Abstract
Complex regulatory networks orchestrate most cellular processes in biological systems. Genes in such networks are subject to expression noise, resulting in isogenic cell populations exhibiting cell-to-cell variation in protein levels. Increasing evidence suggests that cells have evolved regulatory strategies to limit, tolerate, or amplify expression noise. In this context, fundamental questions arise: how can the architecture of gene regulatory networks generate, make use of, or be constrained by expression noise? Here, we discuss the interplay between expression noise and gene regulatory network at different levels of organization, ranging from a single regulatory interaction to entire regulatory networks. We then consider how this interplay impacts a variety of phenomena such as pathogenicity, disease, adaptation to changing environments, differential cell-fate outcome and incomplete or partial penetrance effects. Finally, we highlight recent technological developments that permit measurements at the single-cell level, and discuss directions for future research.
Keywords: expression noise, gene regulatory network, persistence, phenotypic variation, single-cell analysis, differentiation and development
Gene expression noise
Random fluctuations in expression levels of individual proteins are inevitable. These fluctuations are due to the intrinsically stochastic nature of molecular interactions that underlie transcription, translation and post-translational regulation [1–7]. This results in variation in protein expression levels within clonal cell populations, despite a homogenous environment, a phenomenon referred to as gene expression noise or stochasticity [1–7]. In a more general sense, expression noise may also refer to variation in the abundance of biomolecules such as mRNA. Gene expression noise was first described during the second half of the 20th century [3, 4]. However, with recent advancements in single-cell and single-molecule techniques, it has become possible to quantify its extent and better grasp its importance. These approaches have revealed that the expression level of a gene can drastically vary between individual cells, exhibiting stochastic fluctuations that can span up to six orders of magnitude [5].
The last decade witnessed a renewed interest in understanding the significance of expression noise in synthetic gene circuits [5, 8, 9]. These studies revealed that the expression of the genes in such circuits were subject to fluctuations in amplitude, frequency and timing as a result of stochastic effects during the process of gene expression. Subsequent experiments dissected the characteristics of noise and defined its intrinsic and extrinsic components [6, 7] (See Box 1). Research is now also aimed at understanding how expression noise is handled within gene regulatory networks (GRNs), which control the transcriptional, signaling and developmental programmes in cells [5, 10].
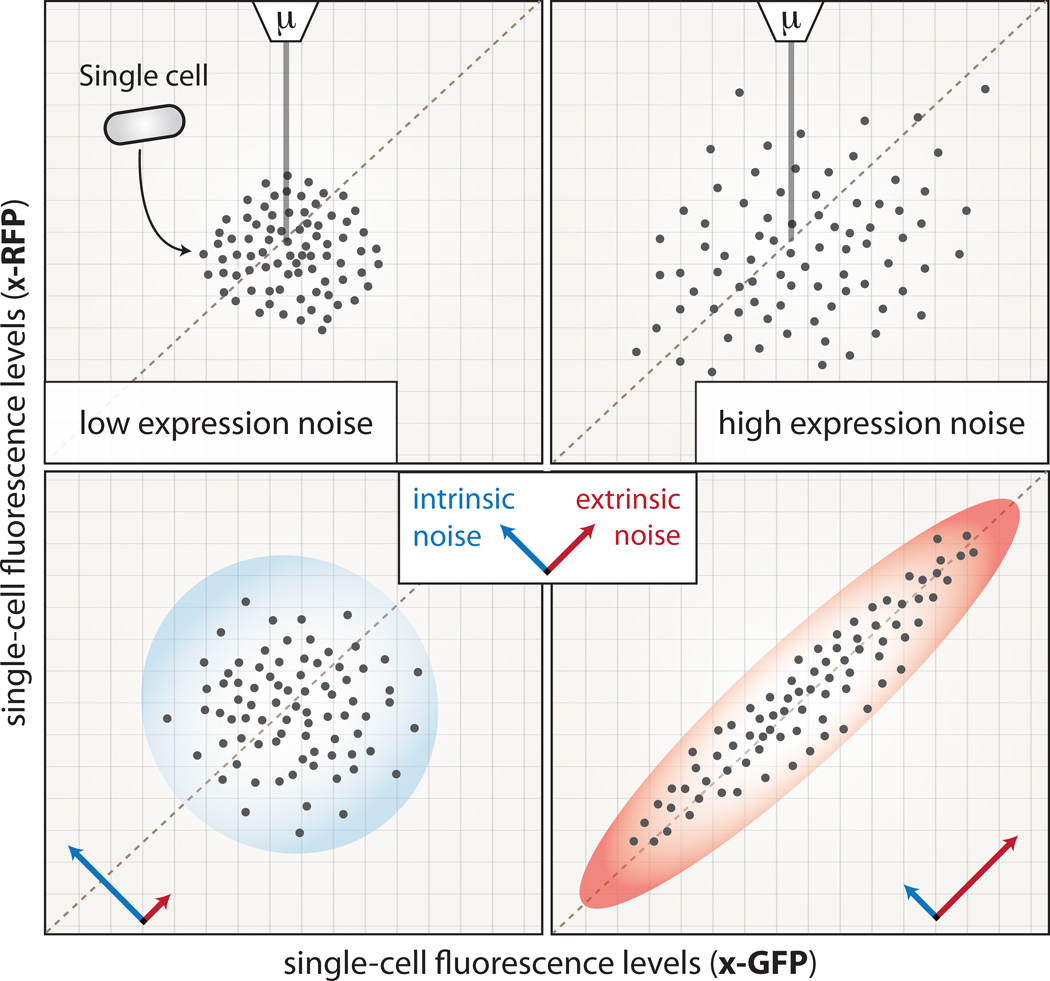
Box 1. Quantification of expression noise.
Expression noise can be quantified using a fluorescently tagged version of a protein of interest, and measuring the distribution of normalized fluorescence intensities across a population of clonal cells in a stable environment (Figure I). Noise is generally defined as the squared coefficient of variation of the fluorescence levels (SCV= (σ/μ)2). This dimensionless value basically reflects how large the standard deviation (σ) of expression levels is compared to the mean expression level (μ). Alternative measures of noise have also been proposed or used, e.g. coefficient of variation (CV=σ/μ), Fano Factor (FΔt= (σ2/μ)Δt) or distance from the median CV (DM) [110]. Expression noise can be decomposed into intrinsic and extrinsic components [7, 111]. The measurement of intrinsic and extrinsic noise of a gene×requires the use of double reporter systems in which two identical promoter regions regulate two distinct fluorescent reporter genes (here x-GFP and x-RFP). In the case of intrinsic noise, each reporter fluctuates independently of the other. In the case of extrinsic noise, the fluctuations in fluorescence of the two reporters co-vary.
Several excellent reviews describe key experiments and concepts pertaining to the molecular mechanisms that generate expression noise in a gene [2, 11, 12] (See Box 2). In this review, we focus on the interplay between gene expression noise and the architecture of gene regulatory networks at different levels of organisation (Figure 1a). This interplay is linked to a fundamental question that the field is just beginning to address: how does expression noise influence the ability of a GRN to accurately and robustly relay information, i.e. convert variations in its inputs into appropriate responses in its outputs? Here, we synthesize the current knowledge on how GRN architecture handles the tradeoff between information transfer and information loss due to expression noise in order to guide cellular outcomes. Further, we review examples demonstrating how this interplay provides considerable advantages to performing a number of fundamental biological processes.
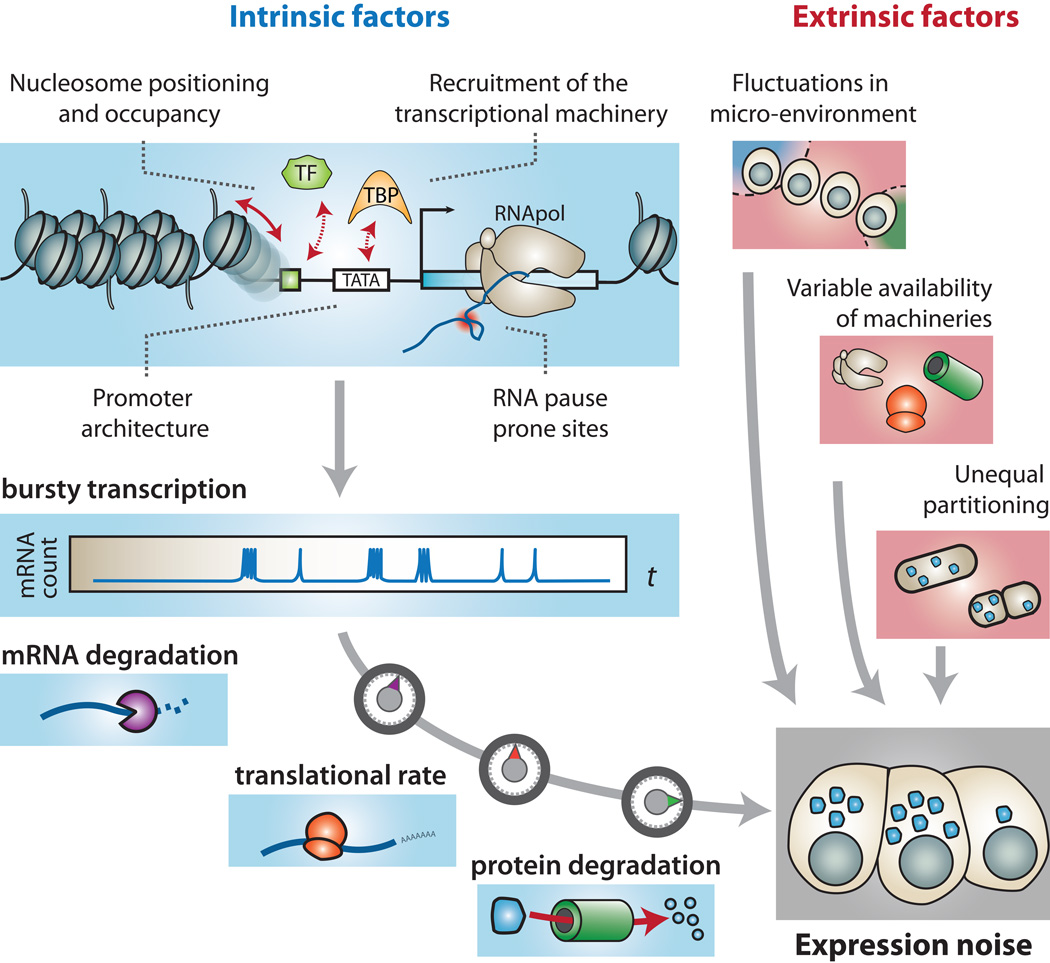
Box 2. Factors contributing to expression noise.
Gene expression is conditioned by the probabilistic occurrence of molecular interactions and reactions. However, all molecular interactions/reactions may not contribute equally to expression noise, which explains the need to identify and characterize the most influent factors contributing to intrinsic and extrinsic expression noise (Figure I).
Intrinsic noise
-
▪Transcriptional bursting – Dynamics and frequency of the pre-initiation complex (PIC) assembly at a promoter site can be perturbed in many ways, thereby causing bursts in transcription initiation [112–114]. In eukaryotes, this particularly depends on:
- ◦
-
◦Nucleosome occupancy and positioning – access to DNA by regulatory proteins depends on local chromatin organization. In a population of cells, this translates to how consistently nucleosomes tend to position and occupy a given genomic coordinate in each individual [118–120], thereby influencing on transcriptional regulation.
-
◦Transcriptional pausing – The presence of polymerase pause-sites can fine-tune noise, by either stalling polymerases or causing premature termination [112].
-
▪
Nuclear architecture – Remote chromosomal segments can come spatially close to form transcription factories. Stochastic recruitment of such segments may lead to noise [121].
-
▪
Chromatin epigenetics – Histone modifications and DNA methylation can be added and removed in a switch-like manner [122, 123].
-
▪
Translation rates [93], mRNA degradation [124] and protein degradation are sources of intrinsic noise for which molecular determinants are yet to be characterized.
Extrinsic noise
-
▪
Availability of basic machinery – Variability in abundance of gene expression machineries can greatly affect the production of all proteins at once in a cell. For example, cell-to-cell variation in ribosome abundance will lead to global variation in protein levels [2].
-
▪
Pathway-specific propagation – Intrinsic noise in the gene expression of regulatory proteins can propagate in the expression dynamics of downstream targets, for which it serves as a source of extrinsic noise [16].
-
▪
Micro-fluctuations in cellular environment – Environments within which cells grow are never completely homogenous. This can affect the behavior of individuals in a cell population and may even be further reinforced by paracrine signaling that alters local cellular environment [15, 125].
-
▪
Cell division or asymmetric partitioning – After cell division, the daughter cells are often approximately of similar size but they can also differ greatly. Such differences will lead to uneven segregation of mRNA and protein material, which globally affects daughter cells [88].
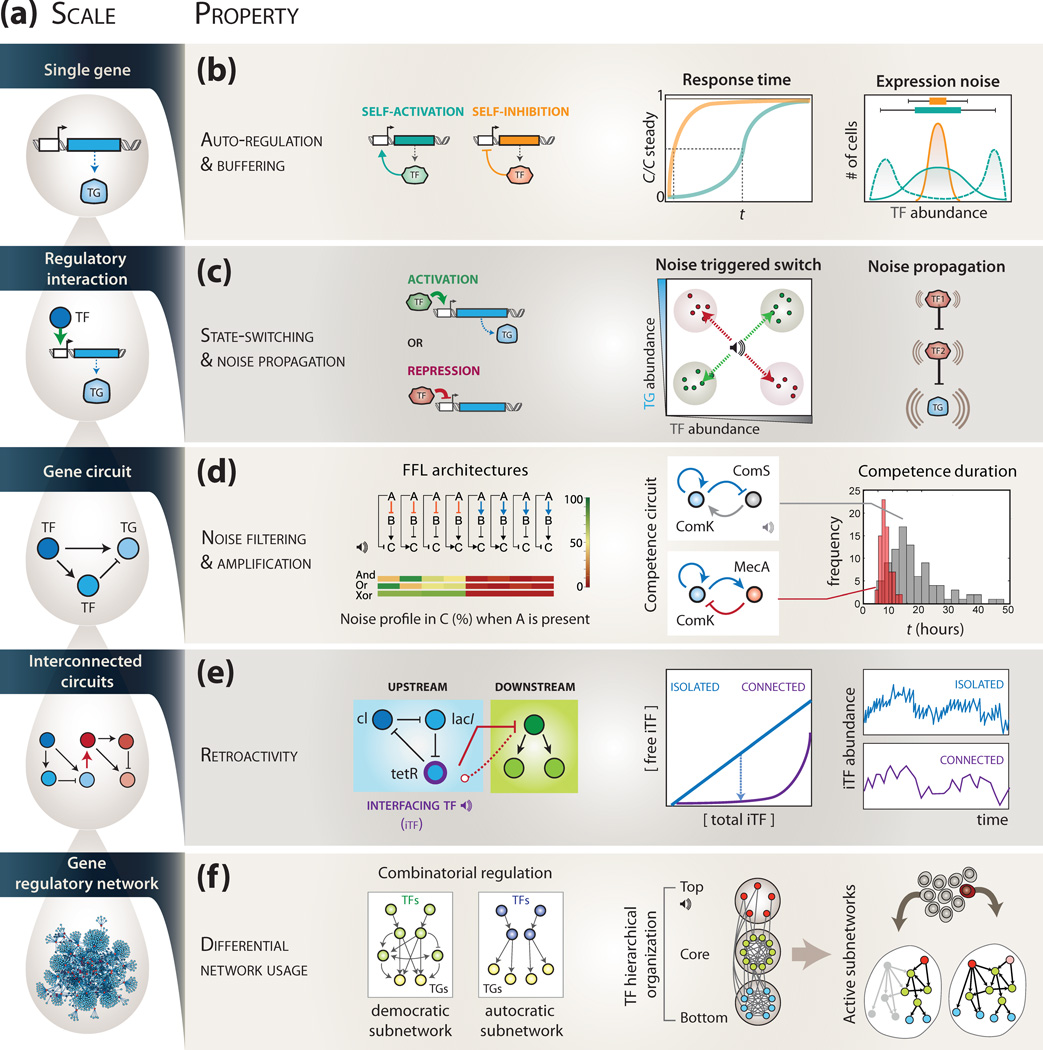
Figure 1. Interplay between expression noise and regulatory network architecture at distinct levels of network organization.
(a) Gene Regulatory Networks (GRNs) consist of distinct levels of organisation, ranging from a single auto-regulatory gene to entire networks. Filled circles represent genes and plain arrows denote regulatory interactions. (b) Self-activating loops (cyan) and self-inhibitory loops (yellow) show opposite noise properties (adapted from [14]). In the first panel, the (normalized) steady state concentration of TF is reached faster in the case of self-inhibition than self-activation. In the second panel, distribution of expression levels is broader in the case of self-activation (cyan, plain curve), and may also be bimodal (cyan, dashed curve). Both behaviors described may be influenced by other factors such as promoter strength, initial conditions and degradation kinetics. (c) Noise-driven state-switching and noise propagation in regulatory interactions. The abundance of a target gene (TG) may be altered by stochastic fluctuations in the abundance of a regulating transcription factor (TF). Noise can also propagate in linear cascades of regulatory interactions. (d) The topology of gene circuits encodes distinct noise susceptibility. For instance, feed-forward loops (FFLs) cluster in two groups of architecture with opposite noise filtering capacity (adapted from [22]). The noise profile for three different logical integration of inputs in the gene C is shown; they include AND (both A and B), OR (A or B) and XOR (either A or B but never both). In B. subtilis, the ComK-ComS circuit controls competence events (upper panel). The use of a synthetic alternative circuit (bottom panel) can achieve higher accuracy in the duration of cycles (red histogram), at the cost of fragility in fluctuating environments (adapted from [25]) (e) The expression dynamics of a gene circuit can be influenced by the presence of a downstream circuit to which it is connected (retroactivity, dashed arrow). The sequestration of the interfacing TF (iTF) on promoters of genes exterior to the upstream circuit increases the autocorrelation time of its expression level, i.e. reduces variation in short time windows, thereby affecting expression noise (adapted from [31]). (f) Combinatorial regulation and hierarchical organisation of GRNs can contribute to adaptability. Distinct noise properties of TFs in different hierarchical layers facilitate sampling of multiple sub-networks, and hence different phenotypes, by the individuals of a clonal cell population (adapted from [42]; see Figure 2a). The volume bell symbol denotes gene expression noise.
The interplay between noise and regulatory network organisation
In order to grasp the extent to which genes remain functional despite variation in their expression levels, we need to understand how cells manage to integrate and transfer information through gene regulatory interactions. Here, we discuss how gene regulatory networks permit the buffering, amplification or toleration of gene expression noise. More specifically, we describe the impact of expression noise at distinct levels of network organisation, ranging from a simple regulatory interaction to genome-scale regulatory networks and sub-networks (Figure 1a).
Auto-regulation and noise
Auto-regulation constitutes the simplest case of feedback mechanism in transcriptional regulation, where a transcription factor (TF) binds to its own promoter region and regulates its own expression. This can occur in two possible scenarios; either self-inhibition (negative auto-regulation) or self-activation (positive auto-regulation), as shown in Figure 1b. Self-inhibitory TFs are particularly widespread in prokaryotic transcription regulatory networks. For example, they are seen in half of Escherichia coli’s TFs [10]. Through this specific type of negative feedback loop, self-inhibitory TFs can maintain a steady expression level by limiting large bursts in their transcription, which minimizes the time needed to reach steady state levels (i.e. the response time) [13, 14]. This favors fast and controlled transcriptional responses, which in turn reduces expression noise [9]. In contrast, positive auto-regulation leads to slower response times and higher expression noise (Figure 1b). In this case, the TF expression is fueled by its own synthesis, of which levels are at first limiting before they reach an activation threshold, manifested by a sigmoidal response over time [14]. In addition, the time taken by a feedback loop influences expression noise [14]; auto-regulatory loops with long delay, as well as feedback loops made of more than one component, can display increased noise.
Regulatory interactions, noise and state switching
While auto-regulation provides a simple way to change the response time and noise sensitivity of a single gene, more complex regulatory programs necessitate higher scales of network architecture. Regulatory interactions between TFs and target genes (TGs) represent the smallest building block of GRNs (Figure 1c). Expression noise in a TG can result from fluctuations in levels of TFs (or of signaling molecules, influencing TF activity); a phenomenon known as noise propagation. In particular, small variations around a critical threshold in TF abundance can lead to a radical switch between the distinct expression states of the TG, and hence of cellular phenotypes. Noise can thus propagate through regulatory interactions, as shown in synthetic regulatory cascades [15]. This effect increases with the size of linear regulatory cascades [16].
Noise propagation is particularly important in assessing the information capacity of a regulatory interaction, i.e. the number of distinguishable stable states in expression level of TG that can be achieved by varying the concentration of a TF [17]. Attempts to estimate the theoretical maximal information transfer possible between a TF and a TG suggest constraints in the dynamic range, input/output relationship, and input expression noise that characterize a regulatory interaction [17–19]. Whether the boundaries of these constraints can be explored in GRNs – and under what circumstances – remains an open, fundamental question. In this context, a recent study utilizing the TNF-NFκB model suggests that in signaling networks with multiple genes, upstream bottlenecks can limit the information gained. Negative feedback to this bottleneck seem to both alleviate and enhance its limiting effect, despite decreasing noise [20].
Gene circuits and expression noise
Coordinated and complex transcriptional responses generally involve multiple regulatory interactions organized into gene circuits. In such an organisation, the information sensed by upstream components of circuits (e.g. concentration of an extracellular ligand) is relayed to activate downstream components (e.g. changes in expression levels of response-related TGs). Interestingly the topology of a gene circuit does not only determines its information-processing capability [14], but also encodes its sensitivity to expression noise [21–23]. For instance, negative feedback loops (FBLs) have been shown to reduce noise [9, 14], which often comes at the expense of signaling sensitivity [24]. However, the implementation of positive feedback loops within particular topologies has been identified to reduce noise without compromising sensitivity to long term changes in inputs. This effect appears to arise due to improved averaging of rapid fluctuations over time [23].
Feed-forward loops (FFLs) constitute another set of network motifs commonly found in GRNs [14]. Typically a FFL comprises of a general TF (A) that regulates two genes: the specific TF (B) and the target gene (C), with C also being regulated by the specific TF (B) (Figure 1d, left panel). Hence, depending on the nature of the regulatory interactions (activation or repression), a total of 23=8 FFL architectures are possible. The reason why in nature certain types of FFLs are much more frequent than others may be related to their noise behavior, which inturn is determined by their topology (Figure 1d). For instance, it was recently shown that FFLs can be classified into two groups, delineated by opposing properties in their noise buffering capacity [22]. Specifically, the steady state noise behavior of FFLs appears to correlate primarily with the type of regulation A has over B. When A represses B, the expression noise in C is high in the presence of A (Figure 1d, green horizontal bars of the noise profile). Conversely, when A activates B, the expression noise in C is low in the presence of A (Figure 1d, red horizontal bars) [22].
Small circuits that are not identified as network motifs can also mediate distinct noise behavior. This is the case of the transcriptional circuit controlling competence in Bacillus subtilis, which is based on two TFs: ComK and ComS. A synthetic circuit, in which the ComS TF is replaced by the protease MecA and wired with a different topology (Figure 1d, right panel), was shown to mimic the competence dynamics of the ComK-ComS circuit. In the native circuit, variability in the duration of competence is a consequence of noise in ComS levels. However, substituting ComS with MecA (thereby altering the topology of the synthetic circuit) allows for similar dynamics of activation but decreases variation in the duration of competence cycles [25]. Cells harboring the synthethic circuit have a lower adaptability to changing environments due to this decreased variation as compared to those harboring the native circuit.
In summary, circuit topology provides a way to locally tune noise propagation, with some topologies favoring noise buffering [24], some limiting information loss (e.g. in signaling networks [20]) and others amplifying it [22]. Thus topologically distinct circuits may have similar dynamic properties but different noise behavior [25]. This distinction might explain why certain network motifs, or combinations thereof, preferentially emerge in biological systems and are retained in evolution [14, 22].
Interconnected gene circuits, information processing and retroactivity
While synthetic circuits can be engineered to operate in isolation, in nature, circuits are predominantly interconnected [26]. Typically, the output of one circuit serves as an input to another circuit. In this context, theoretical models indicate that the dynamic behavior of a gene circuit can be influenced by the downstream components to which it is connected [27]. In this respect, the response time of a given gene circuit is longer when it is serially connected to a downstream circuit, than if it were in isolation (Figure 1e). This phenomenon is termed “retroactivity” [27].
In this setup, the output gene of the upstream circuit is itself a TF that regulates components of a downstream circuit (referred to as an “interfacing TF”). Here, the concentration of the interfacing TF over time serves as input to the downstream circuit. The model predicts that the binding of an interfacing TF to its target promoters can sequester enough TFs from the degradation machinery so that its protein degradation is minimized (Figure 1e). This can increase the time required to readjust the dynamics of the upstream module. At a given expression level, the number of regulated sites occupied by the interfacing TF determines the upstream regulatory module’s susceptibility to retroactivity. Likewise, at a fixed number of regulated sites, the abundance of an interfacing TF will condition the amplitude of retroactivity effects. Therefore, this interplay between TF connectivity and TF abundance may influence the sensitivity to noise of the upstream module. In this context, it should be noted that the number of binding sites (e.g. clusters of binding sites for a same TF) upstream of a gene can potentially influence the nature of the output response [28]. Similarly, interactions of TFs with other proteins, which may lead to sequestration into productive or non-productive complexes, may also influence the kinetics of a regulatory interaction [29, 30] and retroactivity.
Indeed, it was recently demonstrated that the correlation time of an interfacing TF’s expression noise increases when it is allowed to regulate downstream TGs (Figure 1e). This indicates that expression noise increases with retroactivity [31]. In GRNs, one could test the importance of retroactivity effects by comparing the dynamics and expression noise of circuits that have identical architectures but different output connectivities. Quantifying the extent of retroactivity may help to design larger, integrated synthetic gene circuits where the response can be accurately predicted [32].
Sub-networks, redundancies and differential network usage
How is it possible for cells to adapt to changing conditions despite noise propagation in GRNs? Studies on the structural and dynamic properties of entire GRNs have shown that specific topological features such as hierarchical organisation (i.e. vertical stratification) and crossregulation between TFs (i.e. mutual regulation within or among hierarchical layers and combinatorial regulation) can contribute to adaptability [33–37] (Figure 1f, left panel).
Integration of gene expression data with GRN topology has shown that sub-networks that are active in different conditions (e.g. during development or survival) have strikingly different topologies [38–40]. For instance, regulatory programs that drive processes such as cell cycle progression and sporulation have a rather “democratic” structure [41] in which a large number of genes mutually regulate each other, both in a hierarchical and combinatorial manner, when making cellular decisions (Figure 1f, left panel). In contrast, sub-networks that become active under stress conditions show a more “autocratic” topology [41] where a few master regulators control the expression of entire cascades of genes with little combinatorial regulation (Figure 1f, left panel). Therefore, “democratic” networks might limit variation in information flow by facilitating a consensus decision to emerge. This may thereby contribute to increased fidelity in a cellular response. In contrast, “autocratic” networks may permit variation in information flow by allowing expression level differences in key regulatory hubs among individuals in a population to excert distinct phenotypes. This may thereby contribute to phenotypic variability at the population level (see below; Figure 2).
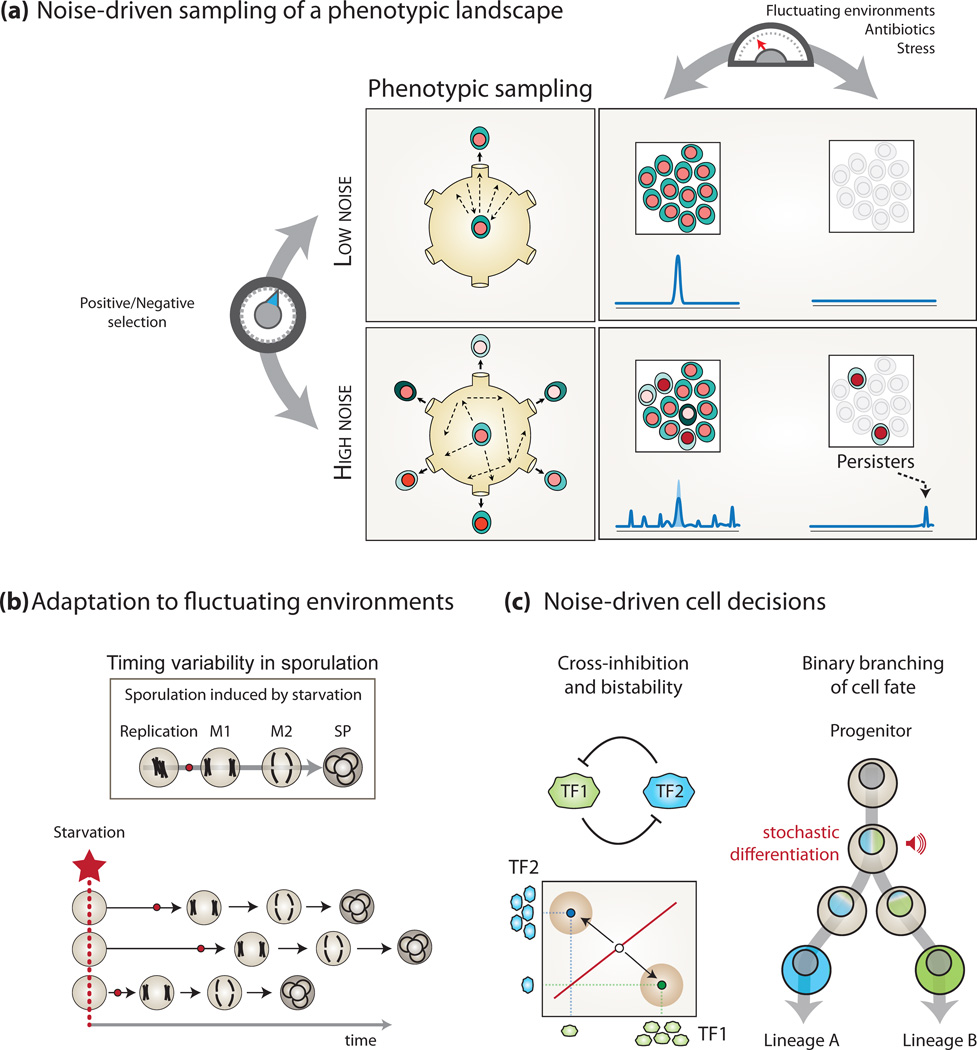
Figure 2. Functional outcomes of the interplay between noise and network topology.
(a) Tuning of expression noise during evolution can occur in a gene-centric manner by positive/negative selection of molecular determinants of intrinsic noise. From low noise to high noise, the phenotypic diversity among individuals of a population increases, which allows for persistence in deleterious environmental fluctuations, stress conditions and antibiotic treatments. (b) Adaptation to fluctuating environments is facilitated by expression noise of key regulatory genes in a clonal cell population. For instance, upon nutrient starvation (red star), individual yeast cells in a population undergo sporulation in an unsynchronized fashion (horizontal profiles). This heterogeneity in sporulation timing is linked to expression noise in the master regulator Ime1p. This favors the maintenance of non-sporulated cells that are pre-adapted in case of reversion to nutrient rich condition (adapted from [59]). The small red circle denotes the point of commitment to the sporulation pathway. (c) Expression noise may determine cellular differentiation, and frequently involves bistable circuit architectures, within which small fluctuations in TF abundance can trigger distinct stable states. This permits the differentiation into distinct lineages from a progenitor cell type.
In fact, most GRNs do not show a strictly autocratic nor democratic topology, but generally show properties that are inbetween [36, 42]. For instance, it has been shown that the TFs in the entire yeast GRN could be organized into three non-overlapping layers: top, core and bottom [42]. In this configuration the top-layer TFs regulate the core- and the bottom-layer TFs, and the core-layer TFs regulate the bottom-layer TFs, essentially forming a feed-forward network structure (Figure 1f, right panel). Additionally, the core-layer was found to carry most of the TF-TF regulatory interactions, and to form most of the feed-forward loops of the yeast GRN. Interestingly, the TFs in each layer display distinct dynamics of gene expression regulation. It was shown that the susceptibility to expression noise could vary across the distinct hierarchical layers of the network [42]. Specifically, TFs in the top-layer have elevated protein abundance and a wide range of protein half-life compared to TFs in downstream layers. Interestingly, these top-layer TFs also show higher expression noise than TFs in other layers. Based on these observations, it was proposed that the inherent hierarchy of the network and stochastic expression of the top-level TFs might permit differential utilization of the same underlying network in distinct members of a clonal cell population. This may be achieved by stochastically (i) activating sub-networks in the GRN, thereby promoting survival in rapidly changing environments [42] or (ii) poising certain individuals to activate different sub-networks when the appropriate input signal is received [42].
Functional outcomes of the interplay of noise and network topology
Expression noise is sometimes viewed as a deleterious yet inevitable burden of gene expression control. This is an idea supported by in silico evolutionary models where for instance suboptimal enzyme concentration resulting from noise was shown to hamper fitness in a constant environment [43]. Consistent with this observation, dosage-sensitive genes appear to have minimized expression noise with selective pressure on their coding sequence and chromosomal location [44–46]. In contrast, however, there is also evidence that the elevated intrinsic noise level of certain genes is under positive selection [47], suggesting that the impact of expression noise is not always deleterious. Beneficial consequences of noise, however, emerge when considering a population of cells in its entirety [48]. In particular expression noise can improve the chances of a clonal population to adapt to variable conditions, by enabling a probabilistic sampling of available states in the phenotypic space; a phenomenon sometimes referred to as “bet-hedging” (See Figure 2a). This strategy permits populations to overcome challenges otherwise costly to implement with deterministic systems [49, 50], hence facilitating adaptive evolution [51]. In the following sections, we discuss how expression noise (i) enables populations of cells to face fluctuating environments, (ii) drive cellular decisions, e.g. in developmental programs and (iii) results in the manifestation of variable phenotypes among genetically identical individuals.
Phenotypic variability in fluctuating environments
Isogenic microbial populations exposed to diverse stress conditions often persist due to a small fraction of surviving individuals who replenish the population. This capacity of survival has been attributed to variable expression levels in key regulatory proteins that are under selection to facilitate random phenotypic switching [52, 53]. In a single cell, expression noise in a set of key master regulators can result in a differential utilization of underlying GRNs, which can allow for phenotypic switching [42]. In a population of cells, such variability can be seen as a source of phenotypic heterogeneity where each possible phenotype may have a higher fitness in different environments (Figure 2a). In this context noise can be considered as a form of “blind” short-term, reversible, adaptation [54–56].
To date, several genes that control phenotypic switching have been identified in different organisms. This is the case of phoU, a gene coding for a negative regulator of general metabolism in E. coli. In rich media PhoU is down regulated, which allows for increased metabolic activity and accelerates growth. However, a low level of PhoU also increases sensitivity to antibiotics and to changes in nutritional environment [57]. Therefore, when exposed to antibiotics only the fraction of cells in which PhoU remains expressed at high levels can drive the survival of the population. These cells are referred to as “persisters” [58], and can limit the eradication of bacterial infections [54]. Similarly, Mycobacterium tuberculosis can switch between a virulent active state and a persisting latent state allowing for post-primary transmissible tuberculosis [56]. This emphasizes the requirement for understanding the molecular origins of phenotypic switching, in order to design efficient treatment strategies that prevent persistent pathogenic subpopulations to replenish. Cell survival in fluctuating environments has also been described in a synthetic system, where the expression level of a master regulator is modulated to promote fast and slow switching between phenotypes [54].
Likewise, noisy expression of Ime1p, a master regulator of meiosis in yeast, is linked to variability in the activation of the sporulation pathway (Figure 2b). Under fluctuating nutrient starvation conditions, some yeast cells sporulate whereas others undergo delayed sporulation [59]. Similarly, the activation of competence in B. subtilis is a probabilistic and transient event linked to expression noise in ComK and ComS, which impact on the frequency and duration of competence cycles, respectively [21, 25, 60, 61].
Although stochastic phenotypic sampling often relies on specific regulatory network architectures that facilitate stochastic switching – e.g. through bistable circuits [54, 62] – evidence suggests that cell populations might also use noise-driven monostable expression to adapt to fluctuating environments without the explicit need of an underlying circuit. In such cases, cells need to reach unstable states of expression to achieve adaptation, as in the case of histidine starvation in E. coli [63]. However, those individual cells tend to revert back to their unfit, yet stable, initial state even in the stress condition. Hence, to achieve adaptation, stochastic exploration of unstable states requires a strong increase in fitness in the unstable state.
Furthermore, an analysis of gene networks derived from chemogenomic studies indicates that a set of rapidly evolving genes with noisy expression plays an important role in natural resistance to harmful chemicals [64]. More specifically, it has been suggested that a balance between (i) the noisy expression of certain key genes required for tolerating specific stress conditions and (ii) robustness conferred by generic stress tolerance genes might be crucial in surviving diverse environmental stress [64].
Differential cell-fate outcome in response to the same uniform stimulus
Fractional survival or cell-death in clonal cell populations upon drug treatment is a well-known phenomenon in some diseases such as cancer. In this respect, cancer cells resemble the bacterial persisters described above. For instance in the case of lung adenocarcinoma, which is commonly treated with the drug Erlotinib (an epidermal growth factor receptor inhibitor), relapse often occurs in patients due to a small fraction of non-dividing persisters. The drug-tolerance seems to originate, at least in the early stages, from altered chromatin configurations due to expression noise in chromatin modifiers [65]. The authors of this study found that the persisters in lung cancer cells (PC9, adenocarcinoma cell line) have an increased expression of a histone demethylase following the upstream activation of an IGF-1 receptor [65]. Improved tumor cell eradication was only possible by use of a combinatorial treatment using an IGF-1 receptor inhibitor, in order to prevent altered chromatin states, followed by Erlotinib treatment [65]. This again highlights the importance of understanding the molecular origins of phenotypic variability in isogenic populations while developing treatment strategies.
Differential response to chemotherapy drugs such as Camptothecin is linked to noisy expression in a small subset of proteins [66]. Some of these noisy proteins were found to be involved in cell-fate decision. In another study, upon induction of apoptosis, it was shown that expression noise of apoptotic proteins is the primary cause of cell-to-cell variability in the timing and probability of cell death of individual members in a cancer cell population [67]. Here noisy expression of cell fate regulatory proteins promotes differential sampling of the same underlying regulatory network (e.g. pro-survival or apoptotic network) by different members in a clonal population, thereby resulting in divergent cell-fate outcomes.
Similarly, a recent study revealed that stochastic receptor expression determines cell fate despite uniform treatment with interferon [68]. When presented with interferon, cells may either adopt an antiviral response or an antiproliferative response. The authors revealed that a low number of receptors is sufficient for mediating an antiviral response and is therefore a robust feature common to all cells. Conversely, only cells with a high number of receptors can mediate the antiproliferative response. Thus for a given cell, the response is binary and dependent on the stochastic expression levels of the receptors on an individual cell [68].
Cellular differentiation and development
During stem cell differentiation, the appropriate expression in space and time of key regulatory proteins dictates lineage specification of progenitor cells (e.g. myeloid lineage commitment from hematopoietic stem cells [69]). They also influence the formation of distinct spatial patterns of celltypes during organ development (e.g. cell fate specification in neural development [70]). In this context, expression noise in TFs might play an important role in development, stem cell maintenance and differentiation, and reprogramming [71–74]. Recent studies report that expression noise of specific TFs controls lineage choices during stem cell differentiation [71, 75] (Figure 2c). While dedicated circuits that filter expression noise might be required for the robustness of certain cellular processes [73, 76], distinct TF expression dynamics across the transcriptional hierarchy, such as observed in the yeast GRN [42] (Figure 1f), might play a role during stem cell differentiation [77]. This can be advantageous for initiating distinct responses and for sampling subnetworks that permit lineage commitment when the appropriate signals are experienced [77].
Noise can also play an important role in cell-fate decisions of multipotent cells, in which independent differentiation programs are executed simultaneously until a commitment point is reached. A recent study in B. subtilis showed that competence and sporulation pathways are both active and “compete” until one of the two reaches its commitment point [78]. In this race, noise in the expression level of competing TFs can influence cellular outcome by providing a head start to one of the regulatory programs. These programs do not cross-regulate each other until either of their respective commitment point is reached, after which cross-regulatory interactions between the competing programs ensure the reliable execution of the chosen regulatory program [78].
While the concepts described here are drawn from studies on unicellular organisms, it holds true for more complex multicellular organisms as well. For instance, during fly eye development, stochastic expression of a single transcription factor (Spineless) results in a mosaic expression of photoreceptors in individual ommatidium that detect different wavelenths required for colour vision [79]. Similarly, mosaic expression of odorant receptors has also been well characterised in the olfactory system of many organisms including humans [80]. Such mosaicism in cell type specificity might not only arise due to stochastic expression of key factors during development but also due to intercellular communication via lateral inhibition [81]. For a review on this topic, please see ref. 49 [49].
Incomplete or partial penetrance effects
While developmental regulatory programs are similar among individuals in a clonal population, all of the individuals of an isogenic population carrying the same mutation may not manifest a mutant phenotype. This phenomenon, known as incomplete or partial penetrance, has been observed in a number of organisms [82, 83] (Figure 2b). This effect is well documented in developmental defects and seem to involve genes that engender distinct phenotypes depending on their expression level [84]. Incomplete penetrance is generally linked to variation in environmental triggers revealing otherwise hidden phenotypic heterogeneity. However, recent studies have shown that intrinsic factors such as the introduction of expression noise by a genetic mutation or stochastic expression of phenotypic buffers and capacitors may also contribute to this phenomenon [82, 83, 85–87]. For example, (i) intestinal differentiation in embryos of Caenorhabditis elegans can be affected by mutation-triggered noise in the expression of components of the skn-1 transcriptional network [83] and (ii) mutations in tbx-9 can be buffered by stochastic expression of Hsp90 [87]. It has also been shown in B. subtilis that noise can facilitate developmental evolution by enabling the initial expression of discrete morphological traits at low penetrance, and allowing their stabilization by gradual adjustment of genetic parameters such as asymmetric segregation of protein molecules during cell division [82, 88].
Concluding remarks and future perspectives
The role of post-translational regulation in gene expression noise
The general view that transcriptional initiation is the most influential step for generating expression noise partially relies on the assumption that variations in mRNA levels explain most variations observed in protein levels. However at the population level, mRNA and protein abundances are weakly correlated [89–91]. Furthermore, it should not be assumed that phenomena observed at a population level remain true at the single cell level. Recent investigations have provided evidence that mRNA and protein levels can be considerably uncorrelated within a single cell at a given time point, as observed in E.coli [92] and inferred in mice [93]. Interestingly, both studies show that protein abundance is predominantly controlled at the post-transcriptional level. This suggests that the control of protein synthesis and degradation likely plays a prominent role in generating noise, a factor that current theoretical models largely overlook. Whether stochasticity in mRNA and protein degradation makes an effective contribution to noise is yet to be addressed.
Average network behavior versus network states within individual cells
The inference of expression levels by measurements made on a whole cell population masks the extent of gene expression heterogeneity between individuals. However, single cell studies are gradually overcoming this limitation [94–96]. Likewise, the inference of regulatory networks with population averages may mask heterogeneity in network usage by subpopulations. Indications that this is the case for pluripotency in mouse embryonic stem (ES) cells have recently been published [77]. In this study, the expression of 8 genes in the mice pluripotency network was studied at a single cell resolution in a population of 83 ES cells. The authors were able to infer that the apparent noise in the expression levels of the pluripotency genes resulted from subpopulations harboring distinct subnetworks. This suggests that heterogeneities in gene expression need not be interpreted as representing different “states” of a single unique network, but as a reflection of the activity of different sub-networks in subpopulations of cells [77]. Though every cell is genetically identical and hence should be governed by the same network topology, active sub-networks may emerge due to considerations such as variation in (i) the expression level of active TFs (e.g. through posttranslational modification or ligand/co-factor availability) or (ii) access to the target gene locus (e.g. via changes in chromatin status or epigenetic modifications). This suggests that phenotypic heterogeneity may also result from stable microstates, reflective of subpopulations rather than continuous expression variability within an entire population [77]. This highlights the need for experiments to study individual circuits and network dynamics at a single cell resolution. Such studies may be possible with the use of microfluidics to measure expression kinetics of TFs in living cells [93, 97].
Single-cell genomics
The ability to better measure expression noise at different scales of network architecture is heavily dependent on technological developments that enable measurements in a high throughput manner, and crucially, at the single-cell level and single-molecule resolution. Technologies that exploit microfluidics, DNA/RNA sequencing, mass cytometry, and mass spectrometry are now giving shape to a new area of “single cell omics” [98, 99]. These should provide the scrutiny and resolution needed to study heterogeneity at the single-cell level [100, 101]. The micro-reaction system [102], an alternative to microfluidic technology, has been introduced and applied to study DNA methylation patterns of individual cells in parallel. RNA FISH data has also been used to reconstruct real-time dynamics of RNA synthesis and degradation to investigate gene expression noise in unsynchronized cells [103]. An improved capacity to measure noise should help in the identification of molecular determinants of gene expression stochasticity (e.g. Box 2). Such an understanding can facilitate the design of synthetic circuits in which noise properties can be finely tuned as desired [104].
The interplay between expression noise and regulatory networks has received a lot of attention at the level of isolated cell populations [5, 10, 74, 105]. In the future, understanding the effects of noise on GRNs that (i) govern intercellular communication to generate synchronized behavior [106], (ii) define cellular patterns during development [107] or (iii) drive host-pathogen interaction [108] should bring important insights in a physiological setup. For instance, it was recently shown that complex behavioral phenotypes of honey bees can be inferred from the rewiring of neuronal GRNs [109]. Further novel insights into behavioral phenotypes may emerge by integrating understanding of noise with GRNs.
Understanding the interplay between gene expression noise and the architecture of gene regulatory networks has implications for a wide range of questions pertaining to biological systems. In the billion years of evolution of cellular systems, expression noise has not only prevailed, but it has also been exploited in many ways. Thus, uncovering principles by which networks have evolved to remain robust and adaptable in the face of gene expression noise should serve as an inspiration to design, engineer and tune gene regulatory networks.
Box 1 Figure I.
Quantification of expression noise.
Box 2 Figure I.
Major factors contributing to expression noise.
Acknowledgements
We thank Marija Buljan, Kai Kruse, Elizabeth Ing-Simmons and Andrew Deonarine for helpful remarks. This work was supported by the Gates Cambridge Scholarship and the Knox Trinity studentship (GC), the AFR Grant Scheme (CNJR), the Medical Research Council (GC, CNJR, and MMB), EMBO young investigator program, HFSP (RGY0073/2010; MMB), European Research Council (AMA) and the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (RJ: 1ZIAES102625-03) and NLM, NCBI (LA).
Abbreviations
- Bistable system
A system for which two alternative stable states exist
- Clonal cell population
Population of cells derived from a single mother cell, which share identical genomic contents (i.e. without mutations)
- Expression noise
Stochastic fluctuations in the protein level of a gene within a clonal cell population maintained in a homogeneous environment. Noise is generally defined as the squared coefficient of variation (SCV), i.e. the ratio between standard deviation and mean value of protein abundance for one gene in a population of cells. In a more general sense, expression noise may also refer to variation in the abundance of biomolecules such as mRNA
- Extrinsic noise
Gene-independent fluctuations that can be attributed to variations in external factors influencing expression of a set of genes (e.g. pathway-specific) or globally. Extrinsic noise can be influenced at a local level by abundance of transcription factors, and at a global level by the abundance of gene expression machinery (e.g. ribosome abundance)
- Fluctuating environment
Environment whose physical, chemical and nutritional components are subjected to frequent variations
- Gene circuit
A small number of genes influencing each other’s expression via regulatory interactions
- Gene regulatory network (GRN)
Network representation of gene regulatory events, in which nodes are genes and regulatory interactions are links. Such interactions include transcriptional regulation and post-translational modifications
- Intrinsic Noise
Gene-specific fluctuations that can be attributed to the stochastic fluctuations in the different steps of the expression process of a specific gene. Factors contributing to intrinsic noise include the gene’s promoter structure, its localization within the chromosome and the nuclear architecture
- Monostable system
A system for which only one stable state exists
- Network motif
Recurrent pattern of interconnections in a network that appear more frequently than expected by chance in random networks of similar size
- Network architecture
Pattern of links connecting the nodes of a network
- Noise propagation
Stochastic expression of a target gene resulting from gene expression noise in upstream regulatory genes, within a given circuit
- Retroactivity
Alteration in the dynamic behavior of a gene circuit due to the connectivity of its output gene with downstream circuits
- Stable/steady state
In the context of a regulatory interaction, the steady state expression level describes a narrow range of expression levels within which target gene abundance is maintained despite small perturbations in the expression of its regulatory transcription factor. This will depend on the kinetics of synthesis and degradation of the target gene
- TATA-box
Consensus DNA sequence 5′-TATAAA-3′ found within the proximal/core promoter of about one-fourth of the genes in eukaryotes and archaea. This sequence is typically bound by the TATA-box binding protein to initiate transcription by the polymerase
- Transcriptional burst
Bursts of mRNA production resulting from a gene's promoter switching between periods of prolonged inactive or "off" state and short-lived active or "on" states
- Transcriptional regulatory network
A network representation of transcriptional events where nodes represent TFs (transcription factors) and TGs (target genes) and links denote regulatory interactions between TFs and TGs, mediated by the binding of a TF to the promoter region of its TG
- Phenotypic buffering or capacitance
Is a phenomenon where individuals in a population produce the same phenotype despite genetic or environmental variation. Hsp90 is an example of a phenotypic buffer that can mask the effect of mutations in their substrate proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lestas I, et al. Fundamental limits on the suppression of molecular fluctuations. Nature. 2010;467:174–178. doi: 10.1038/nature09333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proceedings of the National Academy of Sciences. 1957:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spudich JL, Koshland DE. Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 5.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elowitz MB, et al. Stochastic gene expression in a single cell. Science (New York, N.Y.) 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 7.Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science (New York, N.Y.) 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 9.Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 10.Silva-Rocha R, de Lorenzo V. Noise and robustness in prokaryotic regulatory networks. Annual review of microbiology. 2010;64:257–275. doi: 10.1146/annurev.micro.091208.073229. [DOI] [PubMed] [Google Scholar]
- 11.Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science (New York, N.Y.) 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufmann BB, van Oudenaarden A. Stochastic gene expression: from single molecules to the proteome. Current opinion in genetics & development. 2007;17:107–112. doi: 10.1016/j.gde.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Rosenfeld N, et al. Negative autoregulation speeds the response times of transcription networks. J Mol Biol. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- 14.Alon U. Network motifs: theory and experimental approaches. Nature reviews. Genetics. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 15.Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science (New York, N.Y.) 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 16.Hooshangi S, et al. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3581–3586. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tkacik G, et al. Information flow and optimization in transcriptional regulation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12265–12270. doi: 10.1073/pnas.0806077105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tkacik G, et al. Information capacity of genetic regulatory elements. Physical Review E. 2008;78:1–17. doi: 10.1103/PhysRevE.78.011910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.So LH, et al. General properties of transcriptional time series in Escherichia coli. Nature genetics. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheong R, et al. Information Transduction Capacity of Noisy Biochemical Signaling Networks. Science. 2011;334:354–358. doi: 10.1126/science.1204553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Süel GM, et al. Tunability and noise dependence in differentiation dynamics. Science (New York, N.Y.) 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 22.Kittisopikul M, Süel GM. Biological role of noise encoded in a genetic network motif. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13300–13305. doi: 10.1073/pnas.1003975107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornung G, Barkai N. Noise propagation and signaling sensitivity in biological networks: a role for positive feedback. PLoS Comput Biol. 2008;4:e8. doi: 10.1371/journal.pcbi.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruggeman FJ, et al. Noise management by molecular networks. PLoS Comput Biol. 2009;5:e1000506. doi: 10.1371/journal.pcbi.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cagatay T, et al. Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell. 2009;139:512–522. doi: 10.1016/j.cell.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 26.Dobrin R, et al. Aggregation of topological motifs in the Escherichia coli transcriptional regulatory network. BMC Bioinformatics. 2004;5:10. doi: 10.1186/1471-2105-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Del Vecchio D, et al. Modular cell biology: retroactivity and insulation. Mol Syst Biol. 2008;4:161. doi: 10.1038/msb4100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giorgetti L, et al. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell. 2010;37:418–428. doi: 10.1016/j.molcel.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Buchler NE, Louis M. Molecular titration and ultrasensitivity in regulatory networks. J Mol Biol. 2008;384:1106–1119. doi: 10.1016/j.jmb.2008.09.079. [DOI] [PubMed] [Google Scholar]
- 30.Buchler NE, Cross FR. Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol Syst Biol. 2009;5:272. doi: 10.1038/msb.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim KH, Sauro HM. Measuring retroactivity from noise in gene regulatory networks. Biophys J. 2011;100:1167–1177. doi: 10.1016/j.bpj.2010.12.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chalancon G, Babu MM. Nanobiotechnology: Scaling up synthetic gene circuits. Nat Nanotechnol. 2010;5:631–633. doi: 10.1038/nnano.2010.178. [DOI] [PubMed] [Google Scholar]
- 33.Balaji S, et al. Uncovering a hidden distributed architecture behind scale-free transcriptional regulatory networks. Journal of molecular biology. 2006;360:204–212. doi: 10.1016/j.jmb.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 34.Balaji S, et al. Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. Journal of molecular biology. 2006;360:213–227. doi: 10.1016/j.jmb.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 35.Babu MM. Early Career Research Award Lecture. Structure, evolution and dynamics of transcriptional regulatory networks. Biochem Soc Trans. 2010;38:1155–1178. doi: 10.1042/BST0381155. [DOI] [PubMed] [Google Scholar]
- 36.Bhardwaj N, et al. Analysis of diverse regulatory networks in a hierarchical context shows consistent tendencies for collaboration in the middle levels. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6841–6846. doi: 10.1073/pnas.0910867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhardwaj N, et al. Analysis of combinatorial regulation: scaling of partnerships between regulators with the number of governed targets. PLoS computational biology. 2010;6:e1000755. doi: 10.1371/journal.pcbi.1000755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luscombe NM, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 39.Balazsi G, et al. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7841–7846. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Antonio A, et al. Functional organisation of Escherichia coli transcriptional regulatory network. J Mol Biol. 2008;381:238–247. doi: 10.1016/j.jmb.2008.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bar-Yam Y, et al. Systems biology. Attractors and democratic dynamics. Science. 2009;323:1016–1017. doi: 10.1126/science.1163225. [DOI] [PubMed] [Google Scholar]
- 42.Jothi R, et al. Genomic analysis reveals a tight link between transcription factor dynamics and regulatory network architecture. Mol Syst Biol. 2009;5:294. doi: 10.1038/msb.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Zhang J. Impact of gene expression noise on organismal fitness and the efficacy of natural selection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E67–E76. doi: 10.1073/pnas.1100059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehner B. Selection to minimise noise in living systems and its implications for the evolution of gene expression. Mol Syst Biol. 2008;4:170. doi: 10.1038/msb.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Batada NN, Hurst LD. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet. 2007;39:945–949. doi: 10.1038/ng2071. [DOI] [PubMed] [Google Scholar]
- 46.Becskei A, et al. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nat Genet. 2005;37:937–944. doi: 10.1038/ng1616. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, et al. Positive selection for elevated gene expression noise in yeast. Mol Syst Biol. 2009;5:299. doi: 10.1038/msb.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez-Maury L, et al. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 49.Losick R, Desplan C. Stochasticity and cell fate. Science (New York, N.Y.) 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chalancon G, et al. How do cells adapt to changing environments? Science. 2012 in press. [Google Scholar]
- 51.Espinosa-Soto C, et al. Phenotypic plasticity can facilitate adaptive evolution in gene regulatory circuits. BMC evolutionary biology. 2011;11:5. doi: 10.1186/1471-2148-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balaban NQ, et al. Bacterial Persistence as a Phenotypic Switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 53.Balaban N. Persistence: mechanisms for triggering and enhancing phenotypic variability. Current opinion in genetics & development. 2011;21:768–775. doi: 10.1016/j.gde.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Acar M, et al. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 2008;40:471–475. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- 55.Leibler S, Kussell E. Individual histories and selection in heterogeneous populations. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13183–13188. doi: 10.1073/pnas.0912538107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beste DJ, et al. The genetic requirements for fast and slow growth in mycobacteria. PLoS One. 2009;4:e5349. doi: 10.1371/journal.pone.0005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Zhang Y. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrobial agents and chemotherapy. 2007;51:2092–2099. doi: 10.1128/AAC.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 59.Nachman I, et al. Dissecting timing variability in yeast meiosis. Cell. 2007;131:544–556. doi: 10.1016/j.cell.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 60.Iber D. A computational analysis of the impact of the transient genetic imbalance on compartmentalized gene expression during sporulation in Bacillus subtilis. Journal of molecular biology. 2006;360:15–20. doi: 10.1016/j.jmb.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 61.Süel GM, et al. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 62.Kussell E, Leibler S. Phenotypic Diversity, Population Growth, and Information in Fluctuating Environments. Methods. 2005:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 63.Tsuru S, et al. Adaptation by stochastic switching of a monostable genetic circuit in Escherichia coli. Mol Syst Biol. 2011;7:493. doi: 10.1038/msb.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venancio TM, et al. Robustness and evolvability in natural chemical resistance: identification of novel systems properties, biochemical mechanisms and regulatory interactions. Mol Biosyst. 2010;6:1475–1491. doi: 10.1039/c002567b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohen AA, et al. Dynamic Proteomics of Individual Cancer Cells in Response to a Drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 67.Spencer SL, et al. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levin D, et al. Stochastic receptor expression determines cell fate upon interferon treatment. Mol Cell Biol. 2011;31:3252–3266. doi: 10.1128/MCB.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26:726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 70.Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development (Cambridge, England) 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 71.Kalmar T, et al. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS biology. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arias AM, Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nature reviews. Genetics. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- 74.Hebenstreit D, et al. Dual of the Fates: the roles of transcriptional circuits and noise in CD4+ T-cells. Current opinion in cell biology. 2012 doi: 10.1016/j.ceb.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 75.Chang HH, et al. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barkai N, Shilo B-Z. Variability and robustness in biomolecular systems. Molecular cell. 2007;28:755–760. doi: 10.1016/j.molcel.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 77.Trott J, et al. Dissecting ensemble networks in ES cell populations reveals micro-heterogeneity underlying pluripotency. Mol Biosyst. 2012 doi: 10.1039/c1mb05398a. [DOI] [PubMed] [Google Scholar]
- 78.Kuchina A, et al. Temporal competition between differentiation programs determines cell fate choice. Mol Syst Biol. 2011;7:557. doi: 10.1038/msb.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wernet MF, et al. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 81.Simpson P. Notch signalling in development: on equivalence groups and asymmetric developmental potential. Curr Opin Genet Dev. 1997;7:537–542. doi: 10.1016/s0959-437x(97)80083-4. [DOI] [PubMed] [Google Scholar]
- 82.Eldar A, et al. Partial penetrance facilitates developmental evolution in bacteria. Nature. 2009;460:510–514. doi: 10.1038/nature08150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raj A, et al. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Heyningen V, Yeyati PL. Mechanisms of non-Mendelian inheritance in genetic disease. Hum Mol Genet. 2004;13(Spec No 2):R225–R233. doi: 10.1093/hmg/ddh254. [DOI] [PubMed] [Google Scholar]
- 85.Yeyati PL, et al. Hsp90 selectively modulates phenotype in vertebrate development. PLoS Genet. 2007;3:e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeyati PL, van Heyningen V. Incapacitating the evolutionary capacitor: Hsp90 modulation of disease. Current opinion in genetics & development. 2008;18:264–272. doi: 10.1016/j.gde.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 87.Burga A, et al. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2012;480:250–253. doi: 10.1038/nature10665. [DOI] [PubMed] [Google Scholar]
- 88.Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nature genetics. 2011;43:95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vogel C. Translation's coming of age. Mol Syst Biol. 2011;7:498. doi: 10.1038/msb.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Sousa Abreu R, et al. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maier T, et al. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 92.Taniguchi Y, et al. Quantifying E. coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 94.Tay S, et al. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Larson DR, et al. Real-Time Observation of Transcription Initiation and Elongation on an Endogenous Yeast Gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spiller DG, et al. Measurement of single-cell dynamics. Nature. 2010;465:736–745. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- 97.Ferry MS, et al. Microfluidics for synthetic biology: from design to execution. Methods Enzymol. 2011;497:295–372. doi: 10.1016/B978-0-12-385075-1.00014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bennett MR, Hasty J. Microfluidic devices for measuring gene network dynamics in single cells. Nature reviews. Genetics. 2009;10:628–638. doi: 10.1038/nrg2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li G-W, Elf J. Single molecule approaches to transcription factor kinetics in living cells. FEBS letters. 2009;583:3979–3983. doi: 10.1016/j.febslet.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 101.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kantlehner M, et al. A high-throughput DNA methylation analysis of a single cell. Nucleic Acids Res. 2011;39:e44. doi: 10.1093/nar/gkq1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wyart M, et al. Evaluating gene expression dynamics using pairwise RNA FISH data. PLoS Comput Biol. 2010;6:e1000979. doi: 10.1371/journal.pcbi.1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Murphy KF, et al. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic Acids Res. 2010;38:2712–2726. doi: 10.1093/nar/gkq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Macneil LT, Walhout AJ. Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res. 2011;21:645–657. doi: 10.1101/gr.097378.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Danino T, et al. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Escudero LM, et al. Epithelial organisation revealed by a network of cellular contacts. Nat Commun. 2011;2:526. doi: 10.1038/ncomms1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Snijder B, et al. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- 109.Chandrasekaran S, et al. Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18020–18025. doi: 10.1073/pnas.1114093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Newman JRS, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 111.Swain PS, et al. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ribeiro AS, et al. Dynamical effects of transcriptional pause-prone sites. Computational biology and chemistry. 2010;34:143–148. doi: 10.1016/j.compbiolchem.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 113.Cai L, et al. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 114.Raj A, van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Annu Rev Biophys. 2009;38:255–270. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Choi JK, Kim Y-J. Intrinsic variability of gene expression encoded in nucleosome positioning sequences. Nat Genet. 2009;41:498–503. doi: 10.1038/ng.319. [DOI] [PubMed] [Google Scholar]
- 116.Lam FH, et al. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boeger H, et al. Theory Nucleosome Retention and the Stochastic Nature of Promoter Chromatin Remodeling for Transcription. Cell. 2008:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Blake WJ, et al. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 119.Brown CR, et al. In Vivo Role for the Chromatin-remodeling Enzyme SWI/SNF in the Removal of Promoter Nucleosomes by Disassembly Rather Than Sliding. J Biol Chem. 2011;286:40556–40565. doi: 10.1074/jbc.M111.289918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zaugg JB, Luscombe NM. A genomic model of condition-specific nucleosome behavior explains transcriptional activity in yeast. Genome Res. 2012;22:84–94. doi: 10.1101/gr.124099.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McCullagh E, et al. Coordinate control of gene expression noise and interchromosomal interactions in a MAP kinase pathway. Nature cell biology. 2010;12:954–962. doi: 10.1038/ncb2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lim HN, van Oudenaarden A. A multistep epigenetic switch enables the stable inheritance of DNA methylation states. Nat Genet. 2007;39:269–275. doi: 10.1038/ng1956. [DOI] [PubMed] [Google Scholar]
- 123.Miller-Jensen K, et al. Varying virulence: epigenetic control of expression noise and disease processes. Trends in biotechnology. 2011;29:517–525. doi: 10.1016/j.tibtech.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 124.Hansen KD, et al. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stockholm D, et al. Bistable cell fate specification as a result of stochastic fluctuations and collective spatial cell behaviour. PLoS One. 2010;5:e14441. doi: 10.1371/journal.pone.0014441. [DOI] [PMC free article] [PubMed] [Google Scholar]