Abstract
Gliomas are the most frequent adult primary brain tumor, and are invariably fatal. The most common diagnosis glioblastoma (GBM) afflicts 12,500 new patents in the U.S. annually, and has a median survival of approximately one year when treated with the current standard of care. Alkylating agents have long been central in the chemotherapy of GBM and other gliomas. The DNA repair protein O6-methylguanine-DNA methyltransferase (MGMT), the principal human activity that removes cytotoxic O6-alkylguanine adducts from DNA, promotes resistance to anti-glioma alkylators, including temozolomide and BCNU, in GBM cell lines and xenografts. Moreover, MGMT expression assessed by immunohistochemistry, biochemical activity or promoter CpG methylation status is associated with the response of GBM to alkylator-based therapies, providing evidence that MGMT promotes clinical resistance to alkylating agents. These observations suggest a role for MGMT in directing adjuvant therapy of GBM and other gliomas. Promoter methylation status is the most clinically tractable measure of MGMT, and there is considerable enthusiasm for exploring its utility as a marker to assign therapy to individual patients. Here, we provide an overview of the biochemical, genetic and biological characteristics of MGMT as they relate to glioma therapy. We consider current methods to assess MGMT expression and discuss their utility as predictors of treatment response. Particular emphasis is given to promoter methylation status and the methodological and conceptual impediments that limit its use to direct treatment. We conclude by considering approaches that may improve the utility of MGMT methylation status in planning optimal therapies tailored to individual patients.
Keywords: alkylating agents, biomarker, glioblastoma, glioma, MGMT, chemotherapy resistance
1. Introduction
The protein O6-methylguanine-DNA methyltransferase is one of the most extensively studied DNA repair activities [1,2]. First discovered in bacteria in 1977 [3], the activity was described shortly thereafter in rodent and human cells [1]. Subsequent work revealed MGMT1 to have biochemical and biological properties of translational significance (Table 1), including silencing of expression mediated by methylation of deoxycytidine in CpG dinucleotides in the MGMT promoter [4,5]. Promoter methylation is associated with prolonged progression-free and overall survival in newly diagnosed glioblastomas (WHO grade IV) treated with temozolomide during and after the completion of radiotherapy [6], the present standard of care; hence MGMT is the first DNA repair protein found to be predictive of treatment response in GBM. MGMT promoter methylation status is also predictive in low-grade and anaplastic gliomas treated with other alkylating agent regimens [e.g.,7,8]. While these findings have suggested that MGMT promoter status can be used for treatment stratification of gliomas, important questions about the clinical utility and biological significance of MGMT promoter status remain to be resolved. We will address these questions in this review. We will first describe the biochemical properties and biological activities that suggest MGMT as a biomarker for alkylating agent response in gliomas. We will then summarize the major studies that have sought association of MGMT protein expression, biochemical activity or promoter methylation status with response to alkylator therapy. The discussion will include an examination of the uncertainties that currently limit use of MGMT as a predictive and prognostic marker to guide treatment.
2. Biochemical, genetic and biological characteristics
2.1. Overview
The rational development of individualized treatments for GBM and other gliomas requires identifying tumor-specific biomarkers that predict clinical outcome. As discussed in this section, MGMT possesses biochemical, genetic and biological features that recommend it for such a role.
2.2. Biochemical function
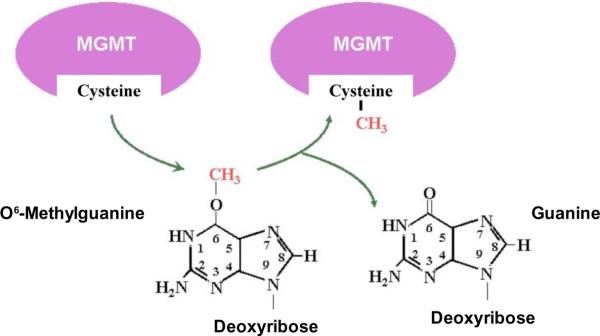
Human MGMT is a monomeric 22 kD protein, and is apparently the sole human repair activity that excises methyl adducts from the O6 position of guanine in DNA [1,2,9]. The preferred substrate is O6-methylguanine in double-stranded DNA, but the protein also removes larger aliphatic and chloroethyl adducts. Structure-function studies indicate that MGMT binds to the minor groove of DNA and detects O6-alkylguanine adducts by a base flipping mechanism that rotates the alkylated base out of the DNA helix and into the active site of the protein. The alkyl adduct is then covalently transferred to an active site cysteine residue (Fig. 1), yielding S-alkylcysteine and guanine [2]. This mechanism differs from the pathways that excise most other DNA adducts in that repair is carried out by a single protein without cleavage of the phosphodiester backbone. MGMT is unique among DNA repair activities in that the active site cannot be regenerated, and alkylation of the protein targets it for ubiquitination and subsequent degradation. This “suicide” mechanism implies that the number of O6-alkylguanine adducts that can be removed from DNA in vivo is limited by the number of MGMT molecules in cells and the rate of de novo synthesis of the protein. This distinctive property has stimulated a search for pseudosubstrate analog inhibitors that could be used to deplete tumor cells of MGMT during alkylator therapy. While a large number of potential inhibitors has been described [2,10], to date none has proved to be unequivocally suitable for clinical use [11].
Figure 1. Repair of O6-alkylguanine adducts by MGMT.
MGMT mediates a stoichiometric reaction in which methyl and other alkyl groups bound to the O6 position of guanine in DNA are via a thioester linkage to a cysteine residue in the active site. Alkylation of MGMT leads to rapid degradation of the protein. Thus, the capacity of a cell to remove O6-alkylguanine from DNA reflects the rate at which adducts are produced by exogenous and endogenous agents and the rate of synthesis of new protein. The “suicide” reaction mechanism of MGMT is unique among human DNA repair enzymes, and suggests the use of substrate analog inhibitors of MGMT in order to increase sensitivity to chemotherapeutic alkylating agents. Image adapted from www.mgmt-agt.net/whatismgmt.htm.
2.3. Physiological function
MGMT prevents the genotoxic effects of O6-alkylguanine adducts produced by exogenous and endogenous alkylators in mammalian and human cells [2,9,12]. MGMT activity varies widely among normal tissues, with brain usually having low levels of expression, and also varies widely among individuals. Notably, MGMT is not necessary for mammalian development as evidenced by the viability of MGMT knockout mice [13]. Knockout animals, however, display heightened sensitivity to the deleterious effects of alkylating agents, including therapeutic alkylators: These findings are immediately relevant to neuro-oncology, demonstrating that MGMT promotes resistance to O6-alkylguanine-induced cytotoxicity in mammalian cells in vivo [e.g., 10,14]. Also relevant is the role demonstrated for MGMT in preventing alkylator-induced carcinogenesis. O6-alkylguanines are potent mutagenic lesions that characteristically produce point mutations, as well as gross chromosomal deletions and rearrangements that are associated with tumor formation and malignant progression [9]. It has long been recognized that O6-methyl and O6-ethylguanine are strong neurocarcinogens in rodents and primates [15,16]. Human exposure to endogenous and exogenous alkylators is believed to be continuous and life-long and there is epidemiologic evidence implicating environmental alkylator exposure with increased risk for human primary brain tumors, including adult gliomas [17 and refs therein]. As discussed below, histologically normal brain of glioma patients is more likely to lack detectable MGMT activity than brain from tumor-free individuals [18,19], suggesting that MGMT acts as a tumor suppressor in the central nervous system.
2.4. Gene structure
The 170 kb human MGMT gene is located on the distal end of the long arm of chromosome 10 at 10q26 and contains 5 exons, the first of which is untranslated [12]. The maximal MGMT promoter extends from position −953 to +202 and contains a minimal promoter (−69 to +19) as well as an enhancer (+143 to +202) that binds the MGMT enhancer binding protein (MEBP). The protein is sequestered in the cytoplasm of MGMT-deficient but not MGMT-proficient cells, suggesting a role in regulating MGMT expression [20].
The MGMT promoter has a CpG island containing 97 CpG sites, the majority of which are in a region extending from ~300 nt 5' to the transcription start site through the first exon [21]. Promoter CpG islands, defined as regions 0.3 to 3 kb in length with CpG content greater than 60%, are hallmarks of genes subject to epigenetic silencing of expression that is mediated by methylation at the 5 position of cytosine in CpG pairs [22]. Ten binding sites for the transcription promoting protein Sp1 are found in the promoter. Binding of Sp1, which is subject to regulation by p53 [23], stimulates transcription, suggesting a mechanism by which MGMT expression may be regulated in response to DNA damage. In addition, the promoter contains two stress-responsive transcription factor (e.g., Jun, Fos) binding AP-1 sites and two glucocorticoid response elements. While these features suggest that MGMT expression is responsive to genomic insult and steroids, only modest (2- to 4-fold), transient elevations of MGMT mRNA content have been reported in rat hepatoma cells exposed to DNA damaging agents, including X-rays and alkylators, and to dexamethasone [24]. Moreover, we believe there is no convincing evidence to date that MGMT is inducible by DNA damaging agents or steroids in human cells.
2.5. Regulation of MGMT expression in human tumor-derived cell lines
Approximately 20% to 30% of cell lines derived from diverse human tumors and 50% of virally transformed cell lines do not have detectable MGMT activity [12]. These cell lines possess an intact MGMT gene as well as the trans-acting factors requisite for transcription, but contain neither MGMT mRNA nor protein, indicating that transcriptional silencing underlies the absence of MGMT activity. Gene silencing in tumor cell lines is commonly accompanied by hypermethylation of promoter CpG islands, suggesting that methylation mediates MGMT silencing in human tumors [25,26]. In accord, an early study found that the level of methylation of 21 of 25 CpG sites in the MGMT promoter was inversely associated with activity in a panel of human-derived cell lines [27]. Additional experiments using methylation-sensitive restriction endonucleases revealed methylation of 8 promoter CpG dinucleotides in MGMT-deficient but not MGMT-proficient glioma cell lines [21,28,29]. Sequence analysis of bisulfite-treated DNA -- bisulfate converts cytosine, but not 5-methylcytosine, to uracil (Fig. 2) -- from MGMT-deficient and -proficient colorectal tumor [21] and myeloma cell lines [28], revealed 3 clusters of CpG sites, one in the first exon and two 5' to the transcription start site, that were heavily methylated in MGMT-deficient lines and essentially unmethylated in MGMT-proficient lines. In agreement, sequencing all MGMT promoter CpGs following bisulfite treatment in 19 MGMT-deficient human tumor cell lines, including 3 from primary brain tumors, showed two heavily methylated CpG clusters, one in the minimal promoter region of exon 1 and the other encompassing the enhancer region 5' to the transcription start site [30]. Subsequent work using methylation-specific PCR of bisulfite-treated DNA (Fig. 3) from glioma cell lines and xenografts substantiated these earlier results [e.g., 31,32]. That promoter methylation has a mechanistic rather than merely correlative association with the MGMT-deficient phenotype is supported by two findings. First, demethylation by long-term exposure to 5-azacytidine induced MGMT expression in one of three MGMT-deficient cell lines [21]. Second, CpG methylation of the MGMT promoter in transfected reporter gene constructs inhibited promoter activity [33]. Additional studies have suggested that methylation-related chromatin structure affects MGMT expression by influencing access of transcription factors to the promoter [34]. More recently, CpG methylation has been shown to promote binding of transcription-repressing methyl-CpG binding proteins in MGMT-deficient human tumor cell lines [30,35]. Binding of these proteins condenses chromatin structure, suppressing gene expression by excluding transcription factors [e.g., 30].
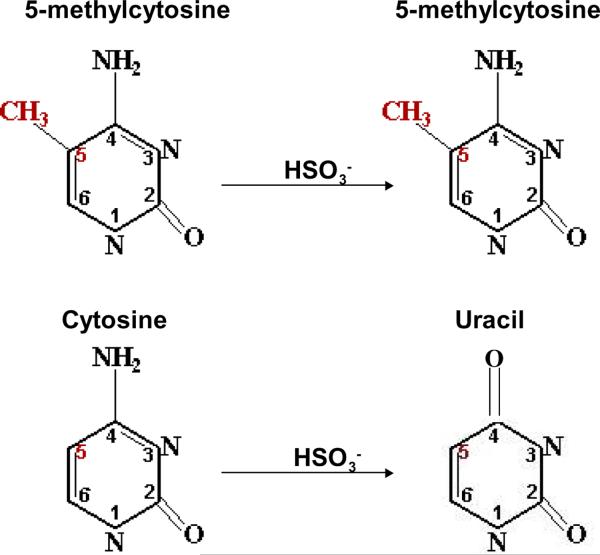
Figure 2. Differential sensitivity of 5-methylcytosine and cytosine to bisulfite-mediated deamination.
5-methylcytosine is insensitive to incubation with bisulfite at mildly acidic pH. In contrast, bisulfite treatment promotes the hydrolysis of cytosine, producing uracil and ammonia. This differential sensitivity has been exploited to detect 5-methylcytosine in DNA. Image adapted from www.imb-jena.de/~sweta/renzymes.
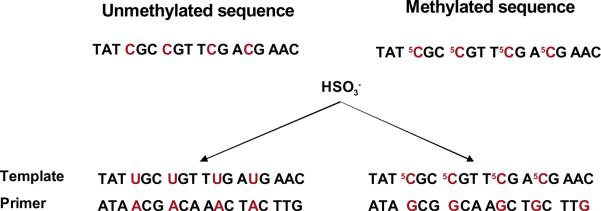
Figure 3. Methylation-specific PCR (MSP).
Bisulfite treatment converts cytosine in DNA to uracil (left) while leaving 5-methylcytosine unaltered (right). The uracil- and 5-methylcytosine-containing template sequences hybridize to unique primers to yield distinct products during PCR amplification.
2.6. MGMT content of human gliomas and normal brain
Most human tumor specimens express MGMT activity [12]. However, early studies revealed that an appreciable fraction of gliomas lack detectable biochemical activity [19,36 and refs therein]. For example, a survey of 174 adult gliomas revealed a 300-fold range of MGMT activity in 76% of samples, with 24% exhibiting no detectable activity, defined as < 0.25 fmol/106 cells or 151 molecules/cell [36]. Another study reported that 7 of 40 (18%) newly operated GBMs had undetectable MGMT activity [37]. In a more recent examination of 149 GBM and anaplastic (i.e., grade 3) gliomas, MGMT activity varied ~220-fold (0.26–57 fmol/106 cells; mean ± SD = 8.4 ± 10 fmol/106 cells or ~5,000 MGMT molecules/cell); 15 tumors (10%) had no detectable activity (Bobola & Silber, in preparation). These data indicate that the MGMT-deficient phenotype is relatively frequent in GBM and may identify a sub-set of tumors that are more sensitive to alkylating agent-based chemotherapy. In accord with this conclusion is the finding that the frequency of MGMT-deficient gliomas was 7-fold lower (4% vs 29%) among tumors recurring after alkylator therapy compared to newly operated tumors. In contrast, prior radiotherapy, which does not cause alkylation, had no effect on the fraction of MGMT-deficient gliomas [19]. More recently, Kaina and colleagues examined the effect of adjuvant therapy on MGMT activity in a sample of 40 GBM [37]. They found that a statistically significant 2-fold elevation of mean MGMT activity accompanied initial recurrence after radiotherapy and alkylating agent chemotherapy compared to newly operated tumors. No statistically significant change in activity accompanied recurrence after treatment with radiation alone. These results strongly suggest that selection for cells expressing higher levels of MGMT accompanies alkylator therapy, and support the hypothesis that MGMT promotes alkylating agent resistance in vivo.
It has been also reported that histologically normal brain adjacent to gliomas displayed MGMT-deficient phenotype in 55%–65% of cases [18,19]. In contrast, normal brain from individuals without glioma had undetectable MGMT activity in 12% of cases, a finding suggesting that absence of MGMT may be a risk factor for human gliomagenesis. Subsequent work indicated that MGMT-deficiency in brain may arise during development [38]. In this study, 76% of 6- to 8-week old embryonic brain specimens lacked detectable MGMT activity. This fraction decreased with developmental age such that 13% of 16–18 week fetal brain specimens were MGMT-deficient, essentially the same fraction as observed in adult normal brain from patients without glioma. These results suggest that lack of MGMT in the normal brain of a minority of individuals represents the persistence of a pre-natal phenotype, and that absent MGMT may be an epigenetically determined susceptibility factor for gliomagenesis. Whether the absence of MGMT activity in normal brain adjacent to gliomas reflects silencing by promoter methylation or atypical regulation by transcription factors during development, and/or other processes, remains to be elucidated.
2.7. MGMT promotes resistance to clinical alkylating agents in glioma cells
It is well established that MGMT promotes resistance to the methylating imidazole derivative TMZ, and to nitrosourea-derived methylating (e.g., procarbazine) and chloroethylating (e.g., BCNU, CCNU) agents in glioma cell lines [e.g., 39–41] and xenograft models of human gliomas [e.g., 29,42]. This conclusion is based on extensive results with cells that either lack MGMT expression, or in which MGMT activity was ablated by treatment with substrate analog inhibitors (e.g., O6-benzylguanine, lomeguatrib) [10,11,32]; in all cases the absence of MGMT was accompanied by greater sensitivity to methylator and chloroethylator cytotoxicity. The extensive findings in glioma cells are concordant with a larger body of evidence obtained in other human and mammalian cells (e.g., [9], and references therein, from Kaina's group). The lethality of O6-methylguanine and O6-chloroethylguanine is emphasized by the fact that both constitute only a small fraction (~7% and 3.5%, respectively) of the base adducts produced by their respective alkylators [43–45]. The repair of these minority lesions by MGMT prevents interruption of DNA replication, thus avoiding the genesis of double-strand breaks, potently cytotoxic lesions that induce apoptosis [9].
The mechanism by which O6-methylguanine induces cytotoxicity has been the subject of intensive investigation [9,46]. The current consensus reflects the observations that replicative DNA polymerases do not stall at O6-methylguanine but incorporate deoxycytidine or thymidine opposite the lesion with approximately equal frequency. In either case, O6-meG does not correctly base pair with the newly incorporated nucleotide, producing a single nucleotide mispair substrate for mismatch repair. Mismatch repair excises a long strand of newly synthesized DNA that encompasses the mispair. Subsequent DNA repair synthesis to fill the gap produces yet another mispair at the adduct remaining in the template strand, thus again eliciting the action of mismatch repair. This futile cycle of abortive attempts to resolve the mispair has the effect of producing a persistent single-strand gap that is converted into an apoptosis-inducing double-strand break during DNA replication in the next S-phase. This model provides a mechanism to account for the resistance to methylating agents that accompanies loss of mismatch repair [46]. Single-strand gap formation can also account for the radiosensitizing effect of O6-methylguanine in DNA, as these gaps are easily converted to DNA double-strand breaks by attack of radiation-induced oxidative free radicals [41].
O6-chloroethylguanine rapidly undergoes rearrangement to form three lesions: O6-hydroxyethylguanine, O6-aminoethylguanine and the exocyclic ethano adduct 1-O6-ethanoguanine [47,48]. The latter adduct slowly undergoes an intramolecular rearrangement to form the 1-(1-guanyl)-2-(3-cytosinyl)ethane inter-strand crosslink, a physical barrier to DNA replication. Stalled DNA replication forks are sites of double-strand break formation during the subsequent round of DNA replication [9]. It is well established that MGMT-mediated resistance to chloroethylating agents is accompanied by reduced crosslink formation. Less certain is whether this is accomplished by the interaction of MGMT with O6-chloroethylguanine or with 1,O6-ethanoguanine. Data from Brent's group indicates that MGMT reacts with the ethano intermediate to produce a DNA-protein crosslink that is excised by nucleotide excision repair [49].
3. MGMT as a predictive and prognostic marker
3.1. Overview
While the contribution of MGMT activity to glioma cell alkylating agent resistance in cell lines and xenografts is firmly documented, evidence that MGMT is a clinical resistance mechanism requires establishment that MGMT expression is inversely associated with progression-free and overall survival following alkylator therapy. At present, this relationship has been established retrospectively for GBM [e.g., [50], and prospective analysis is ongoing (RTOG 0525 trial) [51]. Numerous studies have examined MGMT protein expression visualized by immunohistochemistry, activity measured by biochemical assay, and promoter CpG methylation status determined by a variety of methods that exploit the bisulfite-induced deamination of cytosine, but not 5-methylcytosine, to uracil (Table 2; Fig. 2). However, whether any of these measures of MGMT expression can be used to individualize treatment remains an open question. As discussed below, they have limitations that potentially confound biological interpretation and compromise clinical utility.
3.2. Immunohistochemistry
Examination of paraffin-embedded, formalin-fixed tumor sections by IHC offers a clinically convenient approach to determining MGMT expression in human gliomas. To date, the utility of MGMT expression, assessed by IHC, as a marker of progression-free and overall survival remains to be unequivocally demonstrated. Some studies have found an inverse association between immunopositivity for MGMT and outcome. The earliest of these studies used quantitative immunofluorescence microscopy to evaluate MGMT content in 99 GBM and 47 anaplastic astrocytomas that received BCNU during adjuvant treatment [52]. This method is notable in that it quantifies the number of MGMT molecules per nucleus rather than the fraction of immunopositive cells. Tumors with MGMT expression less than 60,000 MGMT molecules/nucleus (ie., ~100 fmol MGMT /106cells or ~33 fmol MGMT/mg protein, assuming one cell contains 0.3 ng protein) had significantly longer progression-free and overall survival. Comparable results were observed in another study of 64 GBM and anaplastic astrocytomas treated with radiation and BCNU [53]. Several subsequent studies using standard immunostaining techniques found that low MGMT expression (i.e., low fraction of immunopositive cells) was prognostic of longer overall [e.g., 54–57] or progression-free [58] survival following alkylator therapy. The cut-off points for low expression ranged from 5% to 50% and were chosen to reveal the most significant between-group differences. Other studies have not found an association between MGMT immunopositivity and outcome [e.g., 59–62]. The lack of accord among all these reports highlights the intrinsic limitations of IHC [e.g., variability caused by section thickness, fixation and embedding procedures, and antibody dilution as well as poor reproducibility imposed by observer variability (detailed in [61]). Importantly, the IHC studies revealed highly variable intra-tumoral intensity of immunostaining, suggesting heterogeneous MGMT expression. However, unlike the earlier quantitative immunofluorescence microscopy studies, standard IHC cannot quantify this variability. It should be noted that compared to other MGMT assays, IHC is relatively insensitive, with a lower limit of detection ranging from 3,000 to 12,000 molecules/cell [52,63]. By comparison, biochemical assay can detect as few as ~150 molecules/cell [19] and is at least 20-fold more sensitive. As discussed below, median MGMT activity in GBM is ~ 3,000 molecules/cell, suggesting the potential for a high frequency of false negatives using IHC.
3.3. Biochemical activity
The unique reaction mechanism of MGMT, in which an alkyl group is irreversibly bound to the protein (Fig. 1), is the basis for the most commonly used assay to determine activity in tissue samples. Briefly, cleared supernatants of intact tissue homogenates are incubated with DNA containing [3H]-labeled O6-methylguanine. After acid hydrolysis to release the remaining O6-methylguanine from the DNA substrate, [3H]-labeled protein is recovered by filtration. Because only one methyl group is bound per MGMT molecule, the assay determines the number of MGMT molecules in the volume of extract assayed. As noted above, the assay is very sensitive, capable of detecting as few as 150 MGMT molecules/cell [19,36]. Biochemical assay has the advantage of providing an objective, continuous variable, i.e., MGMT activity, which facilitates analysis of the association of activity with clinical outcome. However, in its current form the biochemical assay is not practical for routine clinical laboratory analysis because of the requirement for extract preparation from intact, fresh tissue and the use of a radioactive DNA substrate.
There are only a few studies of the association of glioma MGMT activity with response to alkylating agent-based chemotherapy. In an examination of 62 high-grade gliomas, no difference was reported in progression-free survival following treatment either with BCNU or PCV between 20 MGMT-deficient and 42 MGMT-proficient tumors [19]. A major drawback of this study was the inclusion of diverse diagnoses that vary widely in intrinsic response to adjuvant treatment. In a recent study of 77 de novo GBM, tumors with MGMT activity less than the median value (~3,000 molecules/cell) were significantly less likely to progress than higher activity tumors (hazard ratio = 2.06; P ≤ 0.004); this difference was observed for tumors treated with radiation followed by alkylators and for tumors treated with concurrent radiation and TMZ (Bobola and Silber, in preparation). When entered in Cox regression analyses as a continuous variable, MGMT activity was inversely associated with progression-free survival and showed a greater than 100-fold difference in risk for progression between GBM with the highest and lowest activities. Comparable results were observed for 72 anaplastic gliomas. These findings are in accord with a recent study of 40 GBM showing significantly greater progression-free survival for tumors with MGMT less than the median value of 30 fmol/mg extract protein [37]. It must be kept in mind that MGMT activity measured in tumor tissue represents an average activity of all cells, both neoplastic and normal. For example, vascular endothelial cells in gliomas have been reported to be immunopositive for MGMT (e.g., [64]) and the majority of peripheral blood leukocytes, which are likely to be present in tumor specimens, either express MGMT activity ([18]; Chamberlain and Silber, submitted) or display unmethylated promoters in glioma patients (e.g., [65]). However, the contribution of contaminating nonneoplastic neural cells to tumor specimen activity may be negligible in many instances, as data suggest that brain adjacent to glioma lacks detectable activity in the majority of cases [18,19].
3.4. Promoter CpG methylation status
Numerous studies have shown that methylation of the MGMT promoter CpG island is associated with prolonged progression-free survival and overall survival after alkylator therapy in GBM and other gliomas [4,5]. In the landmark EORTC-NCIC trial, promoter methylation was associated with prolonged progression-free and overall survival in newly diagnosed GBM treated with concurrent TMZ and radiotherapy followed by adjuvant TMZ [6,50]. Patients with methylated tumors had 2-year and 5-year survival rates of 49% and 14%, respectively, compared to 15% and 8% for unmethylated tumors. Additional clinical trials have documented that GBM harboring methylated MGMT promoters show longer survival following concurrent therapy [e.g., 58,62,66–70], adjuvant TMZ together with CCNU [e.g., 71]) and adjuvant BCNU [72]. In addition, the majority of GBM from patients surviving more than 3 years following radiotherapy and various alkylating regimens displayed MGMT promoter methylation [73,74]. Other studies have shown that promoter methylation is associated with better outcome for anaplastic gliomas and grade II gliomas treated with a variety of alkylator-based regimens [e.g., 7,8,75–77]. These findings convincingly demonstrate that promoter methylation status identifies alkylation-sensitive GBM and other gliomas.
4. Determination of methylation status
4.1. Overview
In most assays, the methylation status of CpG residues is determined by the bisulfite-mediated deamination of cytosine, but not 5-methylcytosine, to uracil (Fig. 2; [78]). Considerable effort has been expended developing simple, reliable, and, more recently, quantitative techniques to distinguish uracil from 5-methylcytosine, as well as to identify CpG residues whose methylation status is most tightly associated with treatment outcome. Single molecule sequencing of bisulfite-treated promoter DNA is the gold standard for assaying the frequency of methylation at any given CpG site. While this approach permits determination of the methylation frequency at all 97 CpG residues in the MGMT promoter [e.g., 58], the technique is not clinically tractable, although it has been used to validate other methylation assays [e.g., 79]. Methylation-specific PCR (MSP) assays that discriminate between uracil and 5-methylcytosine at a limited number of promoter CpG residues, as illustrated in Fig. 3, have proved to be more tractable, permitting the analyses of large numbers of tumors that have revealed the association of clinical outcome with promoter methylation status.
4.2. MSP-based techniques
For most major clinical studies, MSP has been used to examine the status of 4–12 CpG sites in the MGMT promoter [4,5]. MSP techniques use two sets of primers to interrogate CpG dinucleotides that are frequently methylated in MGMT-deficient cell lines [e.g., 78,80]; one set contains guanine complementary to 5-methylcytosine and the other contains adenine complementary to uracil (Fig. 3). In its original form, also referred to as nested MSP, the assay examines 9 CpG sites and is designed to produce different-sized PCR products from methylated and unmethylated primers that are resolved by agarose gel electrophoresis. Thus, the result is dichotomous, assigning tumors to one of two categories. The assay was validated by early work that yielded a single, expected band for MGMT-proficient and MGMT-deficient human cell lines [72,80]. However, results with tumor tissue were not as definitive. While MGMT-expressing tumors, evidenced by IHC showing frequent, unambiguous nuclear staining, displayed the unmethylated promoter band, MGMT-non-expressing tumors almost invariably displayed both the methylated and unmethylated products. Appearance of the unmethylated signal has been attributed to the presence of normal, MGMT-expressing cells [e.g., 50,80], and GBM and other gliomas displaying both PCR products are assigned methylated status. The appearance of both PCR products may also reflect the presence of MGMT-deficient and -proficient glioma cells in a tumor, or partially methylated sequences, or even possibly the presence of promoter hemimethylation (i.e., only one MGMT allele is methylated) as recently reported for MGMT-proficient cell lines [32].
Quantitative-MSP (Q-MSP), a variant of MSP utilizing real time PCR techniques [81], attempts to standardize the assay, better differentiate methylated and unmethylated tumors and provide a quantitative estimate of the frequency of methylation. This technique utilizes specific primer sets to simultaneously amplify an MGMT promoter sequence encompassing 8 CpG sites and an unmethylated reference gene, typically Actin B. Amplification of promoter and reference sequences is quantified by generation of fluorescent signals, e.g., by using Taqman- or SYBR-green-based assays. Fluorescence intensity, expressed as CT, the cycle number at which signal is significantly higher than background, is converted to sequence copy number by using standard curves generated with bisulfite-treated plasmids containing the methylated promoter or reference sequence. Promoter copy number is normalized to the reference sequence to allow comparison between tumor samples. These techniques quantify methylation status as either copy number of fully methylated promoters [e.g., 82] or as the percentage of total promoters that are fully methylated [e.g., 62].
Hegi and colleagues used Q-MSP with Actin B as a reference to determine the MGMT promoter methylation status of 134 gliomas [82]. A number of stringent criteria, including Actin B copy number greater than 1,000, had to be satisfied to validate each assay. Interestingly, the normalized copy number of methylated MGMT promoters could be fitted to a bimodal distribution, suggesting that the technique revealed an objective cut-point for distinguishing methylated and unmethylated tumors. In agreement with this hypothesis, the methylation status determined by Q-MSP and MSP in 91 gliomas was concordant in 90% of tumors. While this work suggests that Q-MSP affords an objective basis for assigning methylation status, there is no a priori reason to expect a bimodal distribution of methylated promoters, and the diagnostic heterogeneity of the tumor sample precluded examination of the association of methylated promoter copy number with clinical response. There was also no examination of the relationship of methylated promoter copy number with MGMT expression, limiting insights into the possible biological and biochemical significance of the putative bimodal distribution. A recent study illustrated the potential clinical utility of a variant Q-MSP technique that measured both methylated and unmethylated promoter copy number (semi-quantitative- or SQ-MSP). In an analysis of 81 GBM treated with concurrent TMZ and radiotherapy, overall survival was significantly longer in tumors with a greater than median fraction (35%) of methylated promoters [62]. Fitting the fraction of methylated promoters to a bimodal distribution was reported to yield a cut-point for prolonged survival that was essentially the same as the median (30–35%). Notably, the prognostic value of SQ-MSP was greater than that of gel-based MSP, suggesting that determination of methylation status by real time PCR-based techniques has clinical utility. The methods employed, however, are technically demanding and better suited for dedicated laboratories. In this regard, we note that analysis of methylation status by Q-PCR is commercially available [e.g., 82].
4.3. MSP: Limitations and caveats
Current MSP techniques have shortcomings that limit detection and quantification of CpG methylation. For example, techniques are optimized for primer binding to either completely methylated or unmethylated template sequences. Heterogeneous methylation, either inherent or the result of incomplete bisulfite-mediated deamination, can prevent primer annealing and extension, or foster indiscriminate annealing of primers, producing both methylated and unmethylated products from the same promoter sequence. Methylation-specific pyrosequencing addresses the problem of heterogeneous methylation by providing an average frequency of methylation at all CpG sites assayed [79,81]. Other approaches include digestion with restriction endonucleases to determine CpG methylation status. Combined bisulfite restriction analysis (COBRA; [79]) employs digestion of bisulfite-treated, PCR-amplified DNA with BstUI or TaqI to eliminate unmethylated CpGs. In the case of methylation-specific multiplex ligation-dependent probe amplification (MS-MPLA; [83]), digestion of native DNA with methylation-sensitive HhaI eliminates unmethylated CpG sites from the analysis. The complexity of the latter two methods will likely limit their widespread clinical application.
Current promoter methylation assays are also limited by inability to distinguish signal specific to tumor from that contributed by accompanying normal cells. Most clinical studies circumvent this problem by extracting DNA from tumor cells that have been micro-dissected from formalin fixed, paraffin embedded tissue [e.g., 84]. This approach is attractive in that it allows analysis of archived specimens as well as minimizing contamination by normal cells. However, several caveats should be kept in mind concerning archived material. Very small amounts of DNA, typically no more than several hundred nanograms, are usually recovered, and as much as 90% of DNA can be lost because of strand breaks produced during bisulfite treatment. In addition, fixation promotes DNA-protein cross-links that can compromise template function during PCR [85,86]. As a result of these limitations, MSP can be uninformative in as many as 25% of cases [e.g., 82]. Low yield of poor quality DNA necessitates increasing the number of PCR cycles required to produce an interpretable result. This can introduce uncertainty in the assay by heightening the risk of extending primers indiscriminately bound to heterogeneously methylated templates. Increasing cycle number of PCR assays that can detect fewer than one sequence in a thousand also elevates the risk of detecting signal from contaminating normal tissue. We note that it is commonly assumed that contaminating normal cells invariably express MGMT, as has been reported for endothelial cells in gliomas [64] and therefore contribute only unmethylated promoters that can be discounted in the presence of methylated signal [e.g., 80,87]. However, this is not necessarily the case. Contamination of gliomas with normal cells also results from tumor infiltration of surrounding non-neoplastic brain that frequently lacks detectable MGMT activity [18, 36], raising the possibility that contaminating normal brain cells may also contribute to a methylated signal. Moreover, an appreciable fraction (18% to 25%) of GBM and other gliomas lack detectable MGMT activity [19,32,36], implying that neither tumor nor normal cells express MGMT.
An alternative approach to limiting the contribution of accompanying normal cells would be to isolate cells from fresh tissue prior to analysis. In a recent report [88], Hegi and co-workers compared MGMT mRNA expression, activity and promoter methylation in 10 GBM and paired cultures established from CD133+ cells isolated from the same tumors. All tumors had detectable MGMT activity although seven were methylated by MSP as evidenced by the presence of PCR product for both methylated and unmethylated promoters. Single clone sequencing of bisulfite-treated DNA revealed heavy methylation at 28 CpGs within the minimal promoter region in 25% to 90% of clones of the seven methylated GBM. The CD133+ cultures established from the methylated tumors had little or no MGMT activity, and 100% of bisulfite-sequenced clones showed extensive methylation comparable to that of the tumor of origin. In contrast, the three cultures derived from unmethylated tumors had MGMT activity and displayed little or no promoter methylation. As CD133 expression is a marker of a sub-set of GBM stem cells that are believed to determine the biological and clinical course of GBM [89], these findings suggest that methylation is indicative of low or absent MGMT expression in tumor. The presence of MGMT activity and unmethylated promoters in the seven GBM was attributed to contaminating normal cells. This study represents the most comprehensive effort to date to compare MGMT activity and promoter methylation in GBM stem cells with activity and methylation in the tumor of origin. The authors stated that CD133+ cultures could be established from only about 50% of tumors, raising the possibility that methylation of the MGMT promoter accompanies other traits that promote in vitro proliferation. It should be noted that growth in culture is frequently accompanied by extensive changes in CpG methylation, and the CD133+ cells were cultured for 2 to 12 months prior to analysis. Also, culture conditions can affect MGMT expression, as illustrated by a lymphoblastoid line that expressed MGMT when grown as adherent cells, but lost MGMT expression when grown in suspension [90]. Thus, how representative the methylation pattern of a subset of cultured GBM cells is of tumor cells in vivo remains uncertain.
The fraction of GBM found to have methylated MGMT promoters ranges from 35% to 73%, with similar variability observed for anaplastic and grade II gliomas [4]. This frequency is much greater than the fraction of GBM and other gliomas that lack biochemically detectable activity (~18%–25%; [19,37]). While the discrepancy between the results of promoter methylation and activity assays likely reflects the convention of assigning methylated status to tumors exhibiting both methylated and unmethylated products, the varying estimates of methylation frequency may also reflect, in part, the technical difficulties inherent in all CpG methylation measurements and lack of objective criteria for determining a cutoff point for the fraction of methylated promoters that define the methylated phenotype. The wide range in promoter methylation may also reflect the assumption that contaminating normal cells invariably display unmethylated promoters, an assumption that, as discussed above, may not be valid in all cases.
5. Utilization of methylation status to direct treatment
Identification of biomarkers that predict GBM response to therapy is a critical step in the rational development of treatments tailored to individual patients that produce optimal clinical outcome. Currently, only MGMT promoter methylation assays have been shown to consistently identify GBM that are more likely to respond favorably to alkylating agent-based therapies in multiple independent studies [4,5,50]. The question of whether or not methylation status should be used to allocate treatment, particularly for unmethylated GBM, has been asked repeatedly [e.g., 4,89]. Several considerations and outstanding questions indicate that the answer remains no, and the clinical utility of promoter methylation status is currently limited to providing an estimate of overall prognosis.
Perhaps the greatest impediment to clinical utilization is the lack of a validated, standardized promoter methylation assay. Standardization would permit inter-group comparison of results, facilitating the inclusion of methylation data in the design of clinical trials. Standardization is currently impeded by lack of consensus about what technique is most tractable in the clinical setting, which promoter CpG sites are most informative and what criteria permit an objective assignment of methylation status. Conceivably these impediments can be circumvented by emerging DNA sequencing methods that can detect methylation at all promoter CpG sites without the use of bisulfite-mediated deamination [e.g., 92]. Assay standardization and rigorous criteria for methylation status may also help overcome another impediment, the high backgrounds of false negative and positive results produced by current methylation assays. This lack of specificity is exemplified by the observation that 15% of patients with unmethylated GBM treated with concurrent TMZ-radiotherapy survive 2 years, a 7-fold increase in frequency compared to tumors receiving radiation alone [6]. It is important to keep in mind that this uncertainty, while likely reflecting to some extent the inherent limitations of current assays, may also be indicative of tumor sub-groups in which MGMT is not the primary determinant of treatment outcome.
Another limitation of methylation status as a biomarker is that it does not aid in choosing a particular alternative therapy for unmethylated tumors, other than that the regimen not include alkylating agents. It has been proposed that unmethylated GBM are candidates for MGMT ablative therapy, either using substrate analog inhibitors or TMZ dose intensification [4,11]. However, there is no compelling evidence that methylated GBM would not also benefit from MGMT ablation since assay of these tumors almost invariably reveals evidence of unmethylated promoters, conceivably from MGMT-expressing tumor cells. While, on average, promoter methylation may be associated with no/low MGMT activity in GBM tissue, the correspondence is not absolute, and thus the status of expression in any individual tumor is not unequivocal. For example, methylation status does not consistently correlate with MGMT expression assessed by IHC [e.g., 61,62]. Likewise, MGMT activity of methylated GBM was found to be lower than that of unmethylated tumors in one study [32], but not in another [93]. More recently, we have observed that MGMT activity in 39 unmethylated GBM was significantly higher than that in 15 methylated tumors (10 ± 8.9 vs 4.8 ± 2.6 fmol/106 cells; P ≤ 0.002; Silber unpublished). However, there is considerable overlap of activities between the two groups, reflecting the presence of low activity unmethylated and high activity methylated GBM. This finding reflects a major limitation imposed by the inability of current CpG methylation and biochemical assays to distinguish not only tumor from normal cells but methylated from unmethylated GBM cells within the same tumor.
6. Future considerations and outstanding questions
As set out in this review, current promoter methylation assays suffer from a number of methodological and conceptual limitations that confound assignment of methylation status. Current assays are poor surrogates for MGMT activity and do not reflect intra-tumoral heterogeneity in MGMT expression that is evident by IHC [57]. These factors are problematic if MGMT activity is the exclusive or predominant determinant underlying the predictive power of promoter methylation (See discussion below). Moreover, unlike genetic aberrations that can be unambiguously assigned to tumor cells, promoter methylation assays do not distinguish signal specific to tumor from that contributed by accompanying normal cells. These limitations, together with a number of outstanding questions pertinent to both biological significance and clinical utility (Table 3), need be addressed in order to increase specificity and predictive power such that the assays can identify treatment-responsive individuals.
The clinical utility of MGMT promoter methylation assays would be greatly increased if there were methylation patterns that distinguished tumor from surrounding normal tissue, a possibility yet to be rigorously investigated. Tumorigenesis is frequently accompanied by altered CpG methylation that may change MGMT expression and that may differ from distinctive patterns that are associated with MGMT expression in normal progenitor tissue [26,94]. Moreover, the methylation patterns currently believed to be indicative of MGMT silencing in tumor tissue were derived from GBM cell lines that had been in continuous culture for years, raising the possibility that physiologic patterns of methylation were modified or lost in response to selective pressures exerted by long-term growth in vitro. These considerations suggest that single-molecule bisulfite-sequencing of the promoter and gene body in GBM and normal brain specimens with known MGMT activity may reveal distinctive methylation patterns that distinguish tumor and normal cells, and also allow a quantitative estimation of the fraction of MGMT-non-expressing tumor cells and the distribution of MGMT activity among tumor cells. This information may provide the means to improve stratification by risk for progression among patients, especially if expressed as a continuous variable.
Also complicating interpretation of the significance of promoter methylation status is evidence that it may not be specific for alkylating agent response. Some [e.g., 7,8,95,96], but not all [6], studies have found that methylation is associated with better outcome in gliomas treated with radiation only. In accord are recent reports that radiotherapy as well as alkylator treatment selects strongly against promoter methylation in recurrent tumors [e.g., 32,97]. As radiation sensitivity is not mediated by MGMT activity, these findings suggest that MGMT promoter methylation status may reflect a broader DNA damage sensitivity phenotype. In agreement with this hypothesis, it has been demonstrated that MGMT-non-expressing GBM cell lines display not only greater sensitivity than MGMT-expressing lines to O6-methylguanine, but also to 3-methyladenine, a TMZ-induced cytotoxic adduct that is removed by base excision repair [98]. In addition, other DNA repair proteins are epigenetically regulated [99], including the Werner syndrome helicase (WRN) that promotes TMZ resistance in human GBM cells [100]. Suppression of MGMT together with changes in expression of additional DNA repair proteins could result in sensitivity to alkylator-induced adducts other than O6-methylguanine and to DNA damaging agents in addition to alkylators. Definitive demonstration that MGMT promoter methylation status reflects a global response to therapeutic agents that act by damaging DNA could have an immediate impact on choosing alternative therapies for unmethylated GBM.
Inclusion of additional informative markers together with MGMT promoter methylation status in multivariate regression models may increase the ability to stratify treatment by projected response in GBM. While MGMT is the sole DNA repair protein that removes O6-methylguanine from DNA, additional repair mechanisms promote resistance to the consequences of failure to excise this and other TMZ-induced adducts, i.e., interrupted DNA replication leading to single-strand gaps and double-strand breaks. Human cells possess numerous pathways that promote replication re-start at stalled replication forks [101], gap filling by error-prone DNA polymerases [102] and rejoining of double-strand breaks by homologous and non-homologous recombination [103]. TMZ produces at least two additional cytotoxic lesions, 3-methyladenine and abasic sites, which are not substrates for MGMT. Notably, the abasic site endonuclease activity of Ape1/Ref-1, the DNA repair enzyme primarily responsible for initiating the excision of abasic sites in human cells [104], is inversely associated with progression-free survival in anaplastic gliomas treated with radiotherapy or radiotherapy followed by alkylating agent-based chemotherapy [105]. In addition to DNA repair proteins, a host of genetic and epigenetic molecular markers have been identified in GBM that govern proliferation, response to treatment and survival [e.g., 5,106,107].
As alluded to above, an unresolved question is whether the longer survival following alkylating agent treatment of GBM displaying promoter methylation is solely due to reduced MGMT expression. The genesis of GBM is characterized by changes in gene expression mediated by global DNA hypomethylation accompanied by gene-specific methylation of promoter CpG islands. Ongoing work indicates that GBM can be classified into clinically informative sub-types based on distinctive methylation patterns [e.g., 108,109,110], including one having the hallmarks of a glioma CpG island methylator phenotype (G-CIMP; [106,110]). The observation that the G-CIMP sub-group of GBM has prolonged survival emphasizes the question of whether MGMT promoter methylation accompanies other epigenetic changes associated with better outcome. A recent examination by Etcheverry et al. of 50 adult GBM patients treated with concurrent radiation and TMZ suggests that this may be the case [109]. This study found that depending on the gene, promoter hypermethylation or hypomethylation was associated with better clinical response independent of MGMT methylation status. The promoter methylation of some genes strengthened the prognostic power of MGMT methylation status, suggesting that epigenetic silencing of both contributed to better clinical outcome. However, for other genes, promoter methylation negated the better outcome usually associated with MGMT methylation, suggesting that silencing of these genes promoted resistance to the consequences of not removing O6-methylguanine or to some other, unrelated resistance mechanism(s). While the work of Etcheverry et al. was a small study that requires independent confirmation with a larger sample of GBM, these findings strongly suggest that clinical response to concurrent therapy is multifactorial and that MGMT is not necessarily the predominant determinate of response. They also suggest a mechanistic explanation for the failure of some tumors to respond to therapy as predicted by MGMT promoter methylation status and may identify future targets for therapeutic intervention.
For the variety of reasons discussed in this review, the clinical utility of CpG methylation is presently limited to providing an estimate of overall prognosis for populations of tumors. The ultimate goal is to identify with a high degree of certainty treatment-responsive individuals in order to direct patients to the most efficacious therapy. Ongoing research and advances in technology are likely to increase the usefulness of MGMT promoter methylation status as a tool for personalizing anti-glioma treatment. However, this goal cannot be realized until alternative therapies for newly diagnosed GBM are developed that produce outcomes comparable to concurrent TMZ and radiation for unmethylated tumors. Only with the advent of new, effective therapies can MGMT promoter methylation status truly be used to inform treatment decisions.
Acknowledgements
Supported in part by NIH grant number CA104593 (JR Silber), the Department of Neurological Surgery, University of Washington and a gift to the Brain Tumor Research Fund in memory of Ro Jean Mount. The funding sources did not influence the content of this manuscript. We are grateful to Drs. Lawrence A. Loeb and Eddie J. Fox for critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: BCNU, 1,3-bis(2-chloroethyl)-1-nitrosourea, carmustine; CCNU, 3-(2-chloroethyl)-3-cyclohexyl-1-nitrosouea, lomustine; GBM, glioblastoma multiforme; IHC, immunohistochemistry; MGMT, O6-methylguanine-DNA methyltransferase; MSP, methylation-specific PCR; PCV, procabazine, CCNU, vincristine; TMZ, temozolomide
References
- [1].Mitra S. MGMT: a personal perspective. DNA Repair. 2007;6:1064–70. doi: 10.1016/j.dnarep.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tubbs JL JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair. 2007;6:1100–15. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Samson L L, Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977;267:281–3. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- [4].Weller M, Stupp R, Reifenberger G, Brandes AA, van den Bent MJ, Wick W, Hegi ME. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6:39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- [5].von Deimling A, Korshunov A, Hartmann C. The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations. Brain Pathol. 2011;21:74–87. doi: 10.1111/j.1750-3639.2010.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- [7].van den Bent MJ, Dubbink HJ, Sanson M, van der Lee-Haarloo CR, Hegi M, Jeuken JW, Ibdaih A, Brandes AA, Taphoorn MJ, Frenay M, Lacombe D, Gorlia T, Dinjens WN, Kros JM. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2009;27:5881–6. doi: 10.1200/JCO.2009.24.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wick W, Hartmann C, Engel C, Stoffels M, Felsberg J, Stockhammer F, Sabel MC, Koeppen S, Ketter R, Meyermann R, Rapp M, Meisner C, Kortmann RD, Pietsch T, Wiestler OD, Ernemann U, Bamberg M, Reifenberger G, von Deimling A, Weller M. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27:5874–80. doi: 10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- [9].Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity, and apoptosis induced by alkylating agents. DNA Repair. 2007;6:1079–99. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- [10].Kaina B, Margison GP, Christmann M. Targeting O6-methylguanine-DNA methyltransferase with specific inhibitors as a strategy in cancer therapy. Cell Mol Life Sci. 2010;67:3663–81. doi: 10.1007/s00018-010-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mrugala MM, Chamberlain MC. Mechanisms of disease: temozolomide and glioblastoma--look to the future. Nat Clin Pract Oncol 592008) :476–86. doi: 10.1038/ncponc1155. [DOI] [PubMed] [Google Scholar]
- [12].Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- [13].Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- [14].Hansen RJ, Nagasubramanian R, Delaney SM, Samson LD, Dolan ME. Role of O6-methylguanine-DNA methyltransferase in protecting from alkylating agent-induced toxicity and mutations in mice. Carcinogenesis. 2007;28:1111–6. doi: 10.1093/carcin/bgl218. [DOI] [PubMed] [Google Scholar]
- [15].Kleihues P, Lantos LP, Magee PN. Chemical carcinogenesis in the nervous system. Int Rev Exp Pathol. 1976;15:153–232. [PubMed] [Google Scholar]
- [16].Schlegel J, Stumm G, Mennel HD. Chemical carcinogenesis in the nervous system: past and future. Exp Toxic Pathol. 1994;45:455–466. doi: 10.1016/S0940-2993(11)80504-X. [DOI] [PubMed] [Google Scholar]
- [17].Wrensch M, Minn Y, Chew T, Bondy M, Berger MS. Epidemiology of primary brain tumors: current concepts and review of the literature. Neuro Oncol. 2002;4:278–99. doi: 10.1093/neuonc/4.4.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Silber JR, Blank A, Bobola MS, Mueller BA, Kolstoe DD, Ojemann GA, Berger MS. Lack of the DNA repair protein O6-methylguanine-DNA methyltransferase in histologically normal brain adjacent to primary human brain tumors. Proc Natl Acad Sci USA. 1996;93:6941–6. doi: 10.1073/pnas.93.14.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Silber JR, Blank A, Bobola MS, Ghatan S, Kolstoe DD, Berger MS. O6-methylguanine-DNA methyltransferase-deficient phenotype in human gliomas: frequency and time to tumor progression after alkylating agent-based chemotherapy. Clin Cancer Res. 1999;5:807–14. [PubMed] [Google Scholar]
- [20].Chen ZP, Malapetsa A, McQuillan D, Marcantonio V, Bello G, Mohr J, Remack J, Brent TP, Panasci LC. Evidence for nucleotide excision repair as a modifying factor of O6-methylguanine-DNA methyltransferase-mediated innate chloroethylnitrosourea resistance in human tumor cell lines. Mol Pharmacol. 1997;52:815–20. doi: 10.1124/mol.52.5.815. [DOI] [PubMed] [Google Scholar]
- [21].Qian XC, Brent TP. Methylation hot spots in the 5' flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1997;57:3672–7. [PubMed] [Google Scholar]
- [22].Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- [23].Bocangel D, Sengupta S, Mitra S, Bhakat KK. p53-Mediated down-regulation of the human DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) via interaction with Sp1 transcription factor. Anticancer Res. 2009;29:3741–50. [PMC free article] [PubMed] [Google Scholar]
- [24].Margison GP, Santibáñez Koref MF, Povey AC. Mechanisms of carcinogenicity/chemotherapy by O6-methylguanine. Mutagenesis. 2002;17:483–7. doi: 10.1093/mutage/17.6.483. [DOI] [PubMed] [Google Scholar]
- [25].Jacinto FV, Esteller M. MGMT hypermethylation: a prognostic foe, a predictive friend. DNA Repair. 2007;6:1155–60. doi: 10.1016/j.dnarep.2007.03.013. PMID:17482895. [DOI] [PubMed] [Google Scholar]
- [26].Nagarajan RP, Costello JF. Epigenetic mechanisms in glioblastoma multiforme. Semin Cancer Biol. 2009;19:188–97. doi: 10.1016/j.semcancer.2009.02.005. PMID:19429483. [DOI] [PubMed] [Google Scholar]
- [27].Costello JF, Futscher BW, Tano K, Graunke DM, Pieper RO. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994;269:17228–37. PMID:8006031. [PubMed] [Google Scholar]
- [28].Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. 1997;17:5612–9. doi: 10.1128/mcb.17.9.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Danam RP, Qian XC, Howell SR, Brent TP. Methylation of selected CpGs in the human O6-methylguanine-DNA methyltransferase promoter region as a marker of gene silencing. Mol Carcinog. 1999;24:85–9. [PubMed] [Google Scholar]
- [30].Nakagawachi T, Soejima H, Urano T, Zhao W, Higashimoto K, Satoh Y, Matsukura S, Kudo S, Kitajima Y, Harada H, Furukawa K, Matsuzaki H, Emi M, Nakabeppu Y, Miyazaki K, Sekiguchi M, Mukai T. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene. 2003;22:8835–44. doi: 10.1038/sj.onc.1207183. [DOI] [PubMed] [Google Scholar]
- [31].Kitange GJ, Carlson BL, Mladek AC, Decker PA, Schroeder MA, Wu W, Grogan PT, Giannini C, Ballman KV, Buckner JC, James CD, Sarkaria JN. Evaluation of MGMT promoter methylation status and correlation with temozolomide response in orthotopic glioblastoma xenograft model. J Neurooncol. 2009;92:23–31. doi: 10.1007/s11060-008-9737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Christmann M, Nagel G, Horn S, Krahn U, Wiewrodt D, Sommer C, Kaina B. MGMT activity, promoter methylation and immunohistochemistry of pretreatment and recurrent malignant gliomas: a comparative study on astrocytoma and glioblastoma. Int J Cancer. 2010;127:2106–18. doi: 10.1002/ijc.25229. [DOI] [PubMed] [Google Scholar]
- [33].Harris LC, Remack JS, Brent TP. In vitro methylation of the human O6-methylguanine-DNA methyltransferase promoter reduces transcription. Biochim Biophys Acta. 1994;1217:141–6. doi: 10.1016/0167-4781(94)90027-2. [DOI] [PubMed] [Google Scholar]
- [34].Costello JF, Futscher BW, Kroes RA, Pieper RO. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. 1994;14:6515–21. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Danam RP, Howell SR, Brent TP, Harris LC. Epigenetic regulation of O6-methylguanine-DNA methyltransferase gene expression by histone acetylation and methyl-CpG binding proteins. Mol Cancer Ther. 2005;4:61–9. [PubMed] [Google Scholar]
- [36].Silber JR, Bobola MS, Ghatan S, Blank A, Kolstoe DD, Berger MS. O6-methylguanine-DNA methyltransferase activity in adult gliomas: relation to patient and tumor characteristics. Cancer Res. 1998;58:1068–73. [PubMed] [Google Scholar]
- [37].Wiewrodt D, Nagel G, Dreimüller N, Hundsberger T, Perneczky A, Kaina B. MGMT in primary and recurrent human glioblastomas after radiation and chemotherapy and comparison with p53 status and clinical outcome. Int J Cancer. 2008;122:1391–9. doi: 10.1002/ijc.23219. [DOI] [PubMed] [Google Scholar]
- [38].Bobola MS, Blank A A, Berger MS, Silber JR. O6-methylguanine-DNA methyltransferase deficiency in developing brain: implications for brain tumorigenesis. DNA Repair. 2007;6:1127–33. doi: 10.1016/j.dnarep.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bobola MS, Berger MS, Silber JR. Contribution of O6-methylguanine-DNA methyltransferase to resistance to 1,3-(2-chloroethyl)-1-nitrosourea in human brain tumor-derived cell lines. Mol Carcinog. 1995;13:81–8. doi: 10.1002/mc.2940130204. [DOI] [PubMed] [Google Scholar]
- [40].Bobola MS, Tseng SH, Blank A, Berger MS, Silber JR. Role of O6-methylguanine-DNA methyltransferase in resistance of human brain tumor cell lines to the clinically relevant methylating agents temozolomide and streptozotocin. Clin Cancer Res. 1996;2:735–41. [PubMed] [Google Scholar]
- [41].Bobola MS, Kolstoe DD, Blank A, Silber JR. Minimally cytotoxic doses of temozolomide produce radiosensitization in human glioblastoma cells regardless of MGMT expression. Mol Cancer Ther. 2010;9:1208–18. doi: 10.1158/1535-7163.MCT-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Carlson BL, Grogan PT, Mladek AC, Schroeder MA, Kitange GJ, Decker PA, Giannini C, Wu W, Ballman KA, James CD, Sarkaria JN. Radiosensitizing effects of temozolomide observed in vivo only in a subset of O6-methylguanine-DNA methyltransferase methylated glioblastoma multiforme xenografts. Int J Radiat Oncol Biol Phys. 2009;75:212–9. doi: 10.1016/j.ijrobp.2009.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bodell WJ, Pongracz K. Chemical synthesis and detection of the cross-link 1-[N3-(2'-deoxycytidyl)]-2-[N1-(2'-deoxyguanosinyl)]ethane in DNA reacted with 1-(2-chloroethyl)-1-nitrosourea. Chem Res Toxico. 1993;6:434–8. doi: 10.1021/tx00034a008. [DOI] [PubMed] [Google Scholar]
- [44].Newlands ES, Stevens MF, Wedge SR, Wheelhouse RT, Brock C. Temozolomide: a review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer Treat Rev. 1997;23:35–61. doi: 10.1016/s0305-7372(97)90019-0. [DOI] [PubMed] [Google Scholar]
- [45].Marchesi F, Turriziani M, Tortorelli G, Avisati G, Torino F, De L. Vecchis Triazene compounds: mechanism of action and related DNA repair systems. Pharmacol Res. 2007;56:275–87. doi: 10.1016/j.phrs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- [46].Quiros S, Roos WP, Kaina B. Processing of O6-methylguanine into DNA double-strand breaks requires two rounds of replication whereas apoptosis is also induced in subsequent cell cycles. Cell Cycle. 2010;9:1168–78. doi: 10.4161/cc.9.1.10363. [DOI] [PubMed] [Google Scholar]
- [47].Ludlum DB. DNA alkylation by the haloethylnitrosoureas: nature of modifications produced and their enzymatic repair or removal. Mutat Res. 1990;233:117–26. doi: 10.1016/0027-5107(90)90156-x. [DOI] [PubMed] [Google Scholar]
- [48].Ludlum DB. The chloroethylnitrosoureas: sensitivity and resistance to cancer chemotherapy at the molecular level. Cancer Invest. 1997;15:588–98. doi: 10.3109/07357909709047601. [DOI] [PubMed] [Google Scholar]
- [49].Gonzaga PE, Potter PM, Niu TQ, Yu D, Ludlum DB, Rafferty JA, Margison GP, Brent TP. Identification of the cross-link between human O6-methylguanine-DNA methyltransferase and chloroethylnitrosourea-treated DNA. Cancer Res. 1992;52:6052–8. [PubMed] [Google Scholar]
- [50].Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- [51].Gilbert MR, Kuhn J, Lamborn KR, Lieberman F, Wen PY, Mehta M, Cloughesy T, Lassman AB, Deangelis LM, Chang S, Prados M. Cilengitide in patients with recurrent glioblastoma: the results of NABTC 03-02, a phase II trial with measures of treatment delivery. J Neurooncol. 2011 doi: 10.1007/s11060-011-0650-1. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Belanich M, Pastor M, Randall T, Guerra D, Kibitel J, Alas L, Li B, Citron M, Wasserman P, White A, Eyre H, Jaeckle K, Schulman S, Rector D, Prados M, Coons S, Shapiro W, Yarosh D. Retrospective study of the correlation between the DNA repair protein alkyltransferase and survival of brain tumor patients treated with carmustine. Cancer Res. 1996;56:783–8. [PubMed] [Google Scholar]
- [53].Jaeckle KA, Eyre HJ, Townsend JJ, Schulman S, Knudson HM, Belanich M, Yarosh DB, Bearman SI, Giroux DJ, Schold SC. Correlation of tumor O6-methylguanine-DNA methyltransferase levels with survival of malignant astrocytoma patients treated with bis-chloroethylnitrosourea: a Southwest Oncology Group study. J Clin Oncol. 1998;16:3310–5. doi: 10.1200/JCO.1998.16.10.3310. [DOI] [PubMed] [Google Scholar]
- [54].Nakasu S, Fukami T, Baba K, Matsuda M. Immunohistochemical study for O6-methylguanine-DNA methyltransferase in the non-neoplastic and neoplastic components of gliomas. J Neurooncol. 2004;70:333–40. doi: 10.1007/s11060-004-9170-6. [DOI] [PubMed] [Google Scholar]
- [55].Brell M, Tortosa A, Verger E, Gil JM, Viñolas N, Villá S, Acebes JJ, Caral L, Pujol T, Ferrer I, Ribalta T, Graus F. Prognostic significance of O6-methylguanine-DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clin Cancer Res. 2005;11:5167–74. doi: 10.1158/1078-0432.CCR-05-0230. [DOI] [PubMed] [Google Scholar]
- [56].Chinot OL, Barrié M, Fuentes S, Eudes N, Lancelot S, Metellus P, Muracciole X, Braguer D, Ouafik L, Martin PM, Dufour H, Figarella-Branger D. Correlation between O6-methylguanine-DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. J Clin Oncol. 2007;25:1470–5. doi: 10.1200/JCO.2006.07.4807. [DOI] [PubMed] [Google Scholar]
- [57].Capper D, Mittelbrxonn M, Meyermann R, Schittenhelm J. Pitfalls in the assessment of MGMT expression and in its correlation with survival in diffuse astrocytomas: proposal of a feasible immunohistochemical approach. Acta Neuropathol. 2008;115:249–59. doi: 10.1007/s00401-007-0310-x. [DOI] [PubMed] [Google Scholar]
- [58].Shah N, Lin B, Sibenaller Z, Ryken T, Lee H, Yoon JG, Rostad S, Foltz G. Comprehensive analysis of MGMT promoter methylation: correlation with MGMT expression and clinical response in GBM. PLoS One. 2011;6:e16146. doi: 10.1371/journal.pone.0016146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, Batchelor TT, Futreal PA, Stratton MR, Curry WT, Lafrate AJ, Louis DN. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–45. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rodriguez FJ, Thibodeau SN, Jenkins RB, Schowalter KV, Caron BL, O'neill BP, James CD, Passe S, Slezak J, Giannini C. MGMT immunohistochemical expression and promoter methylation in human glioblastoma. Appl Immunohistochem Mol Morphol. 2008;16:59–65. doi: 10.1097/PAI.0b013e31802fac2f. [DOI] [PubMed] [Google Scholar]
- [61].Preusser M, Janzer R. Charles, Felsberg J, Reifenberger G, Hamou MF, Diserens AC, Stupp R, Gorlia T, Marosi C, Heinzl H, Hainfellner JA, Hegi M. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol. 2008;18:520–32. doi: 10.1111/j.1750-3639.2008.00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Karayan-Tapon L, Quillien V, Guilhot J, Wager M, Fromont G, Saikali S, Etcheverry A, Hamlat A, Loussouarn D, Campion L, Campone M, Vallette FM, Gratas-Rabbia-Ré C. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97:311–22. doi: 10.1007/s11060-009-0031-1. [DOI] [PubMed] [Google Scholar]
- [63].Brent TP, von Wronski MA, Edwards CC, Bromley M, Margison GP, Rafferty JA, Pegram CN, Bigner DD. Identification of nitrosourea-resistant human rhabdomyosarcomas by in situ immunostaining of O6-methylguanine-DNA methyltransferase. Oncol Res. 1992;5:83–6. [PubMed] [Google Scholar]
- [64].Sasai K, Nodagashira M, Nishihara H, Aoyanagi E, Wang L, Katoh M, Murata J, Ozaki Y, Ito T, Fujimoto s., Kaneko S, Nagashima K, Tanaka S. Careful exclusion of non-neoplastic brain components is required for an appropriate evaluation of O6-methylguanine-DNA methyltransferase status in glioma: relationship between immunohistochemistry and methylation analysis. Am J Surg Pathol. 2008;32:1220–7. doi: 10.1097/PAS.0b013e318164c3f0. [DOI] [PubMed] [Google Scholar]
- [65].Sylvester RK, Steen P, Tate JM, Mehta M, Petrich RJ, Berg A, Kolesar J. Temozolomide-induced severe myelosuppression: analysis of clinically associated polymorphisms in two patients. Anticancer Drugs. 2011;22:104–10. doi: 10.1097/CAD.0b013e3283407e9f. [DOI] [PubMed] [Google Scholar]
- [66].Crinière E, Kaloshi G, Laigle-Donadey F, Lejeune J, Auger N, Benouaich-Amiel A, Everhard S, Mokhtari K, Polivka M, Delattre JY, Hoang-Xuan K, Thillet J, Sanson M. MGMT prognostic impact on glioblastoma is dependent on therapeutic modalities. J Neurooncol. 2007;83:173–9. doi: 10.1007/s11060-006-9320-0. [DOI] [PubMed] [Google Scholar]
- [67].Felsberg J, Rapp M, Loeser S, Fimmers R, Stummer W, Goeppert M, Steiger HJ, Friedensdorf B, Reifenberger G, Sabel MC. Prognostic significance of molecular markers and extent of resection in primary glioblastoma patients. Clin Cancer Res. 2009;15:6683–93. doi: 10.1158/1078-0432.CCR-08-2801. [DOI] [PubMed] [Google Scholar]
- [68].Weller M, Felsberg J, Hartman C, Berger H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn JC, Heese O, Krex D, Nikkhah G, Pietsch T, Wiestler O, Reifenberger G, von Deimling A, Loeffler M. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27:5743–50. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- [69].Brandes AA, Franceschi E, Tosoni A, Benevento F, Scopece L, Mazzocchi V, Bacci A, Agati R, Calbucci F, Ermani M. Temozolomide concomitant and adjuvant to radiotherapy in elderly patients with glioblastoma: correlation with MGMT promoter methylation status. Cancer. 2009;115:3512–8. doi: 10.1002/cncr.24406. [DOI] [PubMed] [Google Scholar]
- [70].Stupp R, Hegi ME, Neyns B, Goldbrunner R, Schlegel U, Clement PM, Grabenbauer GG, Ochsenbein AF, Simon M, Dietrich PY, Pietsch T, Hicking C, Tonn JC, Diserens AC, Pica A, Hermisson M, Krueger S, Picard M, Weller M. Phase I/IIa study of cilengitide and temozolomide with concomitant radiotherapy followed by cilengitide and temozolomide maintenance therapy in patients with newly diagnosed glioblastoma. J Clin Onco. 2010;28:2712–8. doi: 10.1200/JCO.2009.26.6650. [DOI] [PubMed] [Google Scholar]
- [71].Herrlinger U, Rieger J, Koch D, Loeser S, Blaschke B, Kortmann RD, Steinbach JP, Hundsberger T, Wick W, Meyermann R, Tan TC, Sommer C, Bamberg M, Reifenberger G, Weller M. Phase II trial of lomustine plus temozolomide chemotherapy in addition to radiotherapy in newly diagnosed glioblastoma: UKT-03. J Clin Oncol. 2006;24:4412–7. doi: 10.1200/JCO.2006.06.9104. [DOI] [PubMed] [Google Scholar]
- [72].Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- [73].Krex D, Klink B, Hartmann C, von Deimling A, Pietsch T, Simon M, Sabel M, Steinbach JP, Heese O, Reifenberger G, Weller M, Schackert G. German Glioma Network Long-term survival with glioblastoma multiforme. Brain. 2007;130:2596–606. doi: 10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- [74].Martinez R, Schackert G, Yaya-Tur R, Rojas-Marcos I, Herman JG, Esteller M. Frequent hypermethylation of the DNA repair gene MGMT in long-term survivors of glioblastoma multiforme. J Neurooncol. 2007;83:91–3. doi: 10.1007/s11060-006-9292-0. [DOI] [PubMed] [Google Scholar]
- [75].Paz MF, Yaya-Tur R, Rojas-Marcos I, Reynes G, Pollan M, Aguirre-Cruz L, García-Lopez JL, Piquer J, Safont MJ, Balaña C, Sanchez-Cespedes M, García-Villanueva M, Arribas L, Esteller M. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–8. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- [76].Everhard S, Kaloshi G, Crinière E, Benouaich-Amiel A, Lejeune J, Marie Y, Sanson M, Kujas M, Mokhtari K, Hoang-Xuan K, Delattre JY, Thillet J. MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006;60:740–3. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- [77].Kesari S, Schiff D, Drappatz J, LaFrankie D, Doherty L, Macklin EA, Muzikansky A, Santagata S, Ligon KL, Norden AD, Ciampa A, Bradshaw J, Levy B, Radakovic G, Ramakrishna N, Black PM, Wen PY. Phase II study of protracted daily temozolomide for low-grade gliomas in adults. Clin Cancer Res. 2009;15:330–7. doi: 10.1158/1078-0432.CCR-08-0888. [DOI] [PubMed] [Google Scholar]
- [78].Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mikeska T, Bock C, El-Maarri O, Hübner A, Ehrentraut D, Schramm J, Felsberg J, Kahl P, Büttner R, Pietsch T, Waha A. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007;9:368–81. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman HG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–7. [PubMed] [Google Scholar]
- [81].Shames DS, Minna JD, Gazdar AF. Methods for detecting DNA methylation in tumors: from bench to bedside. Cancer Lett. 2007;25:187–98. doi: 10.1016/j.canlet.2006.10.014. [DOI] [PubMed] [Google Scholar]
- [82].Vlassenbroeck I, Califice S, Diserens AC, Migliavacca E, Straub J, Di Stefano I, Moreau F, Hamou MF, Renard I, Delorenzi M, Flamion B, DiGuiseppi J, Bierau K, Hegi ME. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn. 2008;10:332–7. doi: 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jeuken JW, Cornelissen SJ, Vriezen M, Dekkers MM, Errami A, Sijben A, Boots-Sprenger SH, Wesseling P. MS-MLPA: an attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest. 2007;87:1055–65. doi: 10.1038/labinvest.3700664. [DOI] [PubMed] [Google Scholar]
- [84].Cankovic M, Mikkelsen T, Rosenblum ML, Zarbo RJ. A simplified laboratory validated assay for MGMT promoter hypermethylation analysis of glioma specimens from formalin-fixed paraffin-embedded tissue. Lab Invest. 2007;87:392–7. doi: 10.1038/labinvest.3700520. [DOI] [PubMed] [Google Scholar]
- [85].Douglas MP, Rogers SO. DNA damage caused by common cytological fixatives. Mutat Res. 1998;401:77–88. doi: 10.1016/s0027-5107(97)00314-x. [DOI] [PubMed] [Google Scholar]
- [86].Gavrilov A, Razin SV. Formaldehyde fixation of cells does not greatly reduce the ability to amplify cellular DNA. Anal Biochem. 2009;390:94–6. doi: 10.1016/j.ab.2009.04.018. [DOI] [PubMed] [Google Scholar]
- [87].Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–99. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- [88].Sciuscio D, Diserens AC, van Dommelen K, Martinet D, Jones G, Janzer RC, Pollo C, Hamou MF, Kaina B, Stupp R, Levivier M, Hegi ME. Extent and patterns of MGMT promoter methylation in glioblastoma- and respective glioblastoma-derived spheres. Clin Cancer Res. 2011;17:255–66. doi: 10.1158/1078-0432.CCR-10-1931. [DOI] [PubMed] [Google Scholar]
- [89].Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat Neurosci. 2011;12:495–508. doi: 10.1038/nrn3060. [DOI] [PubMed] [Google Scholar]
- [90].Cairns-Smith S, Karran P. Epigenetic silencing of the DNA repair enzyme O6-methylguanine-DNA methyltransferase in Mex- human cells. Cancer Res. 1992;52:5257–63. [PubMed] [Google Scholar]
- [91].Stupp R, Hegi ME. Methylguanine methyltransferase testing in glioblastoma: when and how? J Clin Oncol. 2007;25:1459–60. doi: 10.1200/JCO.2006.09.7139. [DOI] [PubMed] [Google Scholar]
- [92].Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–5. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Maxwell JA, Johnson SP, Quinn JA, McLendon RE, Ali-Osman F, Friedman AH, Herndon JE, 2nd, Bierau K, Bigley J, Bigner DD, Friedman HS. Quantitative analysis of O6-alkylguanine-DNA alkyltransferase in malignant glioma. Mol Cancer Ther. 2006;5:2531–9. doi: 10.1158/1535-7163.MCT-06-0106. [DOI] [PubMed] [Google Scholar]
- [94].Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- [95].Vogelbaum MA, Berkey B, Peereboom D, Macdonald D, Giannini C, Suh JH, Jenkins R, Herman J, Brown P, Blumenthal DT, Biggs C, Schultz C, Mehta M. Phase II trial of preirradiation and concurrent temozolomide in patients with newly diagnosed anaplastic oligodendrogliomas and mixed anaplastic oligoastrocytomas: RTOG BR0131. Neuro Oncol. 2009;11:167–75. doi: 10.1215/15228517-2008-073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, Bekele BN, Aldape KD. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12:116–21. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Brandes AA, Franceschi E, Tosoni A, Bartolini S, Bacci A, Agati R, Ghimenton C, Turazzi S, Talacchi A, Skrap M, Marucci G, Volpin L, Morandi L, Pizzolitto S, Gardiman M, Andreoli A, Calbucci F, Ermani M. O(6)-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro Oncol. 2010;12:283–8. doi: 10.1093/neuonc/nop050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Bobola MS, Varadarajan S, Smith NW, Goff RD, Kolstoe DD, Blank A, Gold B, Silber JR. Human glioma cell sensitivity to the sequence-specific alkylating agent methyl-lexitropsin. Clin Cancer Res. 2007;13:612–20. doi: 10.1158/1078-0432.CCR-06-1127. [DOI] [PubMed] [Google Scholar]
- [99].Hegi ME, Sciuscio D, Murat A, Levivier M, Stupp R. Epigenetic deregulation of DNA repair and its potential for therapy. Clin Cancer Res. 2009;15:5026–31. doi: 10.1158/1078-0432.CCR-08-1169. [DOI] [PubMed] [Google Scholar]
- [100].Blank A, Bobola MS, Gold B, Varadarajan S, Kolstoe DD, Meade EH, Rabinovitch PS, Loeb LA, Silber JR. The Werner syndrome protein confers resistance to the DNA lesions N3-methyladenine and O6-methylguanine: implications for WRN function. DNA Repair. 2004;3:629–38. doi: 10.1016/j.dnarep.2004.02.003. [DOI] [PubMed] [Google Scholar]
- [101].Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair. 2007;6:953–66. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- [102].Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Huertasm P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–6. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Abbotts R, Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat Rev. 2010;36:425–35. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- [105].Bobola MS, Emond MJ, Blank A, Meade EH, Kolstoe DD, Berger MS, Rostomily RC, Silbergeld DL, Spence AM, Silber JR. Apurinic endonuclease activity in adult gliomas and time to tumor progression after alkylating agent-based chemotherapy and after radiotherapy. Clin Cancer Res. 2004;10:7875–83. doi: 10.1158/1078-0432.CCR-04-1161. [DOI] [PubMed] [Google Scholar]
- [106].Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SG, Laird PW, Aldape K. Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu IL, Carro MS, Dai F, Tagge MJ, Ferrarese R, Bredel C, Phillips HS, Lukac PJ, Robe PA, Weyerbrock A, Vogel H, Dubner S, Mobley B, He X, Scheck AC, Sikic BI, Aldape KD, Chakravarti A, Harsh GR. 4th NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364:627–37. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Lee Y, Scheck AC, Cloughesy TF, Lai A, Dong J, Farooqi HK, Liau LM, Horvath S, Mischel PS, Nelson SF. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Etcheverry A, Aubry M, de Tayrac M, Vauleon E, Boniface R, Guenot F, Saikali S, Hamlat A, Riffaud L, Menei P, Quillien V, Mosser J. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics. 2010;11:701. doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, Forrest WF, Pujara K, Carrillo JA, Pandita A, Ellingson BM, Bowers CW, Soriano RH, Schmidt NO, Mohan S, Yong WH, Seshagiri S, Modrusan Z, Jiang Z, Aldape KD, Mischel PS, Liau LM, Escovedo CJ, Chen W, Nghiemphu PL, James CD, Prados MD, Westphal M, Lamszus K, Cloughesy T, Phillips HS. Evidence for Sequenced Molecular Evolution of IDH1 Mutant Glioblastoma From a Distinct Cell of Origin. J Clin Oncol. 2011;29:4482–90. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]