SUMMARY
Obesity, type 2 diabetes and heart failure are associated with aberrant cardiac metabolism. We show that the heart regulates systemic energy homeostasis via MED13, a subunit of the Mediator complex, which controls transcription by thyroid hormone and other nuclear hormone receptors. MED13, in turn, is negatively regulated by a heart-specific microRNA, miR-208a. Cardiac-specific over-expression of MED13 or pharmacologic inhibition of miR-208a in mice confers resistance to high fat diet-induced obesity and improves systemic insulin sensitivity and glucose tolerance. Conversely, genetic deletion of MED13 specifically in cardiomyocytes enhances obesity in response to high fat diet and exacerbates metabolic syndrome. The metabolic actions of MED13 result from increased energy expenditure and regulation of numerous genes involved in energy balance in the heart. These findings reveal a role of the heart in systemic metabolic control and point to MED13 and miR-208a as potential therapeutic targets for metabolic disorders.
INTRODUCTION
Maintaining energy homeostasis requires a balance between energy consumption and energy expenditure. Excess food consumption shifts energy balance, causing obesity, a multi-organ disorder that enhances the risk of type 2 diabetes, hypertension, hyperlipidemia and cardiovascular disease (Van Gaal et al., 2006). Impaired metabolism of energy-providing substrates and myocardial lipid accumulation are early abnormalities in obese and insulin-resistant individuals (Harmancey et al., 2008).
The heart adapts to changes in diet and stress by fine-tuning metabolism and gene expression patterns to maximize energy efficiency (Balaban et al., 1986). Thyroid hormone (TH) receptors (TRs) and other nuclear receptors (NRs) are key regulators of energy homeostasis in the heart and other tissues (Huss and Kelly, 2004). Activation of TRs directly through elevation of TH results in enhanced energy expenditure and a lean phenotype (Mitchell et al., 2010; Watanabe et al., 2006). Numerous genes involved in energy balance are controlled by TH responsive elements (TREs) and are directly activated or repressed by TRs (Song et al., 2011). Other nuclear hormone receptors, such as PPARs and retinoid X receptor (RXR), as well as their coactivators and corepressors, also control energy homeostasis by activation of lipogenic genes in the heart and other tissues (Huss and Kelly, 2004).
Recent studies have revealed important roles for microRNAs (miRNAs) in heart disease (Latronico and Condorelli, 2009; van Rooij et al., 2007) and metabolism (Frost and Olson, 2011; Trajkovski et al., 2011). miRNAs inhibit protein expression through base pairing with sequences located predominantly in the 3′ untranslated regions (UTRs) of mRNA targets (Bartel, 2004). The functions of miRNAs often become pronounced under conditions of stress, making them potential targets for disease modulation (Small and Olson, 2011).
MiR-208a is a cardiac-specific miRNA encoded by an intron of the cardiac specific α-myosin heavy chain (MHC) gene. MiR-208a−/− mice do not display overt abnormalities at baseline, but hearts from these mice are resistant to pathological remodeling and fibrosis and do not up-regulate β-MHC, a marker of cardiac stress, in response to pressure overload or hypothyroidism (Callis et al., 2009; van Rooij et al., 2007). Pharmacologic inhibition of miR-208a with Locked Nucleic Acid (LNA)-modified antisense oligonucleotides also enhances cardiac function and prolongs survival in a rat model of diastolic dysfunction (Montgomery et al., 2011). MiR-208a−/− hearts resemble hyperthyroid hearts, which are also protected against pathological hypertrophy and fibrosis (Tancevski et al., 2011; van Rooij et al., 2007). These findings suggest that miR-208a acts, at least in part, by repressing a common component of stress-responsive and TH signaling pathways in the heart. Among the strongest predicted and validated targets of miR-208a is MED13/THRAP1/TRAP240 (Callis et al., 2009; van Rooij et al., 2007), a component of the Mediator complex, which modulates TH-dependent transcription (Ito and Roeder, 2001).
The Mediator is a multiprotein complex that acts as a bridge between DNA-bound transcription factors and RNA polymerase II (Conaway and Conaway, 2011; Malik and Roeder, 2010). Mediator is composed of ~30 subunits, which directly regulate nuclear hormone receptor-dependent transcription of select target genes (Ge et al., 2002; Malik and Roeder, 2005; Park et al., 2005). The activity of the Mediator complex is modulated by association with a tetrameric regulatory complex, called the kinase submodule, comprised of MED12, MED13, Cyclin C and cyclin-dependent kinase 8 (CDK8) (Akoulitchev et al., 2000; Knuesel et al., 2009; Taatjes, 2010).
MED1, a core protein of the Mediator complex, binds directly to NR and is necessary for maintaining energy homeostasis in skeletal muscle (Baek et al., 2002; Chen et al., 2010). Other components of the Mediator complex also regulate pathways involved in fatty acid, cholesterol and lipid homeostasis (Taubert et al., 2006; Wang et al., 2009; Yang et al., 2006). Recently, MED13 was identified in Drosophila as a negative regulator of lipid accumulation, such that its knockdown using a muscle-specific promoter-driven siRNA caused excessive adiposity (Pospisilik et al., 2010).
The potential functions of MED13 or other components of the Mediator complex in the heart have not been explored. Here we used pharmacological and genetic tools to modulate the expression of miR-208a and its target, MED13, in the heart. We show that mice with increased cardiac expression of MED13 are lean and display an increase in energy expenditure and resistance to diet-induced obesity and metabolic syndrome. Remarkably, pharmacologic inhibition of miR-208a confers a similar metabolic phenotype. Conversely, cardiac deletion of MED13 increases susceptibility to diet-induced obesity and metabolic syndrome. Metabolic control by MED13 is associated with altered expression of a collection of TH and NR responsive genes in the heart. Our results delineate a genetic pathway whereby MED13 and miR-208a function in the heart to control whole body metabolism and unveil a role of the heart in systemic energy homeostasis.
RESULTS
Pharmacologic Inhibition of miR-208a Confers Resistance to Obesity
While studying the potential therapeutic effects of miR-208a inhibitors in mice, we noticed that animals treated over a period of several weeks with LNA antimiR-208a were leaner than littermates either untreated or treated with control oligonucleotides. Subcutaneous delivery of antimiR-208a efficiently inhibited miR-208a levels in the heart, as detected by Northern blot analysis (Figure 1A). This antimiR is highly specific for inhibition of the cardiac-specific miR-208a and does not affect miR-208b (Montgomery et al., 2011), which differs from miR-208b by 3 nucleotides. Treatment of mice for 6 weeks with antimiR-208a or a control antimiR had no effect on heart weight, cardiac contractility or heart rate (Figure S1 A–C).
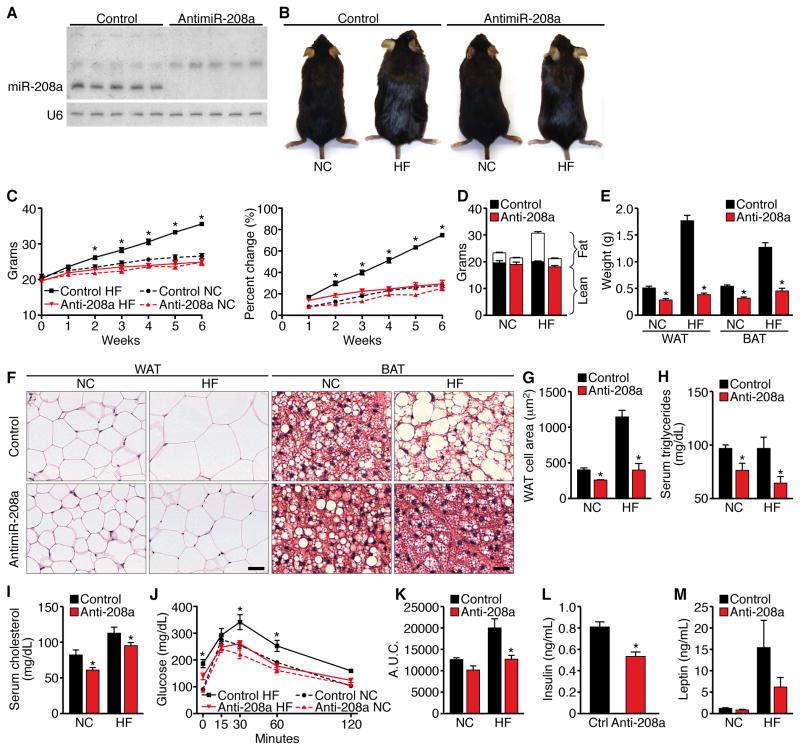
Figure 1. Mice Administered AntimiR-208a are Resistant to Obesity and Glucose Intolerance.
antimiR-208a or control antimiR treated mice on NC or HF diet for 6 weeks n=8–10 unless otherwise stated.
(A) Northern blot analysis of mice treated for 6 weeks with antimiR-208a or control antimiR (control). U6 RNA was detected as a loading control. Hearts from 5 mice from each treatment were analyzed. Note the absence of miR-208a in antimiR-208a-treated hearts.
(B) Pictures of mice treated for 6 weeks.
(C) Growth curves and percentage increase in body weight.
(D) Body composition measured by NMR to determine fat mass and lean tissue mass.
(E) Weight of visceral WAT and subscapular BAT.
(F) H&E stain of visceral WAT and subscapular BAT. Scale bar = 40μm
(G) Cell size of visceral WAT. n=5 Images from 3 sections 200mm apart were analyzed from 7–8 mice in each group representing >500 cells.
(H) Serum triglyceride levels. n=5
(I) Serum cholesterol levels. n=5
(J) Glucose tolerance test (GTT).
(K) Area under the curve for GTT.
(L,M) Fasting insulin and leptin levels. Data are represented as mean ± SEM. n=5 for NC and n=13 for HF diet for all experiments unless stated otherwise. *p<0.05
To investigate the metabolic basis of the lean phenotype, we tested the effect of antimiR-208a on weight gain in response to high fat (HF) diet. Male mice on HF diet treated with control antimiR increased their body weight by 75% within six weeks, whereas antimiR-208a treated mice on HF diet showed only a 29% increase in body weight (Figures 1B and 1C), which was comparable to mice maintained on normal chow (NC) and treated with control oligonucleotide or antimiR-208a. NMR spectrometry revealed that the difference in weight between the treatment groups was due to differences in fat weight (Figure 1D). Consistent with these findings, visceral white adipose and subscapular adipose tissue, containing both white and brown fat, were significantly smaller in antimiR-208a treatment groups on HF diet and NC compared to the control antimiR treated animals, based on fat mass and adipocyte size (Figure 1E–1G and S1D). Serum triglyceride and cholesterol levels were also reduced in antimiR-208a treated mice on HF diet (Figure 1H and 1I). Similarly, hepatic steatosis seen in control animals on HF diet was blunted by treatment with antimiR-208a (Figure S1D).
HF diet-induced obesity causes glucose intolerance. AntimiR-208a treated mice on NC showed a normal glucose response, as measured by glucose tolerance test (GTT) (Figure 1J). On HF diet, control antimiR-treated, obese mice displayed an increase in fat mass and glucose intolerance (Figures 1E and 1J). In contrast, antimiR-208a treated mice showed a normal glucose response after 6 weeks of HF diet as revealed by GTT and the calculated area under the curve (Figures 1J and 1K). Fasting insulin levels from antimiR-208a treated mice were significantly lower than those of control antimiR treated mice (Figure 1L). Similarly, levels of leptin, an adiopocyte-derived circulating hormone that reflects body lipid content (Frederich et al., 1995), were reduced by antimiR-208a compared to control antimiR in animals on NC and HF diet (Fig. 1M). AntimiR-208a had no effect on physical activity or food consumption (Figure S1E and S1F). These findings suggested that miR-208a inhibition improves whole-body insulin sensitivity. Because miR-208a is only expressed in cardiomyocytes (Callis et al., 2009; van Rooij et al., 2007), the beneficial metabolic effects of antimiR-208a suggested a potential influence of the heart on systemic metabolism.
Cardiac Over-expression of MED13 Prevents Obesity and Improves Glucose Homeostasis
The Mediator subunit MED13 is among the strongest predicted and validated targets of miR-208a. Accordingly, MED13 expression was increased in hearts of mice treated with antimiR-208a (Figure S1G). To determine if elevation of MED13 in the heart might evoke systemic metabolic effects analogous to those of miR-208a inhibition, we generated transgenic mice using the αMHC promoter to over-express MED13 specifically in cardiomyocytes. Two independent αMHC-Med13 transgenic (TG) mouse lines, referred to as Lines 1 and 2, showing 9- and 3-fold increases, respectively, in expression of Med13 mRNA and protein compared to wild-type (WT) in the heart were generated (Figure S2A and S2B). Neither αMHC-Med13 TG mouse line showed overt abnormalities. Gross cardiac structure was also normal and cardiac stress markers were not elevated in these mice (Figure S2C and S2D).
Although male αMHC-Med13 TG mice were comparable in length to WT littermates, they were lighter when fed NC, and were resistant to obesity on HF diet, gaining body weight similarly to WT mice on NC (Figures 2A–B, S2E–F). Female αMHC-Med13 TG mice also displayed resistance to weight gain on HF diet (Figure S2G). The αMHC-Med13 TG mice had significantly less fat mass on both the NC and HF diet compared to WT littermates, as measured by NMR (Figure 2C). Visceral WAT and subscapular BAT mass, as well adipocyte size, from the αMHC-Med13 TG mice on HF diet and NC were also reduced compared to WT mice (Figure 2D–F). Gross and histological analysis of subscapular fat from αMHC-Med13 TG mice showed less lipid accumulation on NC and HF diet compared to WT mice (Figure 2E–2F). WT mice accumulate fat with age when maintained on NC. αMHC-Med13 TG mice, at 7 and 12 months of age, also had significantly less fat mass than WT littermates (Figure S2H).
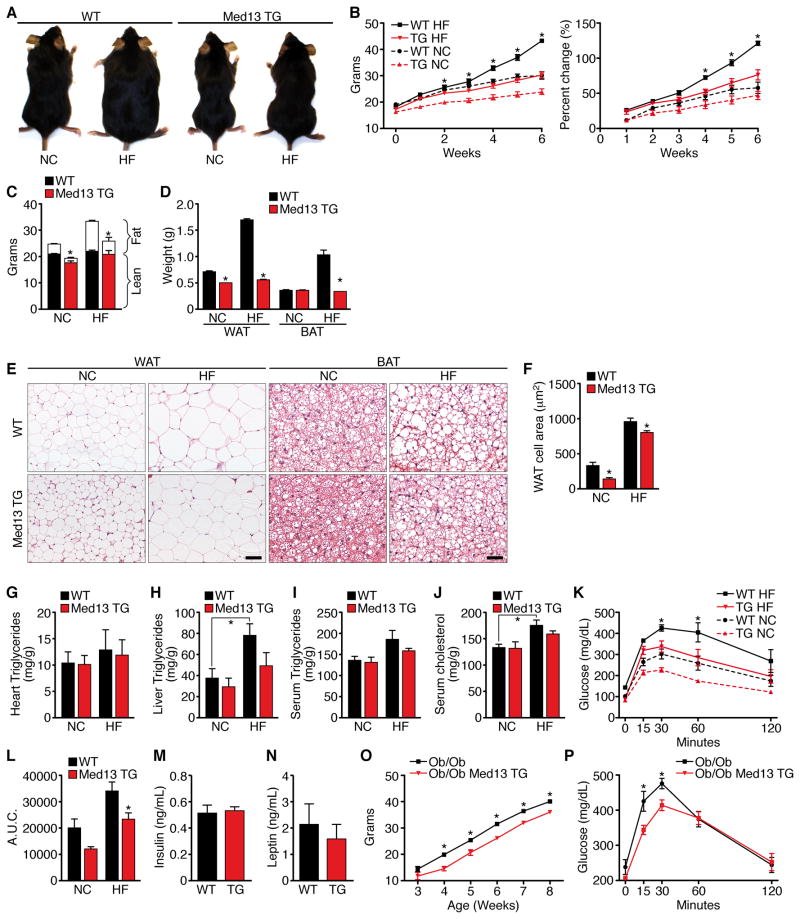
Figure 2. αMHC-Med13 TG Mice are Resistant to Diet-induced Obesity with Improved Glucose Tolerance.
αMHC-Med13 TG mice (Line 1) and WT littermates on NC or HF diet for 6 weeks n=8–10 unless otherwise stated.
(A) Pictures of WT and αMHC-Med13 TG mice after 6 weeks on NC or HF.
(B) Growth curves and percentage increase in body weight.
(C) Body composition measured by NMR to determine fat mass and lean tissue mass.
(D) Weight of visceral WAT and subscapular BAT.
(E) H&E stain of visceral WAT and subscapular BAT. Scale bar = 40μm
(F) Cell size of visceral WAT. Values represent the mean cross section area of each cell. Images from 3 sections 200mm apart were analyzed from 7–8 mice in each group representing >500 cells.
(G–I) Heart, liver and serum triglyceride levels.
(J) Serum cholesterol levels.
(K) Glucose tolerance test (GTT)
(L) Area under the curve for the GTT.
(M–N) Fasting insulin and leptin levels.
(O) Growth curve showing body weight measured weekly for 6 weeks of ob/ob; αMHC-Med13 TG mice compared to ob/ob littermates. n=5–7
(P) GTT of ob/ob; αMHC-Med13 TG mice compared to ob/ob littermates following an overnight fast using 1 mg of glucose/gram bodyweight. n=5–7
Data are represented as mean ± SEM. *p<0.05
Hearts from WT and αMHC-Med13 TG mice on NC or HF diet showed no differences in lipid accumulation based on Oil-Red-O staining (Figure S2I) or triglyceride levels (Figure 2G). However, hepatic lipid and triglyceride accumulation were highly elevated in WT mice on HF diet, but in αMHC-Med13 TG mice were not significantly different from WT mice on NC (Figure 2H). Serum triglyceride and cholesterol levels were also diminished in αMHC-Med13 TG mice compared to WT on HF diet (Figure 2I, J). In addition, αMHC-Med13 TG mice showed a normal glucose response after 6 weeks of HF diet compared to the glucose intolerance seen in WT mice (Figure 2K, L). Fasting insulin and leptin levels in αMHC-Med13 TG mice on NC were comparable to those of WT littermates (Figure 2M, N), together indicating that elevated cardiac expression of Med13 improves whole-body insulin sensitivity following HF diet.
We bred αMHC-Med13 TG mice with leptin deficient ob/ob mice, which become morbidly obese and develop insulin resistance due to the absence of leptin (Tartaglia et al., 1995). Cardiac over-expression of MED13 reduced weight gain in ob/ob mice (Figure 2O and 2P).
MED13 Regulates Energy Consumption
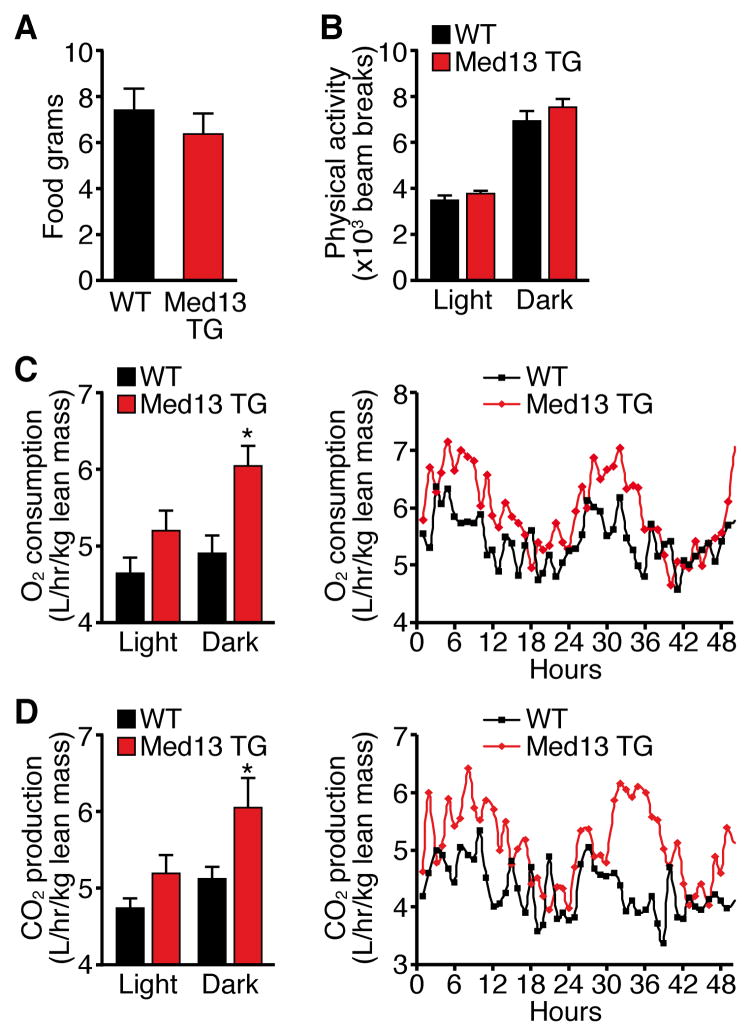
The resistance to weight gain by αMHC-Med13 TG mice was not due to a difference in food consumption or physical activity compared to WT mice, as measured using metabolic chambers (Figure 3A and 3B). Similarly, telemetry revealed no difference in core body temperature of αMHC-Med13 TG mice compared to WT littermate controls (Figure S3). Remarkably, however, lean weight-matched αMHC-Med13 TG mice exhibited an increase in oxygen consumption and carbon dioxide production, suggesting that the observed decrease in fat mass was due to an increase in systemic energy consumption mediated by over-expression of MED13 in the heart (Figure 3C and 3D).
Figure 3. αMHC-Med13 TG Mice have increased Energy Expenditure.
Twelve week old male αMHC-Med13 TG (Line 1) and WT mice on NC were analyzed in metabolic cages over 4.5 days. n=8–9
(A) Food consumption
(B) Physical activity, average beam breaks in the X, Y and Z axis over a 12 hour light/dark cycle.
(C) Average oxygen consumption per hour during the light/dark cycle (left) average traces (right) normalized to lean mass.
(D) Average carbon dioxide production per hour during the light/dark cycle (left) averages traces (right) normalized to lean mass.
Data are represented as mean ± SEM. *p<0.05 vs WT
Down-regulation of Nuclear Hormone Receptor Target Genes by MED13
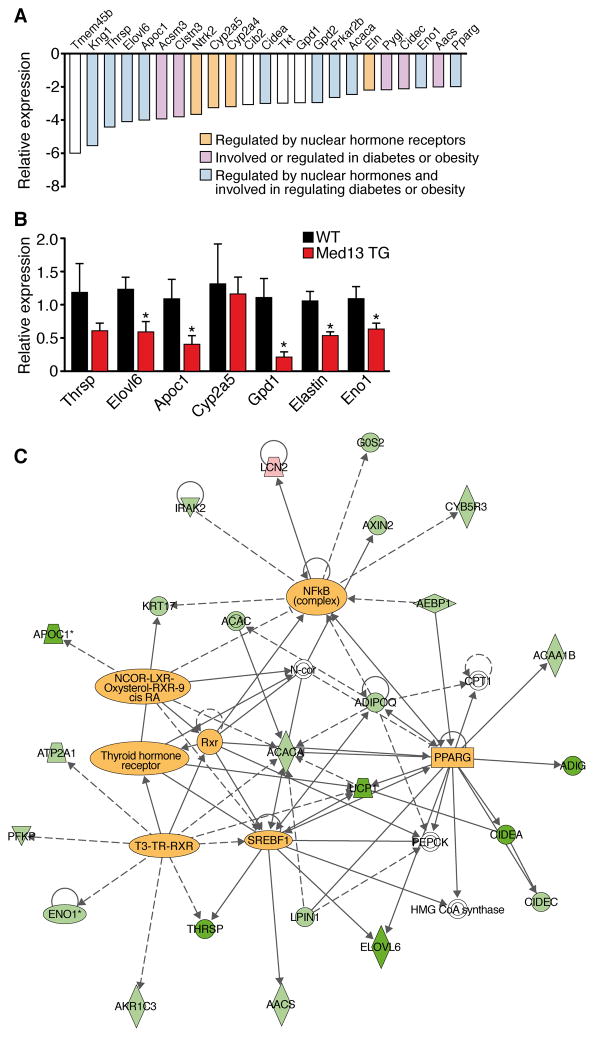
To further explore the molecular basis of the changes in energy homeostasis caused by cardiac over-expression of MED13, we performed microarray analysis on hearts from αMHC-Med13 TG and WT mice. We identified 19 genes up-regulated and 37 genes down-regulated in αMHC-Med13 TG hearts, with a cutoff of 2-fold (Figure 4A). Real-time RT-PCR validated these results (Figure 4B). Up-regulated genes in αMHC-Med13 TG hearts did not appear to fit a specific pattern, whereas the majority of down-regulated genes were involved in metabolism and regulated by NRs (Figure 4A, C). Metabolic genes that were down-regulated in αMHC-Med13 TG hearts were also down-regulated in hearts of mice treated with antimiR-208a (Figure S1H).
Figure 4. Changes in Cardiac Gene Expression in αMHC-Med13 TG Mice.
(A) Microarray analysis of genes down-regulated in hearts from adult αMHC-Med13 TG mice compared to WT mice. Brown bars represent genes regulated by nuclear hormone receptors, purple bars represent genes regulated in diabetes or obesity. Blue bars represent genes regulated by nuclear hormone receptors and are also involved in diabetes or obesity.
(B) Real time qPCR of select genes identified in panel A comparing αMHC-Med13 TG and WT mice. n=4–5 Data are represented as mean ± SEM. *p<0.05 vs WT
(C) Ingenuity pathway analysis of genes regulated over 1.5 fold in the αMHC-Med13 TG mouse array. Highlighted within the diagram are transcription factors that function as nodal points for genes regulated in αMHC-Med13 TG mouse hearts.
The Ingenuity Pathways Analysis application (http://www.ingenuity.com), which identifies molecular relationships among genes, revealed that many of the down-regulated genes in αMHC-Med13 TG hearts are known targets of NRs (TR, RXR, LXR), consistent with the repressive influence of MED13 on NR activity (Figure 4C). Other transcription factors involved in metabolic regulation (PPARγ, NCoR and SREBP) were also identified as regulatory hubs in the control of MED13-responsive genes. Moreover, mice with genetic deletion of genes that were down-regulated in the microarray (Thrsp, Elovl6, Gpd2, Eno1, Cidea, Tkt or PPARγ) display resistance to diet-induced obesity and/or protection against insulin resistance (Anderson et al., 2009; Jones et al., 2005; Xu et al., 2002; Zhou et al., 2003). These findings suggest that MED13 functions in the heart to inhibit expression of select NR-responsive genes, thereby altering whole-body energy homeostasis.
Cardiac over-expression of MED13 reduces fat accumulation in peripheral tissues. We performed microarray analysis on liver, gonadal WAT and BAT from 4 week-old αMHC-Med13 TG mice and WT littermates (Figure S4). Panther analysis of the genes regulated greater than 2-fold revealed that genes associated with metabolic processes were the most strongly affected in cardiac and non-cardiac tissues of αMHC-Med13 TG mice (Thomas et al., 2006).
MED13 Modulates TH-dependent responses
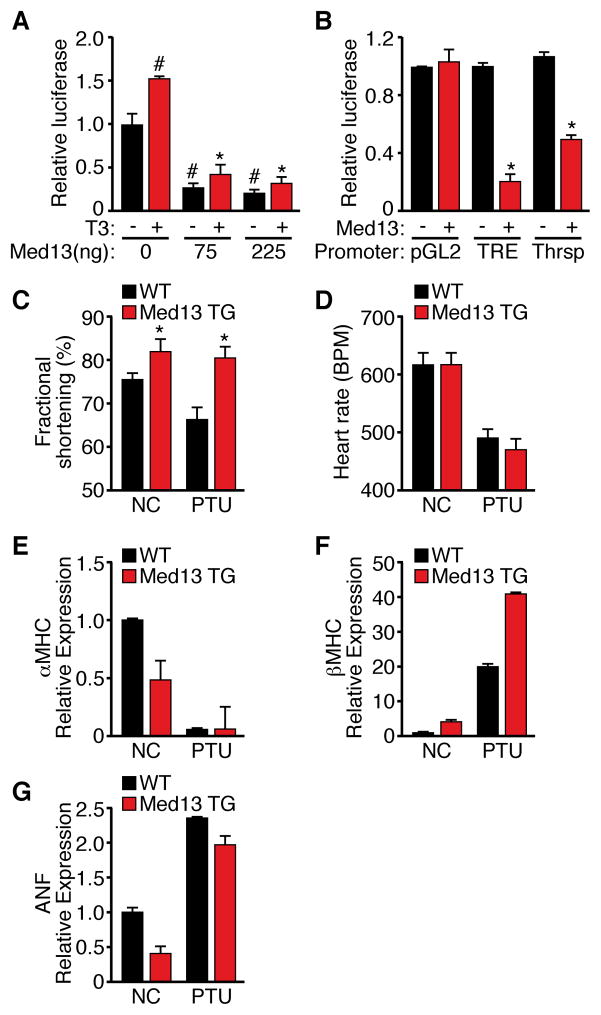
Since MED13 repressed numerous TH responsive genes in the heart, we performed a TH reporter assay to determine if MED13 regulates TR activity. MED13 inhibited activation of a synthetic TH response element (TRE) in the presence of exogenous TRβ by approximately 80% in the presence or absence of TH (T3) and inhibited promoter activity of the TH responsive protein (Thrsp) gene, which was also down-regulated by MED13 in vivo (Figure 5A, B). We conclude that MED13 suppresses TRβ transcriptional activation independent of TH.
Figure 5. Modulation of TH Actions by MED13.
(A) Inhibition of TRE reporter by Med13 in the presence or absence of T3. Data are presented as relative luciferase activation. Each data point represents the average of triplicate transfections from 3 independent experiments. #p<0.05 vs Med13(0ng)–T3, *p<0.05 vs Med13(0ng)+T3
(B) Luciferase assays measuring pGL3, TRE or the Thrsp promoter activity in the presence or absence of Med13. Assays were performed as described in panel A. n=4
(C–D) Cardiac fractional shortening and heart rate as measured by transthoracic echochardiography in αMHC-Med13 TG and WT mice at baseline or following two weeks of PTU treatment. n=8–10
(E–H) Quantitative RT-PCR analysis of cardiac mRNA levels from αMHC-Med13 TG and WT mice on NC or following 2 weeks of PTU treatment. n=3–4
Data are represented as mean ± SEM. * p<0.05
To further investigate the impact of MED13 on TH responsiveness of the heart, we examined whether αMHC-Med13 transgenic mice displayed an altered response to hypothyroidism. Treatment with propylthiouracil (PTU), an inhibitor of T3 biosynthesis, reduces cardiac function in WT mice. In contrast, αMHC-Med13 TG mice were resistant to the PTU-dependent decrease in contractility, as determined by echocardiography (Figure 5C), consistent with a hyperthyroid-like state. Elevated heart rate is a common consequence of enhanced systemic TH signaling, however, αMHC-Med13 TG mice, did not show differences in heart rate compared to WT mice (Figure 5D). In the adult rodent heart, up-regulation of β-MHC is a sensitive marker of hypothyroidism, and is blocked by T3 signaling. In miR-208a null mice, α- to β-MHC switching is blunted in response to PTU treatment (van Rooij et al., 2007), whereas α- and β-MHC expression was altered comparably in WT and αMHC-Med13 TG treated with PTU (Figure 5F–I), indicating that MED13 does not target all TR responsive genes and only a subset of miR-208a functions are mediated by its regulation of MED13. Atrial natruiretic factor (ANF), a cardiac stress marker was elevated to comparable levels in WT and αMHC-Med13 TG mice in response to PTU (Figure 5G). Taken together, these findings suggest that MED13 expression in the heart results in a hyper-metabolic state due in part to altered TH responses.
Cardiac Deletion of Med13 Enhances Susceptibility to Obesity
In light of the dramatic effect of cardiac over-expression of MED13 on global metabolism, we generated mice with a conditional Med13 loss-of-function allele. Deletion of exons 7 and 8 results in a frame shift and loss of a majority of the MED13 coding sequence, including the putative NLS, NR binding motifs, leucine zipper and FoxO-like domain (Figure S5A–B). Cardiac-specific deletion of MED13 using αMHC-Cre had no effect on survival or cardiac function in adult mice (Figures S5C).
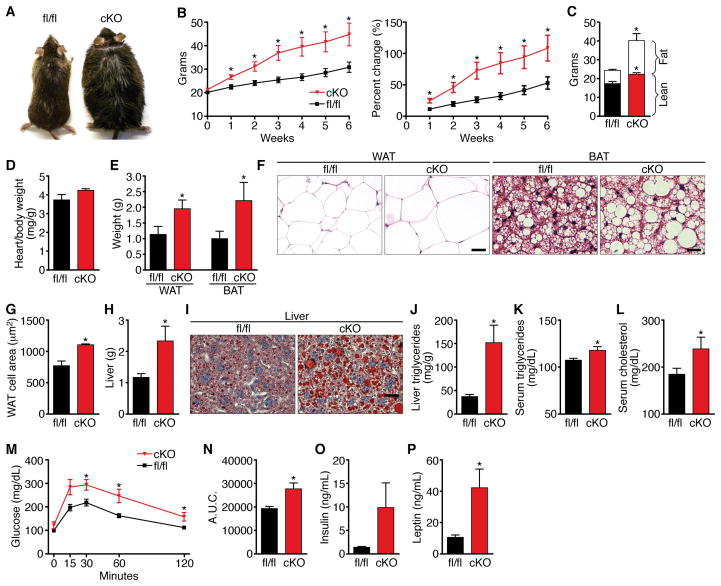
To assess the potential metabolic consequences of cardiac MED13 loss-of-function, we placed Med13 cardiac knockout (cKO) mice on HF diet at 6 weeks of age. The Med13 cKO mice weighed the same as their WT littermates at the onset of the study, but gained significantly more weight beginning within the first week on HF diet and continuing thereafter (Figure 6A and 6B). After 6 weeks on HF diet, we observed a small but statistically significant increase in lean mass and a dramatic increase in fat mass in the Med13 cKO mice compared to Med13fl/fl littermates on HF diet (Figure 6C). Heart weight/body weight ratios were not significantly different in Med13 cKO mice compared to Med13fl/fl controls (Figure 6D).
Figure 6. Cardiac Deletion of Med13 Enhances Susceptibility to Obesity.
Analysis of male Med13fl/fl αMHC-cre (cKO) mice and Med13fl/fl littermates (fl/fl) on HF diet (HF) for 6 weeks.
(A) Pictures of Med13fl/fl αMHC-cre and Med13fl/fl mice.
(B) Growth curves and percentage increase in body weight. n=8–10
(C) Body composition measured by NMR to determine fat mass and lean tissue mass of Med13fl/fl αMHC-cre and Med13fl/fl mice following 6 weeks on HF. n=8–10
(D) Heart weight/body weight ratio. n=8–10
(E) Weight of visceral WAT and subscapular fat. n=8–10
(F) H&E stained visceral WAT and BAT. Scale bar = 40μm
(G) Cell size of visceral WAT. n=5
(H) Liver mass. n=8–10
(I) Oil-red-O stained liver. Scale bar = 40μm
(J–K) Liver and serum triglyceride content. n=6–8
(L) Serum cholesterol levels. n=6–8
(M) Glucose tolerance test (GTT). n=6–8
(N) Area under the curve for the GTT. n=6–8
(O–P) Fasting insulin and leptin levels. n=6
Data are represented as mean ± SEM. *p<0.05
Med13 cKO mice had significantly larger visceral white adipose, subscapular fat and liver mass than controls on HF diet (Figure 6E–6H and S13). Histological analysis of these tissues revealed an increase in lipid accumulation in the Med13 cKO versus Med13fl/fl mice (Figure 6F and 6I). Liver triglycerides as well as serum triglycerides, cholesterol and blood glucose levels were also elevated in the Med13 cKOs (Figure 4J–N). Fasting insulin levels in the Med13 cKO mice were highly variable but were dramatically increased, as were leptin levels (Figure 4O,P). Thus, cardiac deletion of MED13 perturbs whole-body metabolism, enhancing sensitivity to HF diet, a phenotype opposite that of Med13 transgenic or antimiR-208a treated mice.
Discussion
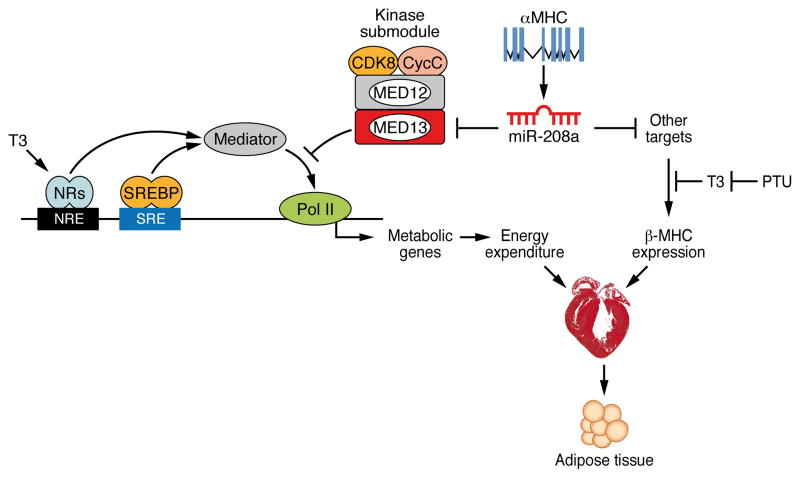
The results of this study demonstrate that MED13, a regulatory subunit of the Mediator complex, functions in the heart to control metabolic homeostasis and energy expenditure in mice. Elevated cardiac expression of MED13 by transgenic over-expression or pharmacologic inhibition of miR-208a enhances metabolic rate, confers resistance to obesity, improves insulin sensitivity and lowers plasma lipid levels. Conversely, cardiac-specific deletion of MED13 causes metabolic syndrome and severe obesity. These findings reveal an important role of the heart in regulation of systemic metabolism and identify MED13 and miR-208a as central components of this metabolic regulatory mechanism (Fig. 7).
Figure 7. Model of Cardiac Dependent Regulation of Energy Homeostasis.
MED13-dependent regulation of select Mediator–dependent metabolic genes in the heart govern global energy expenditure. Modulation of MED13 expression levels in the heart by the cardiac-specific miR-208a results in an altered metabolic state while independently regulating myosin switching in the heart in response to cardiac stress.
Cardiac control of energy homeostasis by MED13
The Mediator complex links gene-specific transcription factors with the basal transcriptional machinery (Ito and Roeder, 2001). The kinase submodule of Mediator, comprised of MED12, MED13, CDK8 and Cyclin c, represses expression of select sets of target genes by interfering with the association of Mediator and RNA polymerase II (Knuesel et al., 2009). There is also evidence for activation of certain genes by the kinase submodule (Alarcon et al., 2009; Belakavadi and Fondell, 2010; Conaway and Conaway, 2011; Donner et al., 2007).
While the importance of the Mediator complex in the control of gene transcription has been well established from in vitro transcription assays (Conaway and Conaway, 2011; Malik and Roeder, 2010), determination of the functions of this complex in adult tissues has been hindered by the fact that all mutant alleles of Mediator subunits analyzed thus far in mice result in early lethality. To date, no functions have been ascribed to MED13 in any mammalian tissues nor have potential functions of any components of the Mediator complex been analyzed in the heart. Intriguingly, humans with mutations in MED12 exhibit hypothyroidism and are predisposed to obesity (Philibert et al., 2002), suggesting that MED12 exerts metabolic functions similar to those we have discovered for MED13.
Through cardiac-specific gain- and loss-of-function of MED13 in mice, our results highlight a central role of this Mediator subunit in governance of metabolism via the control of energy expenditure pathways. Our finding that MED13 deficiency in the heart causes severe obesity in mice on HF diet is consistent with a recent report that MED13 deficiency in muscle of Drosophila causes enhanced lipid accumulation in the fat body (Pospisilik et al., 2010) and suggests an evolutionarily conserved role for MED13 in the control of metabolic homeostasis by striated muscles of metazoans. Deletion of MED1 in skeletal muscle of mice confers resistance to diet-induced obesity (Chen et al., 2010), analogous to the phenotype we observed with over-expression of MED13 in the heart. MED1, a core component of the Mediator complex, controls transcriptional activation by numerous NRs (Ge et al., 2002; Ito et al., 2000; Malik et al., 2004). MED13 has not been shown to directly interact with NRs. MED13 is thought to function as a repressor by sterically hindering the association of RNA polymerase II with the core Mediator complex. Therefore the actions of MED13 on NRs are consistent with the opposing influences of MED13 on MED1, which does directly bind to NRs (Knuesel et al., 2009).
Modulation of cardiac NR signaling by MED13
TH is a major determinant of metabolic rate, energy expenditure and cardiac contractility (Song et al., 2011). The global phenotype resulting from cardiac over-expression of MED13 is similar to that of enhanced TH signaling, which is associated with increased energy expenditure and resistance to fat accumulation in mice and humans (Watanabe et al., 2006). However, only a subset of TH-dependent genes are modulated in response to MED13 over-expression in the heart. TH stimulates and represses TR-dependent genes depending on transcriptional cofactors and intracellular signals (Fondell et al., 1999). Consistent with a repressive influence, MED13 blocked transcription of TRβ reporter genes in vitro, and transgenic over-expression of MED13 in the heart suppressed a broad collection of TR and NR target genes involved in energy balance. Among the genes down-regulated by MED13, deletion of Thrsp, Gpd2, Eno1, Aacs and Pparg in mice results in resistance to diet-induced obesity (Anderson et al., 2009; Jones et al., 2005; Xu et al., 2002; Zhou et al., 2003), as we observed in response to cardiac over-expression of Med13. Elovl6 and Kng1, which are repressed by MED13, modulate insulin and glucose homeostasis (Matsuzaka et al., 2007; Wu et al., 2010).
In addition to regulating a subset of TR targets, gene network analysis indicates a role for MED13 in the control of other transcriptional regulators of lipid biosynthesis and metabolism, including NCoR1, SREBP, LXR, RXR and PPARγ. It is notable in this regard that inhibition of SREBP, RXR, and PPARγ ameliorates diet-induced obesity and type 2 diabetes (Huss and Kelly, 2004; Jones et al., 2005; Yang et al., 2006). Conversely, cardiac-specific over-expression of PPARα causes metabolic cardiomyopathy and hepatic insulin resistance (Finck et al., 2002). Thus, MED13 appears to function in the heart as a master regulator of the transcriptional circuitry responsible for metabolic homeostasis by suppressing the actions of multiple metabolic regulators.
Modulation of energy homeostasis by miR-208a
Pharmacologic inactivation of miR-208a, which is cardiac-specific, through systemic delivery of an antimiR confers an enhanced metabolic phenotype analogous to that of mice with cardiac over-expression of MED13, suggesting that MED13 is a key effector of the metabolic effects of miR-208a inhibition. However, there are also some important distinctions between the phenotypic consequences of miR-208a inhibition and MED13 over-expression. In particular, mice lacking miR-208a are resistant to pathological cardiac remodeling and fail to up-regulate β-MHC in response to stress or hypothyroidism (Callis et al., 2009; van Rooij et al., 2007), whereas MED13 over-expression does not alter β-MHC expression in response to hypothyroidism. These findings suggest that miR-208a acts through multiple downstream targets to control metabolic homeostasis and myosin switching independently (Fig. 7), with MED13 dedicated to the metabolic functions of miR-208a. In this regard, miR-208a, like other miRNAs, has numerous targets. Thus, while our results strongly implicate MED13 in the metabolic actions of miR-208a in the heart, other targets undoubtedly also contribute to this activity of miR-208a.
miR-208a belongs to a family of miRNAs (referred to as MyomiRs), which includes miR-208b and miR-499, encoded by the β-MHC and Myh7b genes, respectively. In contrast to α-MHC and miR-208a, which are expressed specifically in the heart, miR-208b and miR-499 and their host myosin genes are also expressed in slow skeletal muscle (van Rooij et al., 2009). It will be of interest to determine whether these miRNAs act through MED13 in skeletal muscle to control metabolic homeostasis. Especially intriguing is the ability of myosin genes, which encode the major contractile proteins of striated muscles, to modulate such broad ranging processes as cardiac contractility, stress responsiveness and metabolism via the MyomiRs encoded by their introns.
Cardiac regulation of energy homeostasis
The liver, adipose tissue and skeletal muscle are well known to function as the major tissue regulators of whole-body energy expenditure, metabolism and glucose homeostasis. Our findings demonstrate that the heart, which has the highest rate of oxygen consumption per gram of any tissue in the body (Rolfe and Brown, 1997) plays an important role in the control of energy homeostasis and adiposity through a mechanism involving MED13 and miR-208a. Mice with elevated cardiac expression of MED13 show normal food intake, but display increased oxygen consumption and carbon dioxide production, indicative of increased energy expenditure. Conversely, cardiac deletion of MED13 results in a dramatic increase in lipid accumulation without alterations in food intake. Changes in energy consumption can lead to alterations in thermogenesis. However, MED13 cardiac gain-of-function did not alter the ability to maintain core body temperature in mice. The enhanced whole body energy expenditure resulting from cardiac over-expression of MED13 may result from suppression of metabolic genes in the heart and consequent compensatory metabolic activity in non-cardiac tissues. Alternatively, or in addition, MED13 may control systemic energy consumption and storage through a cardiac-derived paracrine factor acting on distal tissues either directly or through secondary targets such as the brain. The heart has been shown to exert endocrine effects by releasing natriuretic peptides, ANP and BNP (Sengenes et al., 2000), however, we did not observe altered expression of ANP or BNP in mice with MED13 gain- or loss-of-function. Gene analysis of non-cardiac tissues, WAT, BAT and liver, suggests activation of a metabolic gene profile consistent with the increase in energy expenditure in response to cardiac MED13 activity. Considering that perturbation of cardiac energy metabolism is among the earliest alterations in the myocardium during the development of diabetes (Harmancey et al., 2008), it will be of interest to investigate the potential involvement of the MED13-miR-208a regulatory circuit in this disorder.
Therapeutic Implications
TH signaling confers numerous beneficial effects, including increased metabolic rate, lipolysis, cholesterol lowering, improved glucose homeostasis, and enhanced cardiac function as seen in antimiR-208a treated and αMHC-Med13 TG mice (Crunkhorn and Patti, 2008; Song et al., 2011; Watanabe et al., 2006). However, hyperthyroidism is also associated with adverse effects, such as tachycardia, arrhythmias, muscle wasting and elevated body temperature, which we did not observe in these mice. There have been numerous efforts to develop TH mimetics for enhancing cardiac function and improving metabolic status, but the adverse side-effects of TH stimulation have limited the usefulness of such compounds (Tancevski et al., 2011). AntimiR-208a seems to display the beneficial activities of thyromimetics, however, the target of antimiR-208a is cardiac-specific, thereby bypassing potential adverse effects on other tissues where TH exerts its effects. The apparent metabolic effects of miR-208a inhibition and MED13 elevation in the heart suggest that miR-208a inhibitors, in addition to providing benefit in the setting of cardiac dysfunction, may have therapeutic usefulness in a variety of metabolic disorders, such as obesity, hypercholesterolemia, type 2 diabetes, hepatic steatosis and hyperlipidemia.
EXPERIMENTAL PROCEDURES
AntimiR Injections
Six week old, male C57Bl6 mice were injected subcutaneously with 10 mg/kg of Locked-Nucleic Acid (LNA) modified antimiR dissolved in saline. The LNA modified antimiR oligonucleotides were produced at miRagen Therapeutics, Inc. and designed to complement miR-208a and a universal control directed against a C. elegans-specific miRNA (Montgomery et al., 2011). The mice were injected for three consecutive days and then given a weekly maintenance injection throughout the experiment.
Generation of αMHC-Med13 TG Mice
The Med13 transcript was amplified from mouse heart total RNA using Med13-specific primers. Med13 cDNA was cloned into a plasmid containing the αMHC promoter and human GH (hGH) poly(A)+ signal (Subramaniam et al., 1991). Transgenic mice were generated by standard techniques and backcrossed into C57Bl6 background for six or more generations.
Generation of Med13 conditional knockout mice
Med13 cKO mice were generated by inserting LoxP sites for Cre-mediated excision flanking exons 7 and 8 as described in the supplemental information (Figure S5).
Plasmids and Transfection Assays
Plasmids containing a TH response element (TRE), TRβ and RXRα were obtained from Dr. David Mangelsdorf. The Thrsp promoter was cloned from genomic DNA by PCR and inserted into the pGL2 vector (Invitrogen). HEK293 cells were transfected using FuGENE 6 (Roche). Transfections were performed with 10ng pCMV lacZ reporter, 50ng of the luciferase reporter, 15ng for each nuclear hormone receptor and 75 ng GFP-MED13 unless otherwise stated. Luciferase activity and β-galactosidase assays were performed on cell lysates.
RNA Analysis
RNA was isolated from mouse tissues using TRIzol reagent (Invitrogen). RT-PCR was performed to generate cDNA. For qPCR, 20 ng of cDNA was used for each reaction with Taqman or Sybergreen probes and normalized to 18S. Sequences for the primers were generated using mouse primer depot software and are listed in Supplemental Information. Microarray analysis was performed by the University of Texas Southwestern Microarray Core Facility using the MouseWG-6 v2.0 BeadChips (Illumina) using RNA extracted from tissues from 3, 4 week old WT or αMHC-Med13 TG mice and subsequently pooled prior to analysis.
Metabolic Cage Studies
Metabolic cage studies were performed by the University of Texas Southwestern Mouse Metabolic Phenotyping Center. Twelve-week old male mice on NC were placed in TSE metabolic chambers for an initial 5-day acclimation period, followed immediately by a 4.5-day experimental period with data collection. Room temperature for all metabolic studies was maintained at 22°C with a 12-h light/dark cycle. Airflow into the chamber was maintained at 0.6 L/min, and exhaust air from each chamber was measured for 3 min every 39 min. Oxygen consumption and carbon dioxide production was measured and was normalized to lean body mass. During this time, ambulatory and rearing activities were also monitored with infrared beams. Food and water intake was monitored.
NMR
Male mice body composition parameters including fat mass, lean tissue mass and water were analyzed using a Bruker Minispec mq10.
Metabolic Assays
Glucose tolerance tests were performed following overnight fasting. Baseline measurements were taken using an Accu-Chek Compact Plus glucometer (Roche). Mice were subsequently injected with 1mg/g glucose intraperitoneally. Glucose levels were then measured at 15, 30, 60 and 120 minutes following glucose injection. Blood, heart and liver were collected from 12-week old male mice for triglyceride, cholesterol, insulin and leptin measurements. Blood was collected using a 1 mL syringe coated in 0.5M K EDTA, incubated for 25 minutes at room temperature followed by centrifugation for 10 minutes at 5000 rpm and serum collected. Insulin and Leptin levels were measured by ELISA assay.
Transthoracic Echochardiography
Cardiac function was evaluated by two-dimensional transthoracic echocardiography on conscious mice using a VisualSonics Vevo2100 imaging system. All measurements were performed by a single experienced operator blinded to the mouse genotypes.
Animal Care
Animals were fed standard chow with 4% kcal from fat ad lib unless otherwise noted. HF diet analyses were done using the 60% kcal from fat diet or the 10% kcal from fat as the NC-control diet (Open Source Diets D12492i and D12450Bi). Propylthiouracil (PTU) was administered in the chow at 0.15% for 2 weeks (Harlan Teklad). All animal procedures were approved by the Institutional Animal Care and Use Committee at UT Southwestern Medical Center.
Statistics
Values are expressed as the mean ± standard error. Student’s t-test was performed for paired analysis. Multiple group analyses were done by using ANOVA with Bonferroni correction applied for appropriate post-hoc analysis. The null hypothesis was rejected if p < 0.05.
Supplementary Material
Figure S1. AntimiR-208a Treatment Effect on Cardiac Function, Lipid Accumulation, Food Intake, Activity and Cardiac Gene Expression, Related to Figure 1.
(A) Heart weight from antimiR-208a and control antimiR treated mice on NC or HF for 6 weeks, n=5–13
(B) Fractional shortening for antimiR-208a and control antimiR treated mice on NC for 6 weeks, n=5
(C) Heart rate in beats per minute for antimiR-208a and control antimiR treated mice on NC for 6 weeks, n=5
(D) Pictures of visceral WAT and liver from antimiR-208a and control antimiR treated mice on NC for 6 weeks
(E) Food consumption n=11
(F) Physical activity, average beam breaks in the X,Y and Z axis over a 12 hour light/dark cycle n=11
(G) Cardiac Med13 mRNA levels from antimiR-208a or control antimiR treated mice on NC. n=5
(H) Altered cardiac metabolic gene expression from antimiR-208a or control antimiR treated mice on NC. n=5
Data is represented as mean ± SEM *p<0.05
Figure S2. Characterization of αMHC-Med13 TG Mice, Related to Figure 2.
(A) Multi-tissue analysis comparing Med13 mRNA levels from αMHC-Med13 TG mice (Line 1) compared to WT mice by quantitative RT-PCR.
(B) Representative immunoblot of Med13 from WT and αMHC-Med13 TG hearts. Anti-tubulin was used as loading control.
(C) H&E stain from αMHC-Med13 TG and WT mouse hearts.
(D) BNP and ANF levels in αMHC-Med13 TG and WT mice on NC
(E) Nose-to-anus length for 12 week old αMHC-Med13 TG and WT mice on NC.
(F) Male 6 week old αMHC-Med13 TG (Line 2, over-expressing Med13 3-fold in the heart)) and WT mice growth curves and percent change in body weight on HF, n=9–14
(G) Female 6 week old αMHC-Med13 TG mice (Line 1) and WT growth curves (left) and percent change in bodyweight on HF and NC for 16 weeks, n=7 or NC, n=3
(H) Body composition analysis of αMHC-Med13 TG (Line 1) and WT mice at 7 and 12 months of age. n=3–5
(I) Oil red O stained hearts and liver from high fat treated αMHC-Med13 TG (Line 1) and WT mice. Scale=40μM
Data are represented as mean ± SEM. * p<0.05 αMHC-Med13 TG vs WT
Figure S3. Core Body Temperature is Comparable in αMHC-Med13 TG and WT mice, Related to Figure 3.
Core body temperature of αMHC-Med13 TG (Line 1) and WT mice measured at room temperature. (n=5)
Figure S4. Analysis of heart, WAT, BAT and liver microarray data comparing αMHC-Med13 TG and WT mice, Related to Figure 4.
Tissue and RNA was isolated from 4 week old mice and analyzed by microarray. Genes that were altered greater than two-fold up or down in the array were further analyzed using Panther software and are displayed as the fraction of changes in biological processes.
Figure S5. Characterization of Med13fl/fl; αMHC-Cre cKO, Related to Figure 6.
(A) Schematic diagram of putative Med13 protein domains, the arrow represents the end of the Med13 resulting from targeted gene deletion. Arms of homology flanking exons 7 and 8 were used to insert loxP sites, a neomycin cassette and DTA for homologous recombination. Cre-mediated recombination of the Med13 cKO removes exons 7 and 8.
(B) PCR from fl/+, +/− and +/+ Med13 genotypes.
(C) Med13fl/fl; αMHC-Cre conditional knockout offspring chart
(D) Expression of Med13 mRNA in heart, n=3.
(E) H&E stain for 6 week old Med13fl/fl; αMHC-Cre and Med13fl/fl hearts.
(F) Representative traces of an echocardiograph from Med13fl/fl; αMHC-Cre and Med13fl/fl.
(G) Cardiac fractional shortening from Med13fl/fl; αMHC-Cre and Med13fl/fl mice. (n=5–7)
(H) Heart rate from Med13fl/fl; αMHC-Cre and Med13fl/fl mice. (n=5–7)
(I–J) Diastolic and systolic internal ventricular diameter from Med13fl/fl; αMHC-Cre and Med13fl/fl mice. (n=5–7)
(K) Med13fl/fl and Med13fl/fl; αMHC-cre cKO images displaying excessive WAT and fatty liver in mice after 12 weeks on HF diet.
Data are represented as mean ± SEM. * p<0.05
HIGHLIGHTS.
Cardiac expression of MED13 controls whole body metabolism, prevents obesity and improves systemic glucose homeostasis
Pharmacologic inhibition of cardiac miR-208a confers resistance to high fat diet-induced obesity
The actions of MED13 and miR-208a are mediated through their control of a metabolic gene program in the heart.
Acknowledgments
We thank Carrie A. Grueter, Jose Cabrera, John McAnally, Evelyn Tennison and Jennifer Brown for their assistance. We appreciate the services of the UT Southwestern Transgenic Core Facility and the UT Southwestern Mouse Metabolic Phenotyping Core supported by National Institute of Health Grants PL1 DK081182 and UL1 RR024923. This work was supported in part by grants from National Institutes of Health, the Donald W. Reynolds Center for Clinical Cardiovascular Research, the Robert A. Welch Foundation (grant I-0025), the Leducq Fondation-Transatlantic Network of Excellence in Cardiovascular Research Program and the American Heart Association-Jon Holden DeHaan Foundation to E.N.O. C.E.G. was supported by a fellowship from the American Diabetes Association (7-09-CVD-04). J.K.E. was supported by RL1DK081185.
Footnotes
Supplemental Information includes Extended Experimental Procedures and five figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GW, Zhu Q, Metkowski J, Stack MJ, Gopinath S, Mariash CN. The Thrsp null mouse (Thrsp(tm1cnm)) and diet-induced obesity. Mol Cell Endocrinol. 2009;302:99–107. doi: 10.1016/j.mce.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek HJ, Malik S, Qin J, Roeder RG. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Kantor HL, Katz LA, Briggs RW. Relation between work and phosphate metabolite in the in vivo paced mammalian heart. Science. 1986;232:1121–1123. doi: 10.1126/science.3704638. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Belakavadi M, Fondell JD. Cyclin-dependent kinase 8 positively cooperates with Mediator to promote thyroid hormone receptor-dependent transcriptional activation. Mol Cell Biol. 2010;30:2437–2448. doi: 10.1128/MCB.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang X, Birsoy K, Roeder RG. A muscle-specific knockout implicates nuclear receptor coactivator MED1 in the regulation of glucose and energy metabolism. Proc Natl Acad Sci U S A. 2010;107:10196–10201. doi: 10.1073/pnas.1005626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. Function and regulation of the Mediator complex. Curr Opin Genet Dev. 2011;21:225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crunkhorn S, Patti ME. Links between thyroid hormone action, oxidative metabolism, and diabetes risk? Thyroid. 2008;18:227–237. doi: 10.1089/thy.2007.0249. [DOI] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Lehman JJ, Leone TC, Welch MJ, Bennett MJ, Kovacs A, Han X, Gross RW, Kozak R, Lopaschuk GD, et al. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell JD, Guermah M, Malik S, Roeder RG. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc Natl Acad Sci U S A. 1999;96:1959–1964. doi: 10.1073/pnas.96.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederich RC, Hamann A, Anderson S, Lollmann B, Lowell BB, Flier JS. Leptin levels reflect body lipid content in mice: evidence for diet-induced resistance to leptin action. Nat Med. 1995;1:1311–1314. doi: 10.1038/nm1295-1311. [DOI] [PubMed] [Google Scholar]
- Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- Harmancey R, Wilson CR, Taegtmeyer H. Adaptation and maladaptation of the heart in obesity. Hypertension. 2008;52:181–187. doi: 10.1161/HYPERTENSIONAHA.108.110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 2001;12:127–134. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- Jones JR, Barrick C, Kim KA, Lindner J, Blondeau B, Fujimoto Y, Shiota M, Kesterson RA, Kahn BB, Magnuson MA. Deletion of PPARgamma in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc Natl Acad Sci U S A. 2005;102:6207–6212. doi: 10.1073/pnas.0306743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nature reviews Cardiology. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- Malik S, Guermah M, Yuan CX, Wu W, Yamamura S, Roeder RG. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol Cell Biol. 2004;24:8244–8254. doi: 10.1128/MCB.24.18.8244-8254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaka T, Shimano H, Yahagi N, Kato T, Atsumi A, Yamamoto T, Inoue N, Ishikawa M, Okada S, Ishigaki N, et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat Med. 2007;13:1193–1202. doi: 10.1038/nm1662. [DOI] [PubMed] [Google Scholar]
- Mitchell CS, Savage DB, Dufour S, Schoenmakers N, Murgatroyd P, Befroy D, Halsall D, Northcott S, Raymond-Barker P, Curran S, et al. Resistance to thyroid hormone is associated with raised energy expenditure, muscle mitochondrial uncoupling, and hyperphagia. J Clin Invest. 2010;120:1345–1354. doi: 10.1172/JCI38793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, Stack C, Latimer PA, Olson EN, van Rooij E. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Li G, Lin YP, Barrero MJ, Ge K, Roeder RG, Wei LN. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell. 2005;19:643–653. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Philibert R, Caspers K, Langbehn D, Troughton EP, Yucuis R, Sandhu HK, Cadoret RJ. The association of a HOPA polymorphism with major depression and phobia. Compr Psychiatry. 2002;43:404–410. doi: 10.1053/comp.2002.33489. [DOI] [PubMed] [Google Scholar]
- Pospisilik JA, Schramek D, Schnidar H, Cronin SJ, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J, et al. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell. 2010;140:148–160. doi: 10.1016/j.cell.2009.12.027. [DOI] [PubMed] [Google Scholar]
- Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiological reviews. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- Sengenes C, Berlan M, De Glisezinski I, Lafontan M, Galitzky J. Natriuretic peptides: a new lipolytic pathway in human adipocytes. FASEB J. 2000;14:1345–1351. [PubMed] [Google Scholar]
- Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yao X, Ying H. Thyroid hormone action in metabolic regulation. Protein Cell. 2011;2:358–368. doi: 10.1007/s13238-011-1046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam A, Jones WK, Gulick J, Wert S, Neumann J, Robbins J. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. The Journal of biological chemistry. 1991;266:24613–24620. [PubMed] [Google Scholar]
- Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancevski I, Rudling M, Eller P. Thyromimetics: a journey from bench to bed-side. Pharmacol Ther. 2011;131:33–39. doi: 10.1016/j.pharmthera.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards GJ, Campfield LA, Clark FT, Deeds J, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- Van Gaal LF, Mertens IL, De Block CE. Mechanisms linking obesity with cardiovascular disease. Nature. 2006;444:875–880. doi: 10.1038/nature05487. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Wang W, Huang L, Huang Y, Yin JW, Berk AJ, Friedman JM, Wang G. Mediator MED23 links insulin signaling to the adipogenesis transcription cascade. Dev Cell. 2009;16:764–771. doi: 10.1016/j.devcel.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- Wu Y, Li Y, Lange EM, Croteau-Chonka DC, Kuzawa CW, McDade TW, Qin L, Curocichin G, Borja JB, Lange LA, et al. Genome-wide association study for adiponectin levels in Filipino women identifies CDH13 and a novel uncommon haplotype at KNG1-ADIPOQ. Hum Mol Genet. 2010;19:4955–4964. doi: 10.1093/hmg/ddq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZP, Wawrousek EF, Piatigorsky J. Transketolase haploinsufficiency reduces adipose tissue and female fertility in mice. Mol Cell Biol. 2002;22:6142–6147. doi: 10.1128/MCB.22.17.6142-6147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Yon Toh S, Chen Z, Guo K, Ng CP, Ponniah S, Lin SC, Hong W, Li P. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat Genet. 2003;35:49–56. doi: 10.1038/ng1225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. AntimiR-208a Treatment Effect on Cardiac Function, Lipid Accumulation, Food Intake, Activity and Cardiac Gene Expression, Related to Figure 1.
(A) Heart weight from antimiR-208a and control antimiR treated mice on NC or HF for 6 weeks, n=5–13
(B) Fractional shortening for antimiR-208a and control antimiR treated mice on NC for 6 weeks, n=5
(C) Heart rate in beats per minute for antimiR-208a and control antimiR treated mice on NC for 6 weeks, n=5
(D) Pictures of visceral WAT and liver from antimiR-208a and control antimiR treated mice on NC for 6 weeks
(E) Food consumption n=11
(F) Physical activity, average beam breaks in the X,Y and Z axis over a 12 hour light/dark cycle n=11
(G) Cardiac Med13 mRNA levels from antimiR-208a or control antimiR treated mice on NC. n=5
(H) Altered cardiac metabolic gene expression from antimiR-208a or control antimiR treated mice on NC. n=5
Data is represented as mean ± SEM *p<0.05
Figure S2. Characterization of αMHC-Med13 TG Mice, Related to Figure 2.
(A) Multi-tissue analysis comparing Med13 mRNA levels from αMHC-Med13 TG mice (Line 1) compared to WT mice by quantitative RT-PCR.
(B) Representative immunoblot of Med13 from WT and αMHC-Med13 TG hearts. Anti-tubulin was used as loading control.
(C) H&E stain from αMHC-Med13 TG and WT mouse hearts.
(D) BNP and ANF levels in αMHC-Med13 TG and WT mice on NC
(E) Nose-to-anus length for 12 week old αMHC-Med13 TG and WT mice on NC.
(F) Male 6 week old αMHC-Med13 TG (Line 2, over-expressing Med13 3-fold in the heart)) and WT mice growth curves and percent change in body weight on HF, n=9–14
(G) Female 6 week old αMHC-Med13 TG mice (Line 1) and WT growth curves (left) and percent change in bodyweight on HF and NC for 16 weeks, n=7 or NC, n=3
(H) Body composition analysis of αMHC-Med13 TG (Line 1) and WT mice at 7 and 12 months of age. n=3–5
(I) Oil red O stained hearts and liver from high fat treated αMHC-Med13 TG (Line 1) and WT mice. Scale=40μM
Data are represented as mean ± SEM. * p<0.05 αMHC-Med13 TG vs WT
Figure S3. Core Body Temperature is Comparable in αMHC-Med13 TG and WT mice, Related to Figure 3.
Core body temperature of αMHC-Med13 TG (Line 1) and WT mice measured at room temperature. (n=5)
Figure S4. Analysis of heart, WAT, BAT and liver microarray data comparing αMHC-Med13 TG and WT mice, Related to Figure 4.
Tissue and RNA was isolated from 4 week old mice and analyzed by microarray. Genes that were altered greater than two-fold up or down in the array were further analyzed using Panther software and are displayed as the fraction of changes in biological processes.
Figure S5. Characterization of Med13fl/fl; αMHC-Cre cKO, Related to Figure 6.
(A) Schematic diagram of putative Med13 protein domains, the arrow represents the end of the Med13 resulting from targeted gene deletion. Arms of homology flanking exons 7 and 8 were used to insert loxP sites, a neomycin cassette and DTA for homologous recombination. Cre-mediated recombination of the Med13 cKO removes exons 7 and 8.
(B) PCR from fl/+, +/− and +/+ Med13 genotypes.
(C) Med13fl/fl; αMHC-Cre conditional knockout offspring chart
(D) Expression of Med13 mRNA in heart, n=3.
(E) H&E stain for 6 week old Med13fl/fl; αMHC-Cre and Med13fl/fl hearts.
(F) Representative traces of an echocardiograph from Med13fl/fl; αMHC-Cre and Med13fl/fl.
(G) Cardiac fractional shortening from Med13fl/fl; αMHC-Cre and Med13fl/fl mice. (n=5–7)
(H) Heart rate from Med13fl/fl; αMHC-Cre and Med13fl/fl mice. (n=5–7)
(I–J) Diastolic and systolic internal ventricular diameter from Med13fl/fl; αMHC-Cre and Med13fl/fl mice. (n=5–7)
(K) Med13fl/fl and Med13fl/fl; αMHC-cre cKO images displaying excessive WAT and fatty liver in mice after 12 weeks on HF diet.
Data are represented as mean ± SEM. * p<0.05