Abstract
Ribonucleotide reductase (RNR) is the enzyme critically responsible for the production of the 5′-deoxynucleoside-triphosphates (dNTPs), the direct precursors for DNA synthesis. The dNTP levels are tightly controlled to permit high efficiency and fidelity of DNA synthesis. Much of this control occurs at the level of the RNR by feedback processes, but a detailed understanding of these mechanisms is still lacking. Using a genetic approach in the bacterium E. coli, a paradigm for the Class Ia RNRs, we isolated 23 novel RNR mutants displaying elevated mutation rates along with altered dNTP levels. The responsible amino-acid substitutions in RNR reside in three different regions: (i) the (d)ATP-binding activity domain, (ii) a novel region in the small subunit adjacent to the activity domain, and (iii) the dNTP-binding specificity site, several of which are associated with different dNTP pool alterations and different mutational outcomes. These mutants provide new insight into the precise mechanisms by which RNR is regulated and how dNTP pool disturbances resulting from defects in RNR can lead to increased mutation.
Keywords: Ribonucleotide Reductase, dNTP pools, DNA replication fidelity, allosteric regulation
1. Introduction
The proper control of the intracellular deoxynucleoside-5′-triphosphates (dNTPs) – the direct precursors for DNA synthesis – is critically important for the efficiency and fidelity of DNA replication and the DNA repair processes needed for genomic stability [1]. Genetically and pharmacologically-induced dNTP pool changes have long been recognized to have genotoxic consequences that can lead to mutagenesis and cell death [2, 3] and are also implicated in disease [4-7].
The controlled production of dNTPs depends on an enzyme termed Ribonucleotide Reductase (RNR), which catalyzes the reduction of ribonucleotides to the corresponding 2′-deoxynucleotides [8]. This reduction is a chemically difficult reaction [9, 10] and requires the presence of a stable organic radical [9, 11, 12]. Based on the precise type of radical, RNRs have been divided into a number of classes [13, 14]. The mammalian RNR and that of many bacteria, yeast, and viruses contain a tyrosyl radical, and this group of enzymes is referred to as Class I RNR. Within this class there is extensive conservation of mechanism and structure [12], and the enzyme from E. coli serves as an important model system [12, 15]. The E. coli enzyme, like the mammalian enzyme, reduces nucleoside diphosphates (NDPs) to the corresponding deoxynucleoside diphosphates (dNDPs) [16]. The quaternary organization of this class of RNR is α2β2, in which the large α (or R1) subunits contain the catalytic site and two allosteric effector-binding sites, and the small β (or R2) subunits contain the tyrosyl radical and a dinuclear iron center, both of which are essential for the enzymatic activity [9, 11]. In E. coli, the R1 and R2 subunits are encoded, respectively, by the nrdA and nrdB genes, which form an operon near 50′ on the E. coli chromosome.
RNR regulation occurs at several levels, including transcriptional during the cell cycle [17-20]. During DNA replication, regulation occurs by intricate allosteric changes, and changes in RNR oligomerization state [21, 22] ensuring adequate levels of the four dNTPs in proper ratios. These allosteric modes of regulation, first described many years ago, are still a subject of intense research [15, 23-26]. One mode of regulation is an on-off switch governed by the ATP/dATP ratio. The site for the type of regulation, the activity site, is located at the N-terminus of the R1 subunit. By monitoring the ATP/dATP ratio, it assures an overall dNTP level that is presumably optimal for efficient DNA replication. A second regulatory site on R1, named the specificity site, is a binding site for dATP, ATP, dGTP, and dTTP. Depending on which dNTP is bound, the nearby catalytic site is conformationally primed to reduce one specific NDP substrate (ADP, CDP, GDP, or UDP). In this manner, this site regulates the specificity of the enzyme such that the four dNTPs are maintained at their desired ratios.
While significant effort has been expended on studying RNR and its regulation, many of the mechanistic details remain to be understood. Also, few studies have been performed addressing how regulation of RNR affects the dNTP pools in vivo and the fidelity of the DNA replication process [27]. One major limitation to these studies has been the overall lack of RNR mutants with altered dNTP pools. In the present work, we use a new genetic system for obtaining E. coli RNR mutants with a mutator phenotype (i.e., elevated mutation rate). In total, we obtained 23 novel single point mutants with a mutator phenotype resulting from altered dNTP pools. These mutants provide new details regarding the precise modes of RNR regulation and the correlations between dNTP pool changes and mutation rates.
2. Materials and Methods
2.1. Strains and media
The E. coli strains used are derivatives of strain NR12470, a Δ(gpt-lac)5 derivative of strain MG1655 [28]. The nrdAB-carrying plasmids pHABcat or pHABamp (see description below) were introduced by transformation, after which the chromosomal nrdAB operon was replaced by the Δ(nrdAB::kan) deletion by P1 transduction. The Δ(nrdAB::kan) allele will be described elsewhere (M. Hung and R. M. S.). The strains were then made recA56, srl-360::Tn10 by P1 transduction, followed by introduction of the series of F’prolacZ episomes originally present in strains CC101 through CC106 [29] by conjugation. Lac plates used for determination of lac reversion frequencies contained 0.2 % lactose as carbon source. XPG plates used for Lac+ papillation studies were minimal medium plates containing X-Gal (40 μg), P-Gal (0.5 mg/ml), and glucose (0.2%), as described [30].
2.2. Plasmids
pHABcat (M. Hung and R. M. S., to be published) is a derivative of the plasmid pHSG576, a chloramphenicol-resistant, low-copy plasmid with the pSC101 origin [31]. It carries a 4,771-bp fragment of E. coli chromosomal sequence spanning the nrdA and nrdB genes including the nrdAB promoter and the downstream yfaE gene. From pHABcat we constructed plasmid pHABamp by replacing its chloramphenicol resistance gene by the bla gene of pBluescript II KS+. The bla gene was amplified with PCR primers containing flanking NcoI and AflII restriction sites, which were used to replace the cat-containing NcoI-AflII fragment of pHABcat. Thus, pHABcat and pHABamp are identical except for the antibiotic resistance gene.
2.3. Isolation of nrdAB mutator mutants
pHABcat DNA was subjected to mutagenesis by hydroxylamine as described [32]. The DNA, treated for various time periods, was electroporated into strain XL1-Blue (Stratagene) to create several libraries, each representing approximately 50,000 to 1,000,000 independent chloramphenicol-resistant clones. DNA prepared from these libraries was then used to transform a set of tester strains. The tester strains were five derivatives of strain NR17519 (recA56 ΔnrdAB::kan pHABamp) containing, additionally, the F’prolac from strains CC101, CC103, CC104, CC105, or CC106 [29]. (Strain CC102 was not used at this stage since the corresponding lac allele is somewhat leaky and not suitable for papillation assays). Aliquots of the transformations were plated on XPG-chloramphenicol plates to yield about 500 chloramphenicol-resistant colonies per plate. The plates were incubated for 5 days at 37°C and inspected daily for colonies that showed evidence of papillation (blue dots within normally colorless colonies). Candidate colonies were restreaked (patched) to confirm the phenotype. To confirm that the mutator phenotype was due to the mutated plasmid, plasmid was extracted and reintroduced. Strains still producing high number of papillae were selected as candidates for further studies, including DNA sequencing of the nrdAB genes. Mutant frequencies in liquid cultures were determined as described [30].
2.4. dNTP pool measurements
dNTPs were extracted using the procedure described by Buckstein et al. [33] with minor modifications. Strains were grown in minimal glucose medium with added casamino acids (Becton-Dickinson) (0.1 %). Overnight cultures were diluted 1:50 in minimal medium and grown at 37° C with shaking to an O.D.600 = 0.5. Quantitation of extracted dNTPs was by ion-pairing reverse phase chromatography [33] on an Agilent 1200 HPLC instrument with a Diode Array Detector. Nucleotides were separated on a Zorbax Eclipse XDB-C18 4.6 × 150 mm 5-micron column. Peaks for the individual dNTPs were identified based on retention times of dNTP standards and confirmed using the peak UV spectra. Quantitation of each dNTP was performed as a percentage of the ATP peak in the same extract. The (d)NTP extraction efficiency as determined by the height of the ATP peak was reproducible within 10-20% among the various samples.
2.5. Modeling methods for ribonucleotide reductase holoenzyme
An E. coli holoenzyme complex structural model was developed through computational docking using the ZDOCK/ZRANK and RDOCK protein-protein computational docking algorithm [34, 35] implemented in the Accelrys Discovery Studio 2.5 software package. We used the E. coli R1 PDB 3R1R structure with AMPNP in the activity site [36] as receptor molecule and the E. coli R2 PDB 2AV8 structure [37] as ligand. Poses were filtered by the ZDOCK program so that docked complexes with residues S39, E42, H46, E60, T61 and A65 from the R1 subunit and E67, E69, R221, G298 and P333 from the R2 subunit at the R1-R2 subunit interface were considered as meeting the docking criteria. Electrostatic and desolvation energy terms were used with a 15 degree angular step. Sixteen out of 3600 poses met the criteria and were used for a refined RDOCK docking calculation using the polar H CHARMm forcefield. All 16 docked poses were reviewed. A low energy docked pose with good zrank and E_Rock scoring was used for analysis. This pose is similar to the docked structure of Uhlin and Eklund [36] (coordinates kindly provided by Dr. J. Stubbe).
Results
The critical role of RNR in furnishing and regulating the cellular dNTPs is well recognized. As noted in the Introduction, at least two major RNR feedback mechanisms have been established to operate for maintaining the dNTP pools at both an appropriate level and the desired ratios among the four nucleotides. However, the precise details of these regulatory steps are still being explored. To obtain new insights into these regulatory processes, we have undertaken an approach of directly isolating E. coli RNR mutants based on their mutator phenotype. The underlying assumption is that RNR mutators, if found, are likely to be characterized by altered dNTP pools, presumably by a defect or alteration in one or more of the regulatory pathways. An important aspect of our approach is that we used random mutagenesis of RNR and, hence, no a priori assumptions were made as to which domains or regions of RNR are involved in regulation, permitting a potentially unbiased view of the in vivo regulatory circuits. The precise approach and the results are described below.
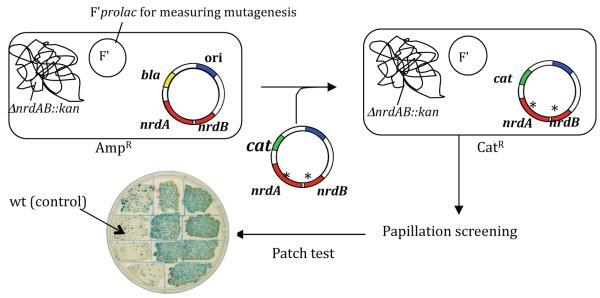
3.1. Isolation of novel nrdAB mutator mutants
To create mutants of E. coli affected in ribonucleotide reductase (RNR), we mutagenized the nrdA and nrdB genes, encoding, respectively, the R1 and R2 subunits of the enzyme. For this purpose, we designed the plasmid shuttle system outlined in Fig. 1. We used a strain in which the chromosomal nrdAB genes are deleted but are complemented by a low-copy plasmid, pHABamp (see Materials and Methods). This plasmid carries the nrdAB operon and provides ampicillin resistance. A near-identical plasmid, pHABcat, differing only in the antibiotic marker (it provides chloramphenicol resistance), was randomly mutagenized in vitro with hydroxylamine [32]. The treated plasmid pool was then used to replace the resident pHABamp plasmid (note, the plasmids belong to the same incompatibility group). This exchange was performed in cells containing F’prolac episomes [29] that permit detection of mutator individuals through their papillation phenotype (Fig. 1). This phenotype is based on the reversion of certain lacZ missense alleles (contained on the F’) within growing colonies. Such reversions can be readily visualized on X-gal-containing media as blue dots (papillae) within the colony or patches (see Fig. 1). The lacZ reversion system has the further advantage that it can address mutational specificity, since a different F’ episome is available to study each of the six possible base-pair substitutions [29]. We used five of the six, scoring lac reversion by A·T→C·G, G·C→C·G, G·C→T·A, A·T→T·A, or A·T→G·C [29]. After visually screening a total of 1.3 million colonies (about equal numbers for each of the base substitutions), we obtained 118 hyper-papillating colonies. Interestingly, all of these were obtained using the screen for lac G·C→T·A and A·T→T·A transversions. Isolation and retransformation of the pHABcat plasmids confirmed the association of the hyperpapillation phenotype with the plasmid. Direct determination of lac mutant frequencies indicated that these mutants were indeed strong mutators (described below).
Fig. 1.
Outline of the genetic system used to obtain mutator mutants of E. coli carrying defects in the nrdAB genes. Plasmid pHABcat is mutagenized (*) in vitro with hydroxylamine and used to transform a strain containing plasmid pHABamp and a chromosomal nrdAB deletion. Replacement of pHABamp by pHABcat then allows scoring for a mutator phenotype based on colony papillation (lacZ reversion). See text and Materials and Methods for details.
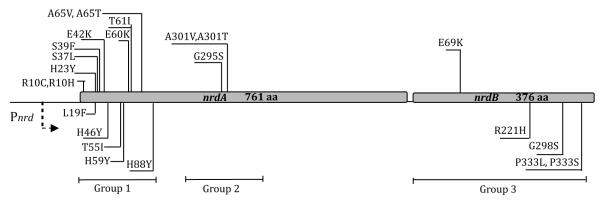
DNA sequencing of the nrdAB operon of the 118 confirmed mutator plasmids revealed a variety of single and multiple amino-acid changes in both nrdA and nrdB. Excluding redundant mutants and those with multiple substitutions, we obtained 23 unique, single amino-acid substitution mutants. As shown in Fig. 2, eighteen were in the nrdA gene encoding R1 and five were in the nrdB gene encoding R2.
Fig. 2.
RNR mutator mutants discovered in this study and their location in the nrdA and nrdB genes. See text for the functional subdivision of mutants in groups 1, 2, and 3.
3.2. Location of mutator mutations at RNR regulatory sites
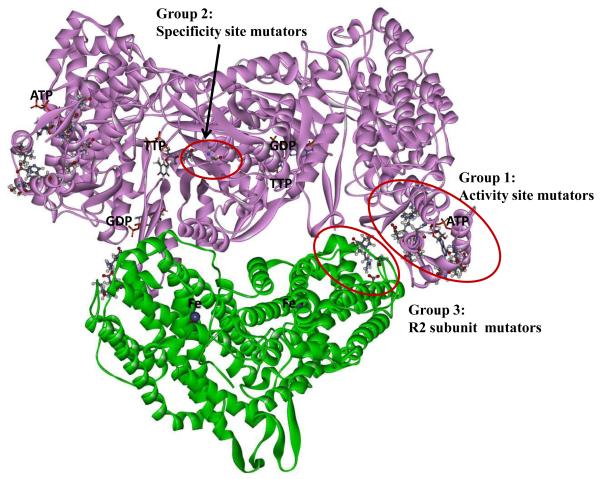
Insight into the mechanisms responsible for the mutator effects may be obtained from the location of the affected amino acid residues. Crystal structures have been described for the isolated E. coli R1 and R2 dimers [36, 38], but no structure is yet available for the R1-R2 holoenzyme. Nevertheless, it is possible to dock the R1 and R2 dimers based on the complementary shapes of the dimers, as performed in other studies [12, 36]. We performed such docking (see Materials and Methods) yielding the structure displayed in Fig. 3.
Fig. 3.
A model of the E. coli NrdAB holoenzyme with locations of discovered mutator mutations. The model is based on the reported crystal structures of the R1 [36] and R2 dimers [37], docked together into the holoenzyme as described in Materials and Methods.
It is clear from Fig. 3 that the mutator residues fall into three discrete groups. Group 1 is formed by the mutants located in and around the R1 activity site. The second group consists of the A301T, A301V, and G295S mutants; these residues are located at or near the RNR specificity site. The third group corresponds to mutants located in the R2 subunit. This is a novel location not implicated in enzyme regulation before. It is also noted that this group is located in close proximity to the group 1 mutants near the activity site. This proximity suggests that the mutator phenotype of the two groups may be based on a common mechanism. This commonality is further supported by further analyses as described below.
3.3. Two types of mutator effects for NrdAB mutators
The isolation of 23 new mutator mutants specifically when assaying for increased lacZ G·C→T·A and A·T→T·A transversions, but not when assaying for increases in other lacZ base substitutions, may reflect the innate nature of the nrdAB mutator effects, but could alternatively be due to higher sensitivity of the particular papillation screens. Therefore, we investigated the mutational specificity of each of the mutants in detail using the complete set of six lac alleles by directly measuring the lac mutant frequencies in liquid cultures. We found that the mutator phenotypes were indeed specific for the G·C→T·A and A·T→T·A transversions; none of the other transitions and transversions were significantly affected (data not shown).
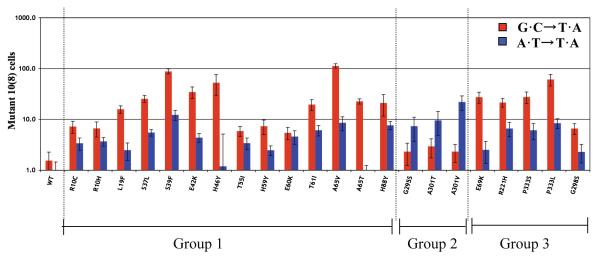
As shown in Fig. 4, the mutator effects for G·C→T·A and A·T→T·A are substantial, increasing the mutation rates an average 10 to 100 times. Furthermore, the mutators can be divided into two classes. One class (the majority) preferentially enhances G·C→T·A transversions, while the other (G295S, A301T, and A301V) preferentially enhance the A·T→T·A transversion. It is also clear from Figs. 3 and 4 that a correlation exists between the mutational specificities and the location of the mutated residues. Specifically, mutators of groups 1 and 3 specifically enhance the G·C→T·A transversions, whereas those of group 2 specifically enhance the A·T→T·A transversions.
Fig. 4.
Mutator effects of nrdAB mutator strains: two classes with differential response for lac G·C→T·A and A·T→T·A transversions and their correlation with group 1, 2, and 3 mutants. The red bar indicates the frequency of G·C→T·A transversions, the blue bar A·T→T·A transversions. The baseline values for the wild-type (wt) control were 1.6 × 10−8 (G·C→T·A) and 0.9 × 10−8 (A·T→T·A).
3.4. dNTP pool imbalances in nrdAB mutators
Quantitation of the dNTP pools in the mutator mutants revealed that, as expected, the pools were altered (see Table 1). Specifically, for group 1 and 3 mutators, there was a large increase in the levels of dCTP (6- to 37-fold) as well as dATP (2.2- to 6.7-fold) [note that in some mutants the dATP concentration reaches 60-70% of the ATP concentrations], with modest increases for dTTP and dGTP (1.5- to 3-fold). The strongest dNTP increases are seen for E42K and A65V (in the R1 subunit) or P333S (in the R2 subunit). The similar pattern of dNTP changes for group 1 and 3, along with their shared mutational specificity (see above), further strengthens the idea that the phenotypes for these groups are due to a similar mechanism. In contrast, the pool changes in the group 2 mutators (R1 specificity site) were different, consistent with a different mutational specificity (Fig. 4). In general, these changes were more modest, with increases for dTTP, dGTP and dCTP, and a decrease in the dATP level (Table 1).
Table 1.
dNTP pools in RNR mutator mutants (% of ATP)a
| RNR | Group | dATP | dTTP | dGTP | dCTP |
|---|---|---|---|---|---|
| Control | – | 10 | 6.2 | 2.3 | 2.0 |
| R10C | 1 | 38 | 14 | 4.5 | 23 |
| L19F | 1 | 24 | 8.2 | 3.4 | 23 |
| S37L | 1 | 46 | 12 | 3.5 | 36 |
| S39F | 1 | 53 | 17 | 7.5 | 31 |
| E42K | 1 | 60 | 15 | 5.3 | 51 |
| H46Y | 1 | 58 | 15 | 6.9 | 74 |
| T55I | 1 | 26 | 14 | 5.2 | 32 |
| H59Y | 1 | 31 | 11 | 3.6 | 24 |
| E60K | 1 | 28 | 8.4 | 4.4 | 24 |
| T61I | 1 | 37 | 15 | 3.8 | 17 |
| A65T | 1 | 22 | 13 | 3.7 | 13 |
| A65V | 1 | 67 | 19 | 7.7 | 41 |
| H88Y | 1 | 34 | 13 | 4.7 | 36 |
| G295S | 2 | 7.8 | 10 | 5.6 | 3.1 |
| A301T | 2 | 7.4 | 8.7 | 1.7 | 5.6 |
| A301V | 2 | 7.9 | 10 | 5.0 | 8.4 |
| E69K | 3 | 28 | 10 | 4.1 | 14 |
| R221H | 3 | 24 | 15 | 2.9 | 33 |
| G298S | 3 | 45 | 17 | 5.6 | 31 |
| P333L | 3 | 38 | 14 | 4.0 | 22 |
| P333S | 3 | 43 | 19 | 7.6 | 51 |
dNTP pools were determined as described in Materials and Methods and are expressed as a percentage of the ATP peak observed within the same HLPC profile. The presented data are based on 2 to 3 determinations for each mutant and 15 to 20 determinations for the wild-type control. The intensity of the ATP peak was reproducible within 10-20 % among the various samples, indicating that the nucleotide recovery efficiency was generally reproducible and that no major changes in ATP level were occurring among the mutants.
4. Discussion
Ribonucleotide reductase (RNR) is a crucial enzyme for DNA synthesis as it serves to provide a controlled supply of dNTPs to the DNA synthesis machinery. Despite this importance, relatively little is known about the precise extent to which RNR controls the cellular mutation rate. Our present results identifying a large, new set of E. coli mutator strains confirm the important control that RNR exerts on the cellular mutation rate and provide novel insights into the RNR feedback regulatory mechanisms.
4.1. Mutator mutants at the RNR activity site
When the mutator mutations are placed onto the model RNR holoenzyme (Fig. 3), 15 are located at the activity site, where ATP and dATP compete and dATP binding inhibits the enzyme [15, 16]. A close-up of the mutated residues within this domain is shown in Fig. 5A. The precise mechanisms by which dATP binding leads to inhibition, and how the enzyme discriminates between structurally similar ATP and dATP, are of active interest [15, 23, 39]. There are several potential mechanisms by which RNR could become refractory to dATP-mediated inhibition: lack of effector binding (dATP and ATP), loss of discrimination between ATP and dATP (recognition of the ribose 2′-OH), or impairment of any conformational changes of the (d)ATP-binding domain that serve to communicate the inhibitory signal to other portions of the enzyme.
Fig. 5.
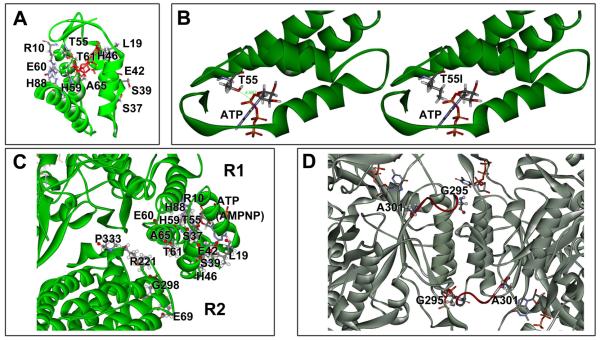
Detail views of RNR features and mutated amino acid residues. (A) Close-up of the RNR activity site (ATP cone) with residues indicated that were found to yield a mutator phenotype. (B) Possible effect of the T55I mutation on (d)ATP binding. The isoleucine side chain intrudes in the nucleotide binding area and may interfere with effector binding. (C) Close-up of the R1-R2 interaction area with R1 and R2 mutator residues indicated. The proximity of R1 and R2 mutants coupled with their similar mutational properties and dNTP pool changes support a functional interaction and common mechanism. (D) A close-up view of the RNR specificity site showing important Loop 2 (red) and the G295 and A301 residues, which yield a mutator phenotype upon mutation.
Previous studies [15, 22] have investigated the effects of the site-specifically introduced H59(A/N) and H88A substitutions, which may correspond to our H59Y and H88Y mutations. The mutant enzymes were not impaired in binding either ATP or dATP; instead, they appeared unable to discriminate effectively between these two effectors [15]. It was also noted that dATP binding is associated with a tightening of the R1-R2 interaction [15], and with formation of a higher order (octameric) oligomeric RNR [22]. Interestingly, the H59 and H88 mutant enzymes were impaired in formation of the tightly-bound state. It was hypothesized that recognition of the 2′-OH of ATP mediates a conformational change in the domain leading to formation of the active enzyme state [15, 22]. However, the precise nature of the conformational switch and downstream effects, are not known.
Surveying our mutants, it is possible that some of our mutants are impaired in (d)ATP binding. For example, as shown in Fig. 5B, the T55I mutation is predicted to restrict the ATP binding pocket by interfering with the binding of the (d)ATP α-phosphate. On the other hand, overall, the combined impact of the multiple mutator residues in this area (see Fig. 5A), encompassing each of the helices (α1, α2, α3, α4) and intervening loops of R1 [12, 36], is strongly suggestive of a broad-based conformational change of the domain upon dATP binding leading to the proposed inhibited R1-R2 complex [15].
Highly relevant to understanding this regulatory step is our discovery of a group of corresponding mutations in the R2 subunit immediately adjacent to the activity domain (E69K, R221H, G298S, P333L, and P333S) (Figs. 3 and 5C). It is reasonable to assume that their mutator effects are mediated by the same mechanism as the R1 activity-site mutants. The similarity of mechanism is supported by the similarity in the dNTP pool alterations, i.e. elevation of all four dNTPs, particularly dATP and dCTP, and by the identical mutational specificity (preferential enhancement of G·C→T·A transversions). The data suggest that this region of R2 is involved with, or responds to, the conformational change in R1, and that this region is further involved in the downstream communication of the inhibitory signal. A conformational change in R2 may disrupt the communication between the tyrosyl radical (Y221) in R2 and the critical cysteine residue (C439) in the R1 catalytic site, which proceeds by a unique long-range proton-coupled electron transfer pathway (PCET) [9, 11, 40]. Alternatively, the R2 residue changes may interfere with the formation of the higher order octameric complex [(R1)4(R2)4], which were shown to represent an inhibited enzyme state [21, 22].
4.2. Mutator mutants near the specificity site
The second important regulatory site is the RNR specificity site, where binding of ATP, dATP, dGTP, and dTTP direct the reduction of CDP, UDP, ADP and GDP, respectively, at the nearby catalytic site [24-26, 41]. Studies have revealed that the communication between the specificity and catalytic site is mediated by conformational changes including Loop 2, which changes its conformation depending upon which dNTP is bound in the specificity site, and, in this manner, determines which NDP can be accommodated in the catalytic site [24, 25, 41]. Interestingly, the three mutator mutants that we have identified in this area (G295S, A301T, A301V) affect residues located at either side of the base of the Loop 2 (see Fig. 5D). While these residues may not be directly involved in the interaction with the dNTP effectors, they likely affect the flexibility of the loop and its ability to respond correctly to the presence of dNTP effectors, disturbing the ratios of the four dNTPs. At the same time, the mutant enzymes are presumably still subject to regulation at the activity site, preventing high-level accumulation of dNTPs. This is consistent with our observation of modest dNTP pool changes in these mutators (Table 1). Recently, pool size disturbances and associated mutator phenotypes have also been observed in the yeast S. cerevisiae using site-specifically created Loop 2 mutants [27].
4.3. Correlation of dNTP pool changes with mutational specificity
One important additional goal of our effort to obtain mutants with altered dNTP pools was to better understand the mechanisms by which the dNTP levels control the mutation rate. Conclusions regarding this issue can already be drawn based on the mutational specificity changes observed with the present lacZ reversion system (Fig. 3).
The frequency of each base-substitution event depends on several factors, including the intrinsic accuracy of the polymerase for a given mispair, as different errors are made at different rates. In addition, for each mispairing, there should increase linearly with the ratio of the incorrect vs. correct dNTP, as the two directly compete for the polymerase active site [27, 43]. Then, following a misinsertion error, extension of the terminal mismatch is required in order for the error to become established as a potential mutation. Because extension of mismatches is generally a difficult (i.e., slow) step, it has to compete with removal of the mismatch by the polymerase-associated proofreading activity, which in most cases will be the predominant event. Nevertheless, at this stage, the extension rate can be specifically promoted relative to the excision rate by increased concentrations of the dNTPs, particularly the ones to be incorporated directly following the mismatch. This stimulatory effect of high dNTPs is often called the “next-nucleotide effect” [27, 43]. Thus, both a high incorrect/correct dNTP ratio and high “next” dNTP concentrations are predicted to increase the observed mutation rate.
In Table 2, we have presented for each of the six lacZ base substitutions and the two potential mispairs for each substitution that can cause them: (i) the relevant incorrect/correct dNTP pair and (ii) the immediately following (correct) dNTP. One critical question at this point is to know which of the two potential mispairs for each base substitution is the predominant (i.e., most frequent) one. Obviously, error-rate increases should be readily observable as increased mutation rate when considering the predominant mispairs, but not necessarily so for the case of the minor mispairs. In a previous study [42] - focused on investigating the differential fidelity of leading and lagging-strand replication in E. coli - certain choices for predominant mispairs were made based on published in vitro misinsertion preferences and mispair extension efficiencies for an array of DNA polymerases. For example, for A·T→G·C transitions the T·G mispair was judged to be more relevant than the A·C mispair [42]. Assuming this same preference for Pol III HE in vivo, it was concluded from the observed differential mutability of the lac allele in the two orientations on the E. coli chromosome that, for this allele, lagging-strand replication is more accurate than leading-strand replication.
Table 2.
Mispairing errors that can lead to reversion of each of the six used lacZ missense alleles and the next (correct) dNTP to be incorporated following the mismatcha.
| lac allele | Substitution | Mispair 1 | Next dNTP | Mispair 2 | Next dNTP |
|---|---|---|---|---|---|
| CC101 | A·T→C·G | A·(G/T) | dATP | T·(C/A) | dATP |
| CC102 | G·C→A·T | G·(T/C) | dCTP | C·(A/G) | dGTP |
| CC103 | G·C→C·G | G·(G/C) | dATP | C·(C/G) | dATP |
| CC104 | G·C→T·A | G·(A/C) | dGTP | C·(T/G) | dCTP |
| CC105 | A·T→T·A | A·(A/T) | dGTP | T·(T/A) | dCTP |
| CC106 | A·T→G·C | A·(C/T) | dATP | T·(G/A) | dATP |
The format for the mispairs is: Template Base·(Wrong Base/Correct Base). In bold, the assumed major mispairs deduced for the G·C→A·T, G·C→T·A, A·T→T·A, and A·T→G·C substitutions, as described in the text. The wild-type DNA sequence at the site of lac reversion is 5′-AAT GAG AGT-3′, while in strains CC101 through CC106 the GAG codon (encoding the essential glutamic acid residue) is replaced by TAG, GGG, CAG, GCG, GTG, or AAG, respectively. Only the single-base change to GAG will restore the Lac+ phenotype (Cupples and Miller 1989). See text for further details.
Importantly, application of this principle to all four lac alleles tested in the study (G·C→A·T, G·C→T·A, A·T→T·A, and A·T→G·C) yielded a consistent result, namely that, in all cases, lagging-strand replication was the more accurate (by several fold) [42]. The consistency of these results, along with the results of several follow-up studies in which inversions of strand specificities were observed (for example, for the case of the SOS mutator effect due to Pol V making lagging-strand replication more error-prone) [44-49], reinforced these choices for dominant mispairs (bolded in Table 2). As shown below, the present results can also be interpreted most plausibly in terms of these particular mispairs.
For further illustration, Fig. 6 displays the lacZ sequence at which the mutations occur. The G·C→T·A transversions (upper line) occur primarily by C·T mispairings (template base underlined), while A·T→T·A (lower line) occur by T·T mispairings [42]. The activity-site mutators (in both R1 and R2) preferentially induce the lac G·C→T·A transversions. The dNTP pool data (Table 1) indicate that their misinsertion rate for this transversion, as driven by the ratio of incorrect vs. correct dNTP (in this case, the dTTP/dGTP ratio), is not significantly altered. On the other hand, their strong increases for dCTP and dATP (up to 25-fold and 4.5-fold, respectively) (Table 1), which are the nucleotides to be incorporated following the misinsertion, are expected to strongly promote the extension (and hence, survival) of the mismatch, avoiding in this manner its removal by exonucleolytic proofreading (next-nucleotide effect). Thus, these data are fully consistent with the observed 10- to 70-fold increase in the lac G·C→T·A frequency. For the A·T→T·A transversions, extension of the T·T mismatch is expected to be promoted, likewise, by the increased dCTP and dATP. However, here, the T·T misinsertion rate is reduced as the relevant incorrect/correct ratio (dTTP/dATP) is decreased by up to 2-fold. Hence, the more modest increase in A·T→T·A transversions for the activity mutators is plausibly accounted for.
Fig. 6.

dNTP pool changes predict mutational specificity. The diagrams show the lacZ sequence at which the G·C→T·A (top) or A·T→T·A (bottom) mutations occur. The red T indicates the misinserted ‘incorrect’ dTTP opposite the relevant template base (underlined), while the blue G or A indicates the ‘correct’ dGTP or dATP competing with the dTTP at the insertion step. The arrows represent the subsequent incorporation of dCTP (next nucleotide) and dATP (next-next nucleotide), which may protect the misinserted base against their exonucleolytic removal. See text for a full description.
The mutational specificity for the specificity-site mutators (G295S, A301V, A301T) is different, A·T→T·A transversions being enhanced preferentially over G·C→T·A (Fig. 3). This finding can also be reconciled with the pool data. For the A·T→T·A, the relevant dTTP/dATP ratio is increased by about 2-fold, while for the G·C→T·A the relevant dTTP/dGTP ratio is decreased by about 2-fold. At the same time, the next nucleotide, dCTP, is enhanced up to 4-fold, promoting mismatch extension.
Note that analysis of the mutator effects in terms of the alternative mispairs (for example, G·A instead of C·T for the G·C→T·A transversions) would not yield a satisfactory result. For the group 1 and 3 mutators, which enhance the G·C→T·A transversions, the incorrect/correct ratio (dATP/dCTP) would be decreased 5- to 10-fold, a clearly antimutagenic effect, while little stimulation would be expected from the small increase in the next nucleotide, dGTP. Table 2 also indicates that dCTP as next nucleotide is predicted to enhance extension of G·T mispairs for the case of the G·C→A·T transitions (CC102 allele). No mutator effect was observed for this allele. However, it should be noted the incorrect/correct ratio for this allele (dTTP/dCTP) is strongly reduced by the highly elevated dCTP level, presumably masking any mutagenic effects.
The correlations described above indicate that alterations of in vivo dNTP pools can be used to study the mechanisms of DNA replication fidelity and mutagenesis. Most importantly, the concentrations of the next correct dNTPs appear to be a main determinant of E. coli replication fidelity. This highlights the important role that exonucleolytic proofreading plays in avoiding mutations resulting from polymerase misinsertion errors [50-52].
4.4. Significance of mutator mutants with respect to other class I RNRs
The amino-acid sequences among the class I RNRs from different species show significant similarities [12]. Alignment of R1 and R2 sequences from E. coli, phage T4, yeast, mouse, and human (Figure S1) shows that 6 out of the 15 R1 residues in our study (R10, T55, A65, G295, and A301) and 3 out of 4 mutated R2 residues (E69, R221, and P333) are fully conserved throughout the species. Thus, the significance of the current findings on RNR, including changes in dNTP pools and mutator effects, is expected to extend to other prokaryotic and eukaryotic systems [5].
Fig. S1. Alignment of RNR R1 (A) and R2 (B) sequences from different species. Different colors indicate different degrees of conservation. Rectangles indicate the residues that were found to yield mutator phenotype when altered. Sequences were obtained from GenBank (NCBI) and alignments were generated using the program VectorNTI (Invitrogen). nrdA and nrdB represent the E. coli ribonucleotide reductase gene.
Supplementary Material
Highlights.
>Mutator mutants of E. coli are described with altered Ribonucleotide Reductase (RNR)
>RNR Mutants display altered dNTP pools and corresponding mutational specificities
>Mutants are found at the enzyme’s allosteric activity and specificity sites
>A novel regulatory important region is discovered in R2 subunit of RNR
Acknowledgements
We thank Drs. W. Copeland, M. Resnick, and S. Wilson of the NIEHS for their helpful comments on the manuscript for this paper and Dr. J. Stubbe and her laboratory for the coordinates of the Uhlin and Eklund model. This work was supported by project Z01 ES065086 of the Intramural Research Program of the National Institute of Environmental Health Sciences (NIEHS).
Footnotes
Conflict of Interest. The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kunz BA. Mutagenesis and deoxyribonucleotide pool imbalance. Mutat. Res. 1988;200:133–147. doi: 10.1016/0027-5107(88)90076-0. [DOI] [PubMed] [Google Scholar]
- [2].Kunz BA, Kohalmi SE, Kunkel TA, Mathews CK, McIntosh EM, Reidy JA. International Commission for Protection Against Environmental Mutagens and Carcinogens. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat. Res. 1994;318:1–64. doi: 10.1016/0165-1110(94)90006-x. [DOI] [PubMed] [Google Scholar]
- [3].Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20:1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- [4].Bornstein B, Area E, Flanigan KM, Ganesh J, Jayakar P, Swoboda KJ, Coku J, Naini A, Shanske S, Tanji K, Hirano M, DiMauro S. Mitochondrial DNA depletion syndrome due to mutations in the RRM2B gene. Neuromuscul. Disord. 2008;18:453–459. doi: 10.1016/j.nmd.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- [6].Nishigaki Y, Marti R, Copeland WC, Hirano M. Site-specific somatic mitochondrial DNA point mutations in patients with thymidine phosphorylase deficiency. J. Clin. Invest. 2003;111:1913–1921. doi: 10.1172/JCI17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Song S, Wheeler LJ, Mathews CK. Deoxyribonucleotide pool imbalance stimulates deletions in HeLa cell mitochondrial DNA. J. Biol. Chem. 2003;278:43893–43896. doi: 10.1074/jbc.C300401200. [DOI] [PubMed] [Google Scholar]
- [8].Nordlund P, Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- [9].Stubbe J, Nocera DG, Yee CS, Chang MC. Radical initiation in the class I ribonucleotide reductase: long-range proton-coupled electron transfer? Chem. Rev. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- [10].Chang MC, Yee CS, Stubbe J, Nocera DG. Turning on ribonucleotide reductase by light-initiated amino acid radical generation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6882–6887. doi: 10.1073/pnas.0401718101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nordlund P, Sjoberg BM, Eklund H. Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature. 1990;345:593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- [12].Eklund H, Uhlin U, Farnegardh M, Logan DT, Nordlund P. Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol. 2001;77:177–268. doi: 10.1016/s0079-6107(01)00014-1. [DOI] [PubMed] [Google Scholar]
- [13].Stubbe J. Ribonucleotide reductases: the link between an RNA and a DNA world? Curr. Opin. Struct. Biol. 2000;10:731–736. doi: 10.1016/s0959-440x(00)00153-6. [DOI] [PubMed] [Google Scholar]
- [14].Sjöberg B, Sahlin M. Encyclopedia of life sciences. John Wiley & Sons, Ltd.; 2005. Radical enzymes. [Google Scholar]
- [15].Birgander PL, Kasrayan A, Sjoberg BM. Mutant R1 proteins from Escherichia coli class Ia ribonucleotide reductase with altered responses to dATP inhibition. J. Biol. Chem. 2004;279:14496–14501. doi: 10.1074/jbc.M310142200. [DOI] [PubMed] [Google Scholar]
- [16].Brown NC, Reichard P. Role of effector binding in allosteric control of ribonucleoside diphosphate reductase. J. Mol. Biol. 1969;46:39–55. doi: 10.1016/0022-2836(69)90056-4. [DOI] [PubMed] [Google Scholar]
- [17].Augustin LB, Jacobson BA, Fuchs JA. Escherichia coli Fis and DnaA proteins bind specifically to the nrd promoter region and affect expression of an nrd-lac fusion. J. Bacteriol. 1994;176:378–387. doi: 10.1128/jb.176.2.378-387.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- [19].Gon S, Camara JE, Klungsoyr HK, Crooke E, Skarstad K, Beckwith J. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006;25:1137–1147. doi: 10.1038/sj.emboj.7600990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Torrents E, Grinberg I, Gorovitz-Harris B, Lundstrom H, Borovok I, Aharonowitz Y, Sjoberg BM, Cohen G. NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J. Bacteriol. 2007;189:5012–5021. doi: 10.1128/JB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fairman JW, Wijerathna SR, Ahmad MF, Xu H, Nakano R, Jha S, Prendergast J, Welin RM, Flodin S, Roos A, Nordlund P, Li Z, Walz T, Dealwis CG. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol. 2011;18:316–322. doi: 10.1038/nsmb.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rofougaran R, Crona M, Vodnala M, Sjoberg BM, Hofer A. Oligomerization status directs overall activity regulation of the Escherichia coli class Ia ribonucleotide reductase. J. Biol. Chem. 2008;283:35310–35318. doi: 10.1074/jbc.M806738200. [DOI] [PubMed] [Google Scholar]
- [23].Reichard P, Eliasson R, Ingemarson R, Thelander L. Cross-talk between the allosteric effector-binding sites in mouse ribonucleotide reductase. J. Biol. Chem. 2000;275:33021–33026. doi: 10.1074/jbc.M005337200. [DOI] [PubMed] [Google Scholar]
- [24].Larsson KM, Jordan A, Eliasson R, Reichard P, Logan DT, Nordlund P. Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase. Nat Struct Mol Biol. 2004;11:1142–1149. doi: 10.1038/nsmb838. [DOI] [PubMed] [Google Scholar]
- [25].Xu H, Faber C, Uchiki T, Fairman JW, Racca J, Dealwis C. Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4022–4027. doi: 10.1073/pnas.0600443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Reichard P. Ribonucleotide reductases: substrate specificity by allostery. Biochem. Biophys. Res. Commun. 2010;396:19–23. doi: 10.1016/j.bbrc.2010.02.108. [DOI] [PubMed] [Google Scholar]
- [27].Kumar D, Abdulovic AL, Viberg J, Nilsson AK, Kunkel TA, Chabes A. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2010;39:1360–1370. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cupples CG, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fijalkowska IJ, Schaaper RM. Effects of Escherichia coli dnaE antimutator alleles in a proofreading-deficient mutD5 strain. J. Bacteriol. 1995;177:5979–5986. doi: 10.1128/jb.177.20.5979-5986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- [32].Miller J. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1992. [Google Scholar]
- [33].Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li L, Chen R, Weng Z. RDOCK: refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- [35].Pierce B, Weng Z. ZRANK: reranking protein docking predictions with an optimized energy function. Proteins. 2007;67:1078–1086. doi: 10.1002/prot.21373. [DOI] [PubMed] [Google Scholar]
- [36].Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- [37].Tong W, Burdi D, Riggs-Gelasco P, Chen S, Edmondson D, Huynh BH, Stubbe J, Han S, Arvai A, Tainer J. Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography. Biochemistry. 1998;37:5840–5848. doi: 10.1021/bi9728811. [DOI] [PubMed] [Google Scholar]
- [38].Nordlund P, Eklund H. Structure and function of the Escherichia coli ribonucleotide reductase protein R2. J. Mol. Biol. 1993;232:123–164. doi: 10.1006/jmbi.1993.1374. [DOI] [PubMed] [Google Scholar]
- [39].Kashlan OB, Cooperman BS. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry. 2003;42:1696–1706. doi: 10.1021/bi020634d. [DOI] [PubMed] [Google Scholar]
- [40].Stubbe J. Di-iron-tyrosyl radical ribonucleotide reductases. Curr. Opin. Chem. Biol. 2003;7:183–188. doi: 10.1016/s1367-5931(03)00025-5. [DOI] [PubMed] [Google Scholar]
- [41].Uppsten M, Farnegardh M, Jordan A, Eliasson R, Eklund H, Uhlin U. Structure of the large subunit of class Ib ribonucleotide reductase from Salmonella typhimurium and its complexes with allosteric effectors. J. Mol. Biol. 2003;330:87–97. doi: 10.1016/s0022-2836(03)00538-2. [DOI] [PubMed] [Google Scholar]
- [42].Fijalkowska IJ, Jonczyk P, Tkaczyk MM, Bialoskorska M, Schaaper RM. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10020–10025. doi: 10.1073/pnas.95.17.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- [44].Maliszewska-Tkaczyk M, Bialoskorska M, Jonczyk P, Schaaper RM, Fijalkowska IJ. The SOS mutator activity: unequal mutagenesis of leading and lagging strands. Proc. Natl. Acad. Sci. U. S. A. 2000;97:12678–12683. doi: 10.1073/pnas.220424697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Banach-Orlowska M, Fijalkowska IJ, Schaaper RM, Jonczyk P. DNA Polymerase II as a fidelity factor in chromosomal DNA synthesis in E. coli. Mol. Microbiol. 2005;58:61–70. doi: 10.1111/j.1365-2958.2005.04805.x. [DOI] [PubMed] [Google Scholar]
- [46].Kuban W, Banach-Orlowska M, Bialoskorska M, Lipowska A, Schaaper RM, Jonczyk P, Fijalkowska IJ. Mutator phenotype resulting from Pol IV overproduction in E. coli: preferential mutagenesis on the lagging strand. J. Bacteriol. 2005;187:6862–6866. doi: 10.1128/JB.187.19.6862-6866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Makiela-Dzbenska K, Jaszczur M, Banach-Orlowska M, Jonczyk P, Schaaper RM, Fijalkowska IJ. Role of Escherichia coli DNA polymerase I in chromosomal DNA replication fidelity. Mol. Microbiol. 2009;74:1114–1127. doi: 10.1111/j.1365-2958.2009.06921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gawel D, Jonczyk P, Fijalkowska IJ, Schaaper RM. The dnaX36 mutator of Escherichia coli: effects of the DNA Polymerase III holoenzyme τ subunit on chromosomal DNA replication fidelity. J. Bacteriol. 2011;193:296–300. doi: 10.1128/JB.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fijalkowska IJ, Schaaper RM, Jonczyk P. DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair. FEMS Microbiol. Reviews. 2012 doi: 10.1111/j.1574-6976.2012.00338.x. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pham PT, Olson MW, McHenry CS, Schaaper RM. The base substitution and frameshift fidelity of Escherichia coli DNA polymerase III holoenzyme in vitro. J. Biol. Chem. 1998;273:23575–23584. doi: 10.1074/jbc.273.36.23575. [DOI] [PubMed] [Google Scholar]
- [51].Gawel D, Hamilton MD, Schaaper RM. A novel mutator of Escherichia coli carrying a defect in the dgt gene, encoding a dGTP triphosphohydrolase. J. Bacteriol. 2008;190:6931–6939. doi: 10.1128/JB.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wheeler LJ, Rajagopal I, Mathews CK. Stimulation of mutagenesis by proportional deoxyribonucleoside triphosphate accumulation in Escherichia coli. DNA Repair (Amst) 2005;4:1450–1456. doi: 10.1016/j.dnarep.2005.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.