Chandra M., Tschirgi M. L., Tardiff J. C. (2005) Increase in tension-dependent ATP consumption induced by cardiac troponin T mutation. Am. J. Physiol. Heart Circ. Physiol. 289, H2112–2119 [DOI] [PubMed] [Google Scholar]
- 21. Harada K., Potter J. (2004) Familial hypertrophic cardiomyopathy mutations from different functional regions of troponin T result in different effects on the pH and Ca2+ sensitivity of cardiac muscle contraction. J. Biol. Chem. 279, 14488–14495 [DOI] [PubMed] [Google Scholar]
- 22. Homsher E., Lee D. M., Morris C., Pavlov D., Tobacman L. S. (2000) Regulation of force and unloaded sliding speed in single thin filaments: effects of regulatory proteins and calcium. J. Physiol. 524, 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gordon A. M., LaMadrid M. A., Chen Y., Luo Z., Chase P. B. (1997) Calcium regulation of skeletal muscle thin filament motility in vitro. Biophys. J. 72, 1295–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clemmens E. W., Entezari M., Martyn D. A., Regnier M. (2005) Different effects of cardiac versus skeletal muscle regulatory proteins on in vitro measures of actin filament speed and force. J. Physiol. 566, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X. E., Holmes K. C., Lehman W., Jung H., Fischer S. (2010) The shape and flexibility of tropomyosin coiled coils: Implications for actin filament assembly and regulation. J. Mol. Biol. 395, 327–339 [DOI] [PubMed] [Google Scholar]
- 26. Lim C. C., Yang H., Yang M., Wang C. K., Shi J., Berg E. A., Pimentel D. R., Gwathmey J. K., Hajjar R. J., Helmes M., Costello C. E., Huo S., Liao R. (2008) A novel mutant cardiac troponin C disrupts molecular motions critical for calcium binding affinity and cardiomyocyte contractility. Biophys. J. 94, 3577–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clarkson M. W., Lee A. L. (2004) Long range dynamic effects of point mutations propagate through side chains in the serine protease inhibitor eglin c. Biochemistry 43, 12448–12458 [DOI] [PubMed] [Google Scholar]
- 28. Süel G. M., Lockless S. W., Wall M. A., Ranganathan R. (2003) Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 10, 59–69 [DOI] [PubMed] [Google Scholar]
- 29. Voet D., Voet J. (2004) Biochemistry, 3rd Ed., pp. 219–227, John Wiley and Sons, Inc., New York [Google Scholar]
- 30. Humphrey W., Dalke A., Schulten K. (1996) VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.