Abstract
Unique peptide-morpholino oligomer (PMO) conjugates have been designed to bind and promote the cleavage of specific mRNA as a tool to inhibit gene function and parasite growth. The new conjugates were validated using the P. falciparum gyrase mRNA as a target (PfGyrA). Assays in vitro demonstrated a selective degradation of the PfGyrA mRNA directed by the external guide sequences, which are morpholino oligomers in the conjugates. Fluorescence microscopy revealed that labeled conjugates are delivered into Plasmodium-infected erythrocytes during all intraerythrocytic stages of parasite development. Consistent with the expression of PfGyrA in all stages of parasite development, proliferation assays showed that these conjugates have potent antimalarial activity, blocking early development, maturation, and replication of the parasite. The conjugates were equally effective against drug sensitive and resistant P. falciparum strains. The potency, selectivity, and predicted safety of PMO conjugates make this approach attractive for the development of a unique class of target-specific antimalarials and for large-scale functional analysis of the malarial genome.
Keywords: RNase P, nuclease resistance
Malaria is the most prevalent and deadliest parasitic disease worldwide (1, 2). It is caused by intraerythrocytic protozoan parasites of the genus Plasmodium. Four species, Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale are known to be infectious to humans, and more recent cases of infection due to Plasmodium knowlesi have also been reported (3). The lack of a universally effective vaccine to combat this disease and the growing spread of resistance to all currently known antimalarials, including those used in combination therapy, emphasize the need for novel approaches to develop new therapies that specifically target essential functions of the parasite to block both infection and transmission.
The genome of P. falciparum has 23 Mb and encodes ∼5,300 proteins, the majority of which are of unknown function and their importance in parasite development remains to be determined. This uncertainty is due mostly to the difficulty of genetically manipulating this organism using forward genetic approaches, lack of a RNAi machinery (4, 5), and absence of regulatable promoters. Current techniques for genetic disruption are mostly useful for those genes that are not essential for blood stage development. Consequently, only a limited number of genes have been shown through genetic means to play an essential role in parasite development and thus are valid targets for development of new antimalarial drugs. Nutritional complementation has recently been used to generate a conditionally lethal P. falciparum mutant lacking the primary purine transporter PfNT1 (6). However, this approach cannot be used widely to create conditionally lethal mutants. Although methods for inducible expression have been reported, they have had limited success in creating stably inducible knockouts (7–12).
Strategies involving the catalytic properties of ribonuclease P (RNase P) targeted to a specific site by an external guide sequence (EGS) complementary to the targeted RNA have been successfully used to inhibit the expression in vivo of bacterial genes from different pathogenic species (13–16). Thus, a unique basis for antimalarial drug design relies on the use of morpholino oligonucleotides (MOs) with a base sequence that acts as an analog of an EGS. A recent study has shown that a basic peptide (cell penetrating peptide, CPP), derived from human T cells, covalently linked to a MO, was very effective in passing through cell walls and membranes in bacteria (17). The resulting complex is cleaved by the resident enzyme RNase P to inactivate the mRNA and thereby reduce its expression. This strategy has been adapted to P. falciparum in which the gene coding for gyrase A, PfGyrA (18), was specifically targeted by the peptide-morpholino oligomer (PMO). Using EGS technology, we report inhibition of P. falciparum development, survival, and PfGyrA expression. The technology also holds the potential for allowing large-scale functional analysis of Plasmodium genomes.
Results
EGS Oligonucleotide Directs Cleavage of the P. falciparum gyrA mRNA in Vitro by Human RNase P.
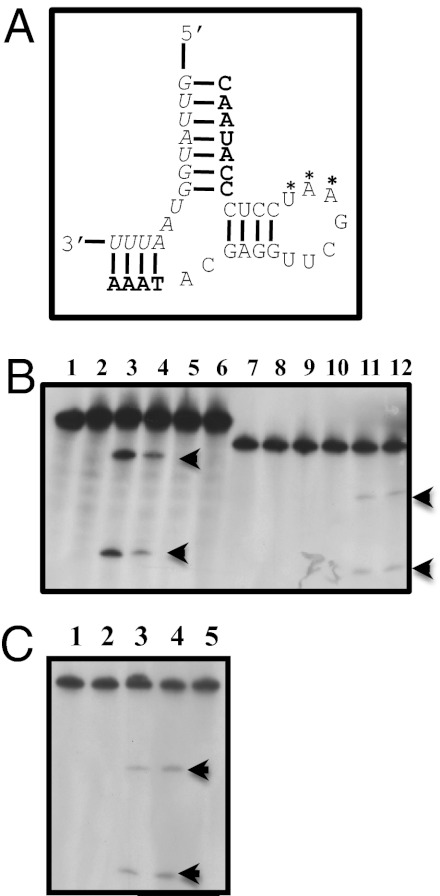
The gyrase A gene from Escherichia coli harbors a short sequence, which is highly conserved across many bacteria, and has successfully been targeted for RNase P-mediated degradation (17). This sequence is also partially conserved in the gyrA gene of P. falciparum, which should play an essential function in parasite replication (Table 1). The sequence used in the experiments reported here is a target for complementary EGS and as part of a chemically derived MO. Interestingly, this sequence is different from that in the human topoisomerase II TopoII gene by three nucleotides and two of them are contiguous in the region important for the action of the EGS (Table 1). An EGS RNA (Fig. 1A), with a structure required for recognition by an eukaryotic RNase P, was cloned (13) and incubated in vitro in the presence of a 32P-labeled mRNA segment of PfGyrA in the absence or presence of RNase P (Materials and Methods). The reaction products were subsequently separated by electrophoresis on the basis of their molecular size. As expected for a successful EGS (13), the EGS complementary to PfGyrA produced the anticipated fragments with a relatively crude fraction of RNase P from HeLa cells, whereas an EGS targeting the E. coli gyrase mRNA did not cleave this construct (Fig. 1 B and C). Consequently, conjugates were synthesized with the corresponding MO (Materials and Methods) and tested for their ability to be transported inside the parasite and to inhibit the growth of P. falciparum during its intraerythrocytic development.
Table 1.
Sequence alignment of the EGS-targeted regions in the E. coli, P. falciparum, P. yoelli, and P. berghei gyrase A genes and the H. sapiens TOP2A (topo II) gene
| Organism | Target DNA sequence |
| E. coli | CGGTCAGGGTAACTTCGGT330 |
| P. falciparum | AGGTTATGGTAATTTTGGT825 |
| P. yoelli | TGGATATGGTAATTTTGGG870 |
| P. berghei | CGGATATGGTAATTTTGGA786 |
| H. sapiens | TGATTATTATAATTTTGAG6457 |
Numbers at right are the positions in the sequence alignment of the gene of the last 3′ nucleotide in this particular segment. Bold fonts are the complements of the nucleotides in the EGSs that were made in this study.
Fig. 1.
Identification of a gyrase mRNA target, design of an EGS, and cleavage in vitro of gyrA mRNA. (A) Schematic of EGS-mRNA binding. mRNA of Plfgyr is in italics; nucleotides in the EGS involved in hydrogen bonding with the mRNA are in bold; stem-loop for RNase P recognition is in normal font; A*A*U* in Plfgyr2 are replaced with TGC, respectively. (B) Cleavage in vitro by RNase P of gyrA mRNA. HeLa RNase P (DEAE purified) was used on E. coli gyrA mRNA (Ecgyr mRNA) or P. falciparum gyrA mRNA (Plfgyr mRNA). Lane 1, Ecgyr mRNA alone; lane 2, Ecgyr mRNA + enzyme; lane 3, Ecgyr mRNA + enzyme + 50× Ecgyr full-length (FL) EGS; lane 4, Ecgyr mRNA + enzyme + 50× Ecgyr EGS; lane 5, ECgyr mRNA + enzyme + 50× Plfgyr FL EGS; lane 6, Ecgyr mRNA + enzyme + 50× Plfgyr EGS; lane 7, Plfgyr mRNA alone; lane 8, Plfgyr mRNA + enzyme; lane 9, Plfgyr mRNA + enzyme + 50× Ecgyr FL EGS; lane 10, Plfgyr mRNA + enzyme + 50× Ecgyr EGS; lane 11, Plfgyr mRNA + enzyme + 50× Plfgyr FL EGS; lane 12, Plfgyr mRNA + enzyme + 50× Plfgyr EGS. Arrows indicate the positions of cleavage products. (C) Cleavage of P. falciparum gyrA mRNA (Pflgyr mRNA) by RNase P using different EGSs. HeLa RNase P (purified) was used for cleavage in vitro. Lane 1, Plfgyr mRNA alone; lane 2, Plfgyr mRNA + enzyme; lane 3, Plfgyr mRNA + enzyme + 50× Plfgyr EGS; lane 4, Plfgyr mRNA + enzyme + 50× Plfgyr2 EGS; lane 5, Plfgyr mRNA + enzyme + 25× PlfgyrΔ2bp EGS. EGS are listed as described in Fig. 1B legend. The sequences of conjugates are shown in Table S1.
Specific Fluorescent-Labeled Conjugate Is Transported into Malaria Parasites.
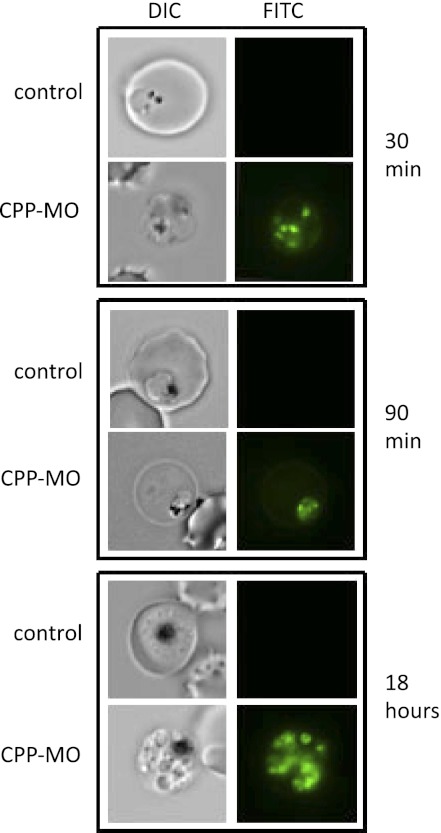
To assess the ability of the conjugates to enter Plasmodium-infected erythrocytes and to examine the effect of d- versus l-amino acid on the delivery of the conjugates, a fluorescein derivative of the CPP-MO was used. Fluorescent conjugates containing either five d- or l-arginines (Table 2) and in which the dye is linked to the amino terminus of the conjugates, were added to a culture of ring stage parasites and parasites were examined at different time points during the intraerythrocytic life cycle of the parasite. Fluorescence microscopy analysis revealed that within 30 min, the conjugates could be readily detected within infected erythrocytes and inside the parasite (Fig. 2). After 90 min, most of the fluorescence was detected inside the parasite. Cultures maintained for 18 h, in which parasites evolved to an early schizont stage also exhibited fluorescence suggesting a high stability of the conjugate. No fluorescence could be detected in uninfected red blood cells. This result suggests that the trafficking of the conjugates is mediated by permeation pathways or selective transporters on the plasma membrane of the infected erythrocyte and parasite plasma membrane. Interestingly, the fluorescence signal of the conjugate containing five d-arginine residues was at least 1,000-fold stronger than that of a similar conjugate with all l-arginine residues. The stability of the fluorescent conjugate is a remarkable indicator of the utility of such compounds as drug candidates.
Table 2.
Peptide sequence used in the conjugates
| Tyr Ala Arg Val Arg Arg Arg Gly Pro Arg Gly Tyr Ala Arg Val Arg Arg Arg Gly Pro Arg Arg |
Residues marked in bold were D forms that were used in the fluorescent conjugate mentioned in the text. In all other experiments, all residues were L forms.
Fig. 2.
Localization of CPP-MO conjugates in P. falciparum-infected erythrocytes. A fluorescein labeled CPP-MO was added at 3 μM to a highly synchronized late ring stage culture and the incorporation into the parasite was followed by fluorescent microscopy. Images were taken from samples mixed with a conjugate containing five d-arginine (Table 2).
CPP-MO Conjugate Inhibits P. falciparum Development and Reduces PfgyrA mRNA Expression.
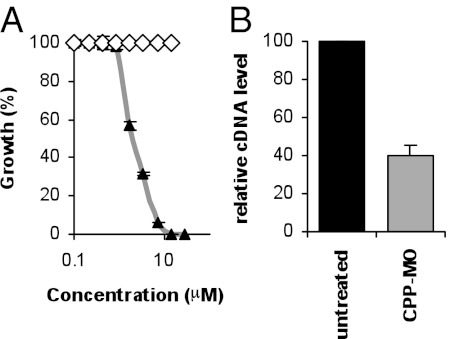
To determine the efficacy of the conjugate against the parasite, a synchronized culture of the P. falciparum 3D7 clone at the early ring stage of development was maintained in the absence or presence of increasing concentrations of the conjugate, and parasite proliferation was monitored using a SYBR Green I assay (Materials and Methods). Consistent with results obtained in vitro (Fig. 1 B and C), the conjugate inhibited the growth of 3D7 parasites with IC50 values ranging between 0.8 and 2 μM depending on the batch of the compound (Fig. 3A). The IC90 and IC100 of the conjugate were reached at ∼6.5 and 10 μM, respectively. As a control, a conjugate that targets the E. coli ampicillin resistance gene had no effect on parasite proliferation (Fig. 3A). To further demonstrate that the CPP-MO mediates degradation of the PfGyrA mRNA in the parasite, cultures containing primarily late rings and early trophozoites were maintained in the absence or presence of 4 μM of the Plfgyr EGS-based CPP-MO and total RNA was extracted 4 h posttreatment and analyzed by qRT-PCR using primers specific to the PfGyrA mRNA. Compared with untreated parasites, conjugate treatment resulted in a 60% decrease in the amount of PfGyrA mRNA (Fig. 3B). As a control, qRT-PCR analysis using primers specific to the P. falciparum actin gene showed equal amounts of mRNA in both treated and untreated parasites.
Fig. 3.
Effect of CPP-MO conjugates in P. falciparum-infected erythrocytes. (A) Effect of treatment with CPP-MO targeting either the P. falciparum PfGyrA or the E. coli bla mRNAs on the growth of the parasite during its intraerythrocytic life cycle. Growth assays were performed using a SYBR Green-I assay (Materials and Methods) after culturing parasites for 72 h at 37 °C in the presence of the conjugates. Results are a mean of triplicate experiments ± SD. (B) Effect of Plfgyr EGS-based CPP-MO on PfGyrA mRNA levels. PfGyrA mRNA levels of PfgyrA in the absence (black bar) or presence (gray bar) of the conjugate were measured using qRT-PCR. The Pf-β-actin1 gene was used as an internal control and the relative expression level of PfgyrA in the absence of the conjugate was set to 100. Results are mean of triplicate experiments ± SD; *P < 0.01.
CPP-MO–Mediated Stage Specificity of Inhibition.
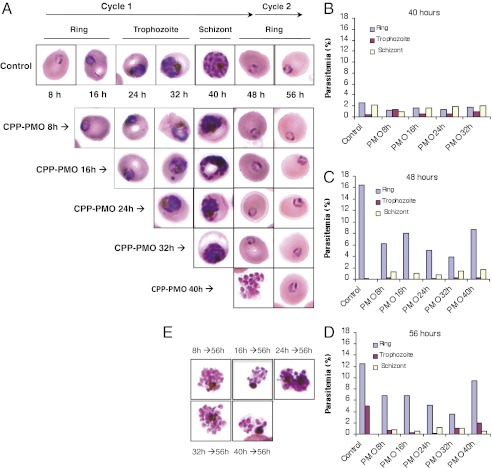
Previous studies have shown that PfGyrA is expressed throughout the intraerythrocytic life of the parasite (18). Therefore, an examination of the addition of the PfGyrA specific CPP-MO at different stages of parasite development was carried out to see whether parasite progression from one stage to the next could be altered. The conjugate was added to a highly synchronized ring stage culture of the parasite at its IC50 value at different time intervals and the growth and morphology of the parasites were monitored every 8 h (Fig. 4). Microscopy analyses revealed that addition of the conjugate at the ring stage resulted in both a delay in parasite progression from one stage to another as well as alteration in the morphology of the parasites. By 40 h, whereas untreated cultures were predominantly at the schizont stage or have already produced new daughter parasites resulting in increased overall parasitemia, CPP-MO–treated parasites were blocked at all stages of intraerythrocytic development with fewer new daughters produced and the total parasitemia did not significantly increase (Fig. 4 A and B). By 48 h postinvasion, untreated parasites reached 16% parasitemia with virtually all parasites at the ring stage, whereas the total parasitemia of cultures treated with the CPP-MO conjugate was half that of untreated cultures. From the 3% starting population of ring stage parasites, half remained at the schizont stages (Fig. 4C).
Fig. 4.
Effect of Plfgyr EGS-based CPP-MO on P. falciparum development and egress. Parasite intraerythrocytic development was assessed in the absence or presence of 2 μM of the CPP-MO used in Fig. 1B after 8, 16, 24, 32, and 40 h following merozoite invasion. (A) Morphology of parasites during the course of the assay. (B–D) Percentage of each parasite developmental stage 40 h (B), 48 h (C), and 56 h (D) after merozoite invasion. (E) Images of abnormal schizonts unable to rupture 56 h postmerozoite invasion.
When the conjugate was added at the trophozoite (24 h) or early schizont (32 h) stages, the number of new daughter ring stage parasites produced was much more reduced compared with early treatment (Fig. 4 C and D), consistent with increased expression of PfGyrA during the trophozoite and schizont stages (18). When these parasites were examined 56 h postinvasion, up to 30% of the original population remained at the schizonts. Parasite cultures treated at 40 h postinvasion (2% schizonts and 2.5% rings) and analyzed 8 h later produced 8% rings and contained the same number of schizonts, whereas untreated cultures were made exclusively of ring stage parasites (16% parasitemia). Examination of these parasites 16 h posttreatment, showed that the number of schizonts has decreased, whereas the number of new daughter parasites produced did not increase, suggesting that the merozoites were either inviable or did not invade new red blood cells. In comparison, control cultures moved from 2% schizonts at 40 h postinvasion to 16% rings 56 h postinvasion.
We have also examined the morphology of parasites following treatment with the conjugates at its IC50 value. The majority of trophozoites and schizonts observed in treated cultures showed abnormal morphology (Fig. 4E).
PfGyrA Specific PMO Is Effective Against both Drug-Sensitive and Drug-Resistant Strains.
Two CPP-MO conjugates were examined against drug-resistant isolates of P. falciparum (Table 3), one pyrimethamine-resistant strain Hb3 and two chloroquine-resistant strains W2 and Dd2. As shown in Table 3, both conjugates were equally effective against drug-sensitive and drug-resistant strains. As expected, the 3D7 strain was sensitive to all drugs, the HB3 strain was resistant to pyrimethamine, and Dd2 and W2 strains were resistant to chloroquine.
Table 3.
Efficiency of two CPP-MO conjugates against chloroquine and pyrimethamine sensitive or resistant strains
| % of inhibition |
||||
| 3D7 | Hb3 | Dd2 | W2 | |
| Plfgyr EGS | 95.9 ± 0.6 | 100 | 100 | 100 |
| Plfgyr2 EGS | 63.1 ± 0.4 | 73.0 ± 2.8 | 77.2 ± 0.5 | 75.3 ± 1.5 |
| PYR 50 nM | 71.5 ± 0.05 | 0 | 0 | 8.6 ± 2.3 |
| CQ 25 nM | 99.0 ± 0.83 | 100 | 0 | 12.3 ± 2.3 |
| ART 25 nM | 91.5 ± 4.8 | 99.5 ± 0.2 | 18.8 ± 2.7 | 98.9 ± 0.6 |
| AQ 25 nM | 100 ± 0.0 | 100 | 97.8 ± 0.5 | 97.1 ± 0.4 |
Growth inhibition was determined using a SYBR Green-I assay after culturing parasites for 3 d at 37 °C in the presence of the CPP-MOs or antimalarials. Results are mean of triplicate experiments ± SD. The concentration of CPP-MO was 2 μM.
Discussion
The basic peptide-morpholino oligonucleotide method has been successfully used in treatment of muscular dystrophy (19) and mouse infection by bacteria (20). Some success has also been achieved with a therapy against highly pathogenic viruses (21). Because RNA-based methods such as RNAi are not employable in P. falciparum (4, 5), the RNase P-based target-specific mRNA degradation described herein is an ideal alternative approach for both functional analysis of the Plasmodium genome as well as malaria therapy.
The data presented here showed effective use of the PMO method to target the PfGyrA gene of P. falciparum. This gene is predicted to play an essential function in parasite replication and thus was an ideal target for validation of this method in P. falciparum. The specificity of the technology used here has already been demonstrated in tissue culture cells with respect to the flu virus, RNase P protein subunits, and thymidine kinase activity, and in bacteria with respect to drug resistance, gyrase enzyme, and type III transport (16, 22–27).
As with other oligonucleotide-based technologies, the challenge resides in the ability of the construct to be delivered into the infected erythrocyte. Our studies using fluorescently labeled conjugates indicated that they rapidly traffic into the infected erythrocyte. No fluorescence was detected in uninfected erythrocytes, suggesting that these constructs do not readily enter red blood cells. Infection of erythrocytes by P. falciparum is accompanied by major changes in the erythrocyte membrane, which result in increased permeability to a large number of substrates, many of which are essential nutrients such as sugars, purine nucleosides and nucleobases, vitamins, and amino acids (28, 29). Once inside the red blood cytoplasm, these precursors are transported inside the parasite by specific permeases expressed on the parasite plasma membrane (30–32). The selective delivery of the conjugates into infected erythrocytes but not uninfected erythrocytes, suggest that they may likely use the new permeation pathway (33) on the red blood cell membrane and possibly an oligopeptide permease on the parasite plasma membrane.
Current antimalarial therapy efforts supported by Medicines for Malaria Venture and the World Health Organization have focused on the development of new drugs against validated targets (34). However, because of the limited amenability of P. falciparum to genetic manipulation, only a handful of such targets have been validated genetically (6, 35–38). For most of the proposed validated targets, the validation is inferred from in silico analyses (39) or pharmacological studies using inhibitors. However, this latter approach is not reliable because of off-target effects associated with most of those compounds. Large scale efforts to assess the importance of each of the P. falciparum 5,300 genes is urgently needed and will help not only identify novel drug targets but also better advance our understanding of the biology of this important parasite and its pathogenesis. The success of our CPP-MO approach using the PfGyrA gene suggests that it can be applied to characterize the function of many other P. falciparum genes and establish a list of the validated targets, which can be effectively pursued for drug-based therapy.
With the widespread resistance to chloroquine and recent reports of resistance to artemisinin in Southeast Asia (1), efforts to develop new antimalarial therapies that use chemical classes that have not previously been used is urgent. Our finding that specific PfGyrA conjugates are equally potent against drug sensitive and resistant strains, and the unique specific features of the PMO approach in targeting specific mRNA suggest that this strategy may find a wider use not only as a tool for functional analysis of P. falciparum genes but also as a selective therapeutic approach. The reliability of any new antimalarial therapy is an important feature of its development. More than three mutation steps are needed to inactivate the conjugate, providing the base changes in the morpholino oligonucleotide are not next to each other. In the unfortunate case where the mutations are contiguous, another part of the mRNA to be attacked can simply be chosen as the target. This has already been achieved in E. coli (14).
The amino acid composition of the peptide and the overall sequence and size of the compounds used in this assay can easily be altered to improve the efficiency of the conjugates. The unique properties of these compounds will make them valuable in areas of the world where resistance to commonly used antimalarials is widespread.
Materials and Methods
Strains and Culture Conditions.
P. falciparum strains 3D7, Hb3, W2, and Dd2 were used and cultured using standard growth condition (40) modified by replacing human serum with 0.5% Albumax I (Invitrogen).
Synthesis of the Conjugates.
The method used was similar to that described in ref. 17.
Preparation of RNase P and Substrates.
The preparation of human RNase P is described in ref. 13. Substrates were prepared as described in refs. 13, 14.
Localization of a Fluorescent Conjugate in P. falciparum.
A highly synchronized late ring stage culture was mixed with 3 μM of CPPFL-MO (conjugate covalently bound to fluorescein) or RPMI (negative control). The culture was incubated for 18 h and samples were collected at different time points. Cells were harvested by centrifugation and washed with RPMI. Cells were placed on slides and covered with glass coverslips. Cells were immediately viewed under a Nikon Eclipse TE2000-E microscope equipped with 100× objective and a Roper CoolSNAP HQ camera. The images were analyzed using Meta Imaging.
SYBR Green-Based Parasite Growth Assay.
This proliferation assay was adapted from the malaria SYBR Green I-based fluorescence assay (41). CPP-MOs were added to a 96-well plate with final concentrations stated above. A highly synchronized early ring stage parasite culture was added to the plate containing conjugate. Controls were performed using noninfected erythrocytes, infected erythrocytes without conjugate, and infected erythrocytes treated with 1 μM CQ (3D7 strain) or 2.5 μg/mL of blasticidin (Hb3, W2, and Dd2 strains) as controls. Plates were incubated for 72 h at 37 °C in a gas chamber. After 72 h, erythrocytes were lysed with 20 mM Tris (pH 7.5), 5 mM EDTA, 0.008% saponin, 0.08% Triton-X 100, 1× SYBR Green I and incubated for 1 h in the dark at room temperature. Plates were read at 497/520 nm on a Synergy MX, Biotek fluorescent plate reader.
Analysis of Gene Expression by qPCR.
The Plfgyr EGS-based CPP-MO was added at 4 μM to late ring/early trophozoite synchronized cultures at 10% parasitemia (2% hematocrit) and the cultures were incubated for 4 h at 37 °C. Total RNA extraction from untreated and treated parasite cultures was performed as previously described (42) and the concentration of RNA was determined using a nanodrop. RNA samples (800 ng of total RNA) were treated with 1 unit of RQ1 DNase (Promega) and the absence of DNA contamination was checked by real-time PCR. cDNA were then synthesized from total RNA (250 ng) using iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was carried out using Quantitect SYBR Green PCR kit (Qiagen) using the Applied Biosystems 7500 real-time PCR system. Data were analyzed using the comparative critical threshold (ΔΔCt) method in which the amount of target RNA (gyrA) was compared with Pf-β-actin1, which served as an internal control as previously described (43). Primers used for qPCR are listed in Table S2.
Determination of the EGS Stage Specificity of Inhibition.
The experiment was initiated using a highly synchronous ring stage culture (8 h following merozoite invasion) at a parasitemia of 3%. Parasites were loaded into a 96-well plate and incubated under standard conditions. At different time intervals, the culture was mixed with 2 μM of the conjugate. Every 8 h the growth and morphology of the parasite was monitored by light microscopy. Images were collected using a Zeiss microscope equipped with 100× objective and an Infinity camera.
Supplementary Material
Acknowledgments
We are grateful to Dr. N. Gandotra who assisted with the preparation of some conjugates and S. Usmani-Brown for her assistance with qPCR analyses. Y.A. is supported by the Burroughs Welcome Fund (1006267). C.B.M. is supported by Grants AI51507 from the National Institute of Allergy and Infectious Diseases and 1006267 from the Burroughs Welcome Fund. S.A. is the recipient of a subaward from a National Institutes of Health grant to F. Liu of the University of California, Berkeley.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203516109/-/DCSupplemental.
References
- 1.Anonymous . World Malaria Report. Geneva: World Health Organization; 2010. [Google Scholar]
- 2.Murray CJ, et al. Global malaria mortality between 1980 and 2010: A systematic analysis. Lancet. 2012;379:413–431. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- 3.Singh B, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 4.Kolev NG, Tschudi C, Ullu E. RNA interference in protozoan parasites: Achievements and challenges. Eukaryot Cell. 2011;10:1156–1163. doi: 10.1128/EC.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ullu E, Tschudi C, Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 6.El Bissati K, et al. The plasma membrane permease PfNT1 is essential for purine salvage in the human malaria parasite Plasmodium falciparum. Proc Natl Acad Sci USA. 2006;103:9286–9291. doi: 10.1073/pnas.0602590103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong CM, Goldberg DE. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat Methods. 2007;4:1007–1009. doi: 10.1038/nmeth1132. [DOI] [PubMed] [Google Scholar]
- 8.Meissner M, et al. Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood stages using Toxoplasma gondii transactivators. Proc Natl Acad Sci USA. 2005;102:2980–2985. doi: 10.1073/pnas.0500112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muralidharan V, Oksman A, Iwamoto M, Wandless TJ, Goldberg DE. Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag. Proc Natl Acad Sci USA. 2011;108:4411–4416. doi: 10.1073/pnas.1018449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russo I, Oksman A, Vaupel B, Goldberg DE. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc Natl Acad Sci USA. 2009;106:1554–1559. doi: 10.1073/pnas.0806926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dvorin JD, et al. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science. 2010;328:910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrell A, et al. A DOC2 protein identified by mutational profiling is essential for apicomplexan parasite exocytosis. Science. 2012;335:218–221. doi: 10.1126/science.1210829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrier-Takada C, Altman S. Inactivation of gene expression using ribonuclease P and external guide sequences. Methods Enzymol. 2000;313:442–456. doi: 10.1016/s0076-6879(00)13028-9. [DOI] [PubMed] [Google Scholar]
- 14.Guerrier-Takada C, Salavati R, Altman S. Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc Natl Acad Sci USA. 1997;94:8468–8472. doi: 10.1073/pnas.94.16.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundblad EW, Altman S. Inhibition of gene expression by RNase P. New Biotechnol. 2010;27:212–221. doi: 10.1016/j.nbt.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Shen N, et al. Inactivation of expression of several genes in a variety of bacterial species by EGS technology. Proc Natl Acad Sci USA. 2009;106:8163–8168. doi: 10.1073/pnas.0903491106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wesolowski D, et al. Basic peptide-morpholino oligomer conjugate that is very effective in killing bacteria by gene-specific and nonspecific modes. Proc Natl Acad Sci USA. 2011;108:16582–16587. doi: 10.1073/pnas.1112561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dar MA, Sharma A, Mondal N, Dhar SK. Molecular cloning of apicoplast-targeted Plasmodium falciparum DNA gyrase genes: Unique intrinsic ATPase activity and ATP-independent dimerization of PfGyrB subunit. Eukaryot Cell. 2007;6:398–412. doi: 10.1128/EC.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cirak S, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: An open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tilley LD, Mellbye BL, Puckett SE, Iversen PL, Geller BL. Antisense peptide-phosphorodiamidate morpholino oligomer conjugate: Dose-response in mice infected with Escherichia coli. J Antimicrob Chemother. 2007;59:66–73. doi: 10.1093/jac/dkl444. [DOI] [PubMed] [Google Scholar]
- 21.Warren TK, et al. Advanced antisense therapies for postexposure protection against lethal filovirus infections. Nat Med. 2010;16:991–994. doi: 10.1038/nm.2202. [DOI] [PubMed] [Google Scholar]
- 22.Guerrier-Takada C, Li Y, Altman S. Artificial regulation of gene expression in Escherichia coli by RNase P. Proc Natl Acad Sci USA. 1995;92:11115–11119. doi: 10.1073/pnas.92.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko JH, Izadjoo M, Altman S. Inhibition of expression of virulence genes of Yersinia pestis in Escherichia coli by external guide sequences and RNase P. RNA. 2008;14:1656–1662. doi: 10.1261/rna.1120508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu F, Altman S. Inhibition of viral gene expression by the catalytic RNA subunit of RNase P from Escherichia coli. Genes Dev. 1995;9:471–480. doi: 10.1101/gad.9.4.471. [DOI] [PubMed] [Google Scholar]
- 25.McKinney JS, Zhang H, Kubori T, Galán JE, Altman S. Disruption of type III secretion in Salmonella enterica serovar Typhimurium by external guide sequences. Nucleic Acids Res. 2004;32:848–854. doi: 10.1093/nar/gkh219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plehn-Dujowich D, Altman S. Effective inhibition of influenza virus production in cultured cells by external guide sequences and ribonuclease P. Proc Natl Acad Sci USA. 1998;95:7327–7332. doi: 10.1073/pnas.95.13.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trang P, Lee J, Kilani AF, Kim J, Liu F. Effective inhibition of herpes simplex virus 1 gene expression and growth by engineered RNase P ribozyme. Nucleic Acids Res. 2001;29:5071–5078. doi: 10.1093/nar/29.24.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirk K, Saliba KJ. Targeting nutrient uptake mechanisms in Plasmodium. Curr Drug Targets. 2007;8:75–88. doi: 10.2174/138945007779315560. [DOI] [PubMed] [Google Scholar]
- 29.Saliba KJ, Kirk K. Nutrient acquisition by intracellular apicomplexan parasites: Staying in for dinner. Int J Parasitol. 2001;31:1321–1330. doi: 10.1016/s0020-7519(01)00258-2. [DOI] [PubMed] [Google Scholar]
- 30.Kirk K, Martin RE, Bröer S, Howitt SM, Saliba KJ. Plasmodium permeomics: Membrane transport proteins in the malaria parasite. Curr Top Microbiol Immunol. 2005;295:325–356. doi: 10.1007/3-540-29088-5_13. [DOI] [PubMed] [Google Scholar]
- 31.Martin RE, Ginsburg H, Kirk K. Membrane transport proteins of the malaria parasite. Mol Microbiol. 2009;74:519–528. doi: 10.1111/j.1365-2958.2009.06863.x. [DOI] [PubMed] [Google Scholar]
- 32.Martin RE, Henry RI, Abbey JL, Clements JD, Kirk K. The ‘permeome’ of the malaria parasite: An overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 2005;6:R26. doi: 10.1186/gb-2005-6-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirk K. Membrane transport in the malaria-infected erythrocyte. Physiol Rev. 2001;81:495–537. doi: 10.1152/physrev.2001.81.2.495. [DOI] [PubMed] [Google Scholar]
- 34.Anonymous malERA Consultative Group on Drugs A research agenda for malaria eradication: Drugs. PLoS Med. 2011;8:e1000402. doi: 10.1371/journal.pmed.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joet T, Eckstein-Ludwig U, Morin C, Krishna S. Validation of the hexose transporter of Plasmodium falciparum as a novel drug target. Proc Natl Acad Sci USA. 2003;100:7476–7479. doi: 10.1073/pnas.1330865100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joët T, Krishna S. The hexose transporter of Plasmodium falciparum is a worthy drug target. Acta Trop. 2004;89:371–374. doi: 10.1016/j.actatropica.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Slavic K, et al. Life cycle studies of the hexose transporter of Plasmodium species and genetic validation of their essentiality. Mol Microbiol. 2010;75:1402–1413. doi: 10.1111/j.1365-2958.2010.07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Witola WH, et al. Disruption of the Plasmodium falciparum PfPMT gene results in a complete loss of phosphatidylcholine biosynthesis via the serine-decarboxylase-phosphoethanolamine-methyltransferase pathway and severe growth and survival defects. J Biol Chem. 2008;283:27636–27643. doi: 10.1074/jbc.M804360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crowther GJ, et al. Identification of attractive drug targets in neglected-disease pathogens using an in silico approach. PLoS Negl Trop Dis. 2010;4:e804. doi: 10.1371/journal.pntd.0000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 41.Johnson JD, et al. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob Agents Chemother. 2007;51:1926–1933. doi: 10.1128/AAC.01607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol Biochem Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- 43.Ferreira ID, Rosário VE, Cravo PV. Real-time quantitative PCR with SYBR Green I detection for estimating copy numbers of nine drug resistance candidate genes in Plasmodium falciparum. Malar J. 2006;5:1. doi: 10.1186/1475-2875-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.