Abstract
With rising rates of drug-resistant infections, there is a need for diagnostic methods that rapidly can detect the presence of pathogens and reveal their susceptibility to antibiotics. Here we propose an approach to diagnosing the presence and drug-susceptibility of infectious diseases based on direct detection of RNA from clinical samples. We demonstrate that species-specific RNA signatures can be used to identify a broad spectrum of infectious agents, including bacteria, viruses, yeast, and parasites. Moreover, we show that the behavior of a small set of bacterial transcripts after a brief antibiotic pulse can rapidly differentiate drug-susceptible and -resistant organisms and that these measurements can be made directly from clinical materials. Thus, transcriptional signatures could form the basis of a uniform diagnostic platform applicable across a broad range of infectious agents.
Keywords: antibiotic resistance, genomics, tuberculosis
Successful treatment of infectious diseases requires not only the rapid identification of the infecting organism but also prompt determination of which antibiotics will be effective in treating the infection. Delays in detecting drug-resistant infections in turn delay the implementation of effective therapy and are associated with increased mortality (1, 2). Currently, hospitals still rely largely on an assortment of cultures, immunoassays, and molecular techniques (3, 4) for diagnosis of infectious disease; these assays can be slow and require specialized training. A prime example of the shortcomings of current diagnostics is tuberculosis: culture-based methods require a minimum of several days to detect the presence of Mycobacterium tuberculosis, and drug susceptibilities may not be known for weeks to months after the initial visit to the clinic (5).
One approach to increase the speed, precision, and generality of infectious-disease diagnostics is to use DNA-based techniques, which largely involve enzymatic amplification of specific DNA sequences (4, 6–8). To date, such approaches have been deployed only in limited settings, as in identifying specific viral pathogens or fastidious organisms that are challenging to culture, such as the causative agents of gonorrhea and Chlamydia (6, 9). Identification of pathogens using DNA-based approaches requires knowledge only of the pathogen genome sequence; however, DNA-based detection of antibiotic resistance presents a much greater challenge, requiring precise knowledge of the genomic features that confer resistance, thus limiting the method to known resistance genotypes. Examples of these features include resistance-conferring genetic elements [e.g., the mecA gene in methicillin-resistant Staphylococcus aureus (MRSA) (10, 11)] or known resistance-associated polymorphisms [e.g., mutations in rpoB in rifampin-resistant M. tuberculosis (12, 13)]. However, a fundamental limitation is that the known genetic elements or mutations typically constitute only a fraction of the genetic basis for clinically relevant resistance.
Here we explore an alternative approach: using RNA detection to obtain both genotypic and phenotypic information. Like DNA, RNA contains abundant genomic information to allow accurate identification of pathogens. However, unlike DNA, the RNA transcriptome also provides critical dynamic phenotypic information. Brief antibiotic exposure can trigger transcriptional responses in susceptible, but not in resistant, microbes within a few minutes (14), providing a means to couple pathogen identification directly with antibiotic-susceptibility testing.
In this work, we provide a proof-of-principle for RNA-based detection and drug-resistance testing of pathogens. Specifically, we demonstrate that detection of a set of pathogen RNA transcripts can provide ample specificity to diagnose precisely the presence of a broad range of pathogens and, more critically, to distinguish antibiotic-susceptible and -resistant organisms. The natural abundance of RNA transcripts, along with improved RNA detection methods, allows us to measure these expression signatures directly from patient samples without the need for organism isolation or nucleic acid purification and amplification.
In our experiments, we used a simple commercially available method for RNA quantitation (nCounter analysis, NanoString Technologies) that involves minimal sample processing and no enzymatic manipulation (15). RNA in crude culture lysates or patient specimens is hybridized with a pool of fluorescently bar-coded oligonucleotide probes designed to target specific transcripts from a broad collection of organisms of interest. For a given organism, the designed probes target transcripts that identify the organism uniquely at the species level and simultaneously allow its transcriptional response to antibiotic exposure to be measured. Because large numbers of transcripts are investigated in a pool, organism identity and drug sensitivity are determined in a single assay. Although this assay is well suited for these proof-of-principle studies, continued innovation in this and other quantitative nanosensor technologies likely will lead to tools for even more rapid RNA detection.
Results
RNA-Based Pathogen Detection.
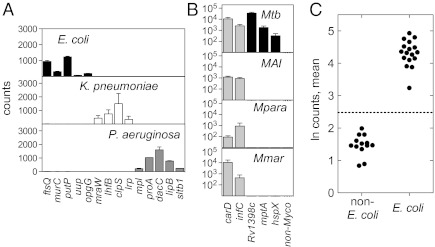
We first sought to identify pathogens through direct detection of RNAs specific to the species. As an initial test, we targeted three Gram-negative pathogens: Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae. To identify organism-specific sequences for probe design, we analyzed publicly available coding sequences from multiple isolates of each of these three pathogens to find genes that are highly conserved within but not between species. We generated a pool of probes targeting these species-specific mRNAs, including four or five probes for each organism to obtain desired levels of specificity. (Probe lists are given in SI Appendix, Table S1.) We were able to detect and distinguish each organism in pure culture (Fig. 1A) and in a complex mixture including eight additional Gram-negative pathogens by directly probing crude lysates (SI Appendix, Fig. S1). Because microbiologic diagnosis of M. tuberculosis is among the slowest diagnosis made in a clinical setting, we then sought to extend our findings to this pathogen. We designed probes to both genes conserved throughout the genus Mycobacterium and genes specific to M. tuberculosis and were able to distinguish M. tuberculosis from other pathogenic mycobacteria (Fig. 1B).
Fig. 1.
Identification of bacteria in culture with direct RNA detection. (A and B) Bacterial cultures were lysed and analyzed with nCounter probes for species-specific transcripts. y axis: transcript raw counts; x axis: gene. Each bar represents the average and SD of two biological replicates. (A) Detection from culture of Gram-negative bacteria. Probes are shown for E. coli (black bars), K. pneumoniae (white bars), and P. aeruginosa (gray bars). (B) Genus- and species-specific detection of mycobacteria in culture: M. tuberculosis (Mtb), M. avium subsp. intracellulare (MAI), M. paratuberculosis (Mpara), and M. marinum (Mmar). Gray bars show genus-wide probes; black bars show M. tuberculosis-specific probes. (C) Synthesis of four E. coli probes into a single organism ID score. The natural logs of the observed counts for each transcript were averaged. Each point represents a single strain. The dashed line indicates three SDs from the mean of the non-E. coli samples (P = 0.0027).
To transform these data into a binary determination of the presence or absence of a pathogen in a given sample, we condensed the counts from multiple transcripts into a single metric that assesses the presence or absence of an organism, using the mean of the natural logarithm of the raw counts from each probe, normalized for hybridization efficiency using internal controls. When applied to a set of 17 clinical E. coli isolates, every isolate was differentiated easily from a set of 13 non-E. coli samples (Z score > 6.5 relative to non-E. coli controls; Fig. 1C). This method also was effective when applied across a wider range of bacterial pathogens, including K. pneumoniae, P. aeruginosa, M. tuberculosis, Proteus mirabilis, Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes, and Stenotrophomonas maltophilia (SI Appendix, Fig. S2 and Table S2). Of note, the specificity of the probes allowed us to distinguish between species within a genus easily (the two streptococcal species were not detected with the other's probes, and Klebsiella oxytoca strains were not detected with K. pneumoniae probes).
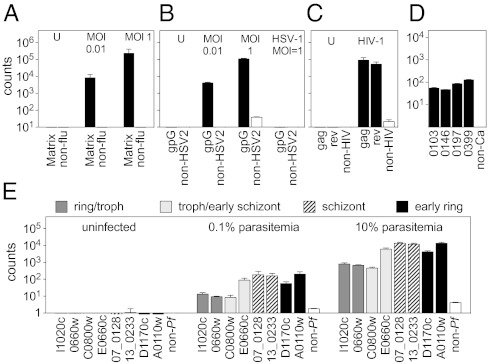
Because RNA is universal in pathogens ranging from bacteria, viruses, and fungi to parasites, RNA detection can provide a common diagnostic platform applicable across a broad range of infectious agents. Using a large pool of mixed pathogen probes, we were able directly and specifically to detect signals to identify influenza virus, herpes simplex virus-2 (HSV-2), and HIV-1 in cell culture in a dose-dependent manner (Fig. 2 A–C); the fungal pathogen Candida albicans (Fig. 2D); and the different stages of the Plasmodium falciparum life cycle in infected erythrocytes (Fig. 2E). As with bacterial detection, viral detection was highly specific; HSV-2 probes did not detect HSV-1 virus even at relatively high multiplicity of infection (MOI) (Fig. 2B). Current clinical laboratory practice uses a range of techniques from standard culture to fluorescent microscopy to diagnose infections with these organisms; however, RNA-based direct detection has the potential to streamline identification of almost all potential infectious agents into a single platform.
Fig. 2.
RNA-based identification of viruses, a fungus, and a parasite by direct analysis of lysates using nCounter probes. Shaded bars indicate organism-specific probes, white bars indicate average of non-organism probes. (A) 293T cells infected with influenza A at the indicated MOIs and harvested after 8 h. U, uninfected; non-flu, average of 42 non-influenza probes. Error bars in A indicate the SD of two biological replicates. (B) HeLa cells infected with HSV-1 or HSV-2 at the indicated MOIs and harvested after 24 h. U, uninfected; non-HSV2, average of 44 non-HSV-2 probes. (C) Human peripheral blood mononuclear cells infected with HIV-1 and harvested after 36 h. U, uninfected; non-HSV2, average of 41 non-HIV probes. (D) Candida albicans grown to log phase lysed. Non-Ca, average of 80 non-Candida albicans probes. Error bars in B–D indicate SD of two technical replicates from one of two representative independent experiments. (E) Primary red blood cells infected with P. falciparum and harvested at the indicated levels of parasitemia. Non-Pf, average of 32 non-Plasmodium falciparum probes. Error bars in E indicate the SD of two biological replicates.
RNA-Based Determination of Drug Susceptibility.
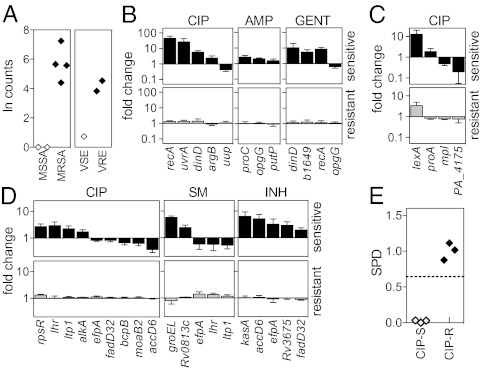
In the clinical setting, determining appropriate therapy requires knowing not only which pathogens are present but also their antibiotic susceptibilities. We thus sought to demonstrate that RNA detection can be used to determine antibiotic resistance rapidly. In the case of mobile genetic elements that encode drug-resistance genes, detection of transcripts from known elements would enable us to determine resistance genotypically, in a manner analogous to PCR-based approaches that detect resistance-encoding genes in some bacteria (16–18). To demonstrate that an RNA-based approach is able to detect such resistance genes, we probed S. aureus isolates for mecA mRNA, which confers resistance to methicillin, and Enterococcus isolates for vanA mRNA, which confers resistance to vancomycin. In both cases, we could detect the relevant transcript (Fig. 3A), thus allowing rapid identification of MRSA and vancomycin-resistant Enterococcus (VRE), with the potential to match PCR-based methods while bypassing the need for nucleic acid isolation, purification, and amplification. In principle, this method can be extended to other genetic determinants of pathogenicity, such as virulence factors acquired through horizontal genetic exchange in food-borne pathogens [e.g., Shiga toxin in enterohemorrhagic or Shiga toxigenic E. coli (19)].
Fig. 3.
RNA signature-based determination of antimicrobial susceptibility. (A) Detection of resistance genes on mobile genetic elements. For MSSA and MRSA: detection of mecA transcript upon cloxacillin exposure. For vancomycin-sensitive Enterococcus (VSE) and VRE: detection of vanA upon vancomycin exposure. Each point represents a different clinical isolate. (B–D) Transcriptional responses to drug exposure. Log-phase cultures were exposed to antibiotic, lysed, and analyzed using nCounter probe sets. Counts were normalized to the mean of the middle 50% of all counts for a sample, and induction was determined by comparing drug-exposed and unexposed samples. Genes contributing to the signature are on the x axis. Shown are the responses of a panel of susceptible (black bars, Upper) or resistant (gray bars; Lower) strains of E. coli to ciprofloxacin (CIP), ampicillin (AMP), or gentamicin (GM) (B); of P. aeruginosa to ciprofloxacin (C); and of M. tuberculosis to isoniazid (INH), streptomycin (SM), or ciprofloxacin (D). Error bars represent SD of three to seven isolates tested. See SI Appendix, Table S3 for a full list of strains tested. (E) SPD of E. coli laboratory and clinical isolates exposed to ciprofloxacin. Transcriptional responses of each tested isolate were condensed into a single metric, SPD. The dashed line indicates three SDs from the mean of the resistant samples. Each point represents one strain performed in two to four biological replicates.
We then sought to extend RNA-based determination of antibiotic resistance beyond the detection of specific known genotypes. Antibiotic exposure can trigger a stereotypical transcriptional response in susceptible microbes as rapidly as within a few minutes (14). For example, when treated with ciprofloxacin, P. aeruginosa up-regulates both recA and lexA, part of the canonical SOS response (20). These transcriptional programs represent one of the earliest detectable cellular changes that distinguish susceptible and resistant organisms. For many organism/antibiotic pairings, these responses have been well characterized using microarrays. We hypothesized that sensitive organisms exposed to drug would display a stereotypical transcriptional signature, but resistant organisms, regardless of the mechanism of resistance, would show no response, thus providing a basis for rapidly distinguishing antibiotic-susceptible from antibiotic -resistant strains based on phenotype.
Using published data from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) or ArrayExpress (http://www.ebi.ac.uk/arrayexpress/), we identified genes that are regulated differentially upon exposure to various antimicrobial agents and thus can be used to indicate the presence or absence of a response. We generated a pooled antibiotic-sensitivity probe set to detect uniquely these genes for several pathogens of interest (14, 20, 21). Following a 10-min exposure of E. coli K-12 to ciprofloxacin, we observed changes in transcript levels of a subset of genes that together define a ciprofloxacin-susceptibility expression signature (Fig. 3B; probes are listed in SI Appendix, Table S7). A similar signature was detected in an additional laboratory isolate and in five clinical isolates tested (SI Appendix, Table S3). In contrast, this signature was not elicited in the seven resistant clinical strains tested (Fig. 3B and SI Appendix, Table S3). Similarly, on exposure of antibiotic-susceptible E. coli strains to gentamicin or ampicillin or on exposure of antibiotic-susceptible P. aeruginosa strains to ciprofloxacin, we observed stereotypic transcriptional responses specific to each antibiotic/microbe combination (Fig. 3 B and C and SI Appendix, Table S3) that were not elicited in resistant strains.
Some genes (e.g., recA, involved in the SOS response) are up-regulated in several species in response to multiple antibiotics, whereas other transcriptional changes are organism and/or mechanism specific. For example, exposure of E. coli to the β-lactam ampicillin [which blocks cell-wall synthesis and also hinders proline transport (22)] induces opgG, involved in the osmoregulated synthesis of periplasmic glucan (23), and proC, which encodes a proline synthetic enzyme. In contrast, the transcript levels of the majority of genes remain unchanged in response to antibiotic exposure, underscoring the sufficiency of following only a small subset of genes, i.e., a signature, rather than the entire genome. It should be noted that genes selected in this work were chosen based on a limited set of previously published data, and additional genomewide monitoring of a broader range of drug-susceptible isolates might improve these signatures and thus strengthen this approach.
Rapid phenotypic testing for antibiotic susceptibility would have a particularly profound impact in tuberculosis, because established methods for phenotypic testing take weeks to months. During this time, patients with unsuspected resistance who are treated with standard therapy remain inappropriately treated, potentially resulting in the development of further drug resistance and worse outcomes. Moreover, until these patients receive adequate therapy, they remain infectious and can spread drug-resistant disease (24). We therefore defined expression signatures in response to the anti-tubercular agents isoniazid, ciprofloxacin, and streptomycin (25) and showed that susceptible and resistant laboratory and clinical isolates could be distinguished after 3–6 h of exposure to the antibiotic (Fig. 3D). Some genes in the transcriptional profiles are mechanism specific (e.g., alkA and lhr for ciprofloxacin; groEL for streptomycin; and kasA and accD6 for isoniazid) (25, 26). Other genes, particularly those involved in mycolic acid synthesis or intermediary metabolism, are down-regulated in response to multiple antibiotics, indicating a shift away from growth toward damage control.
As with organism identification, we transformed the transcriptional signatures elicited by drug exposure into a binary outcome indicative of sensitivity or resistance. To condense the transcriptional responses into a single, quantitative metric, we calculated the squared projected distance (SPD) of the expression response from each experimental sample along a vector from the centroid of control, antibiotic-susceptible samples to the centroid of resistant controls (a detailed description of this metric is provided in the SI Appendix). Strains susceptible to a given antibiotic show closely clustered transcriptional responses, resulting in small SPDs (SI Appendix, Table S3). Conversely, antibiotic-resistant strains have larger SPD values, the result of the signature genes failing to respond to antibiotic. Using this metric, we can clearly separate sensitive and resistant laboratory and clinical isolates of E. coli (Fig. 3E and SI Appendix, Fig. S3), P. aeruginosa (SI Appendix, Fig. S4), and M. tuberculosis (Fig. 4B and SI Appendix, Fig. S5).
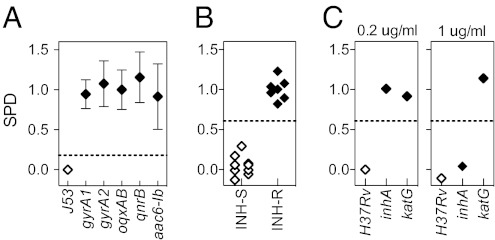
Fig. 4.
Determination of antibiotic susceptibility is independent of the mechanism of resistance. (A) Ciprofloxacin-sensitive E. coli J53 and derivatives containing either a chromosomal mutation in gyrA or plasmid-mediated quinolone resistance determinants [aac(6′)-Ib, qnrB, or oqxAB] were exposed to ciprofloxacin. Error bars indicate mean and SD of two biological duplicates from one of two independent experiments. (B) M. tuberculosis laboratory and clinical isolates exposed to 1 μg/mL isoniazid. INH-R, isoniazid-resistant; INH-S, isoniazid-sensitive.(C) Isoniazid-sensitive and high- or low-level resistant M. tuberculosis strains were exposed to isoniazid at 1 μg/mL or 0.2 μg/mL. Dashed lines indicate three SDs from the mean of the resistant samples.
Because transcription represents a phenotypic response to antibiotic exposure, we hypothesized that the sensitive signature should not be elicited in resistant strains regardless of the mechanism of resistance. To evaluate this principle, we engineered a set of isogenic strains with distinct, well-characterized resistance mechanisms and compared their transcriptional responses to antibiotic exposure. We tested five ciprofloxacin-resistant mutants of E. coli strain J53, each engineered to carry a different mechanism of resistance: two with single mutations in the fluoroquinolone target gene topoisomerase gyrA (G81D or S83L) and three carrying plasmid-mediated quinolone-resistance genes with distinct mechanisms of action (27) including aac(6′)-Ib, which encodes an acetylating, inactivating enzyme; qnrB, which encodes a protein that blocks the active site of GyrA; and oqxAB, which encodes an efflux pump (Fig. 4A). The susceptible parent strain, J53, demonstrated the expected signature upon ciprofloxacin exposure and a small SPD similar to that of other drug-sensitive isolates (SI Appendix, Table S3). However, none of the ciprofloxacin-resistant mutants showed the sensitivity signature upon ciprofloxacin exposure, and all had large SPDs, thus confirming that an expression signature can distinguish resistant from sensitive strains, independent of the mechanism of resistance.
To demonstrate the applicability of this principle beyond E. coli, we tested M. tuberculosis isolates with known distinct mechanisms of resistance to the first-line drug isoniazid. The first resistant strain has a mutation in katG, which encodes a catalase necessary for prodrug activation (28); the second has a mutation in the promoter of inhA, which encodes the target of isoniazid (29). Because of these distinct mechanisms, these two strains have differing levels of resistance to isoniazid: The katG mutant shows high-level resistance [minimum inhibitory concentration (MIC) >6.4 μg/mL], whereas the inhA promoter mutant has lower-level resistance (MIC = 0.4 μg/mL). Exposure of the sensitive laboratory strain H37Rv to either low (0.2 μg/mL) or high (1 μg/mL) concentrations of isoniazid elicited the expected transcriptional signature. Exposure to the lower isoniazid concentration failed to elicit a transcriptional response in either resistant strain, but at the higher isoniazid concentration, the inhA mutant responded in a susceptible manner, in contrast to the katG mutant (Fig. 4C). This result confirms that the loss of the transcriptional signature is independent of mechanism. Moreover, it suggests that the concentrations required to elicit responses can be used to differentiate high- and low-level resistance.
Application of an RNA-Based Diagnostic to Simulated and Real Patient Specimens.
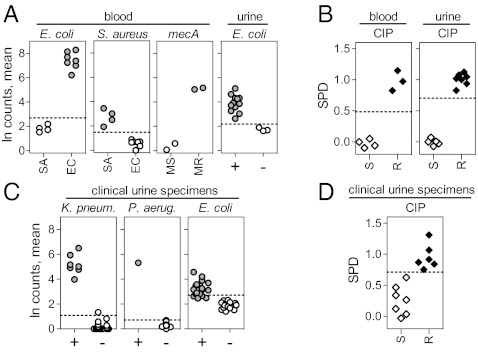
To show that an RNA-based diagnostic platform can be used to probe clinical specimens directly, we first sought to identify pathogens and their drug susceptibilities in blood samples spiked with bacteria. We introduced MRSA or methicillin-sensitive S. aureus (MSSA) or ciprofloxacin-sensitive (CIPS) or -resistant (CIPR) E. coli into human blood samples and then probed for pathogen RNA. From these samples, we were able to identify the spiked organism directly (Fig. 5A) and simultaneously to detect both genotypic and phenotypic antibiotic resistance. Using probes to mecA transcript, we were able to distinguish MRSA from MSSA easily (Fig. 5A), and, using our defined expression signatures of antibiotic exposure, we were able to distinguish CIPS from CIPR E. coli (Fig. 5B). We were similarly successful using urine into which we had introduced E. coli. We grew CIPS and CIPR isolates of E. coli in clean-caught urine from healthy donors and probed directly for and detected species-specific transcripts (Fig. 5A) as well as drug-sensitive transcriptional responses upon CIP exposure (Fig. 5B).
Fig. 5.
Identification of bacteria and determination of antibiotic susceptibility directly from spiked samples and clinical urine specimens. (A) Detection of species-specific transcripts in blood or urine spiked with S. aureus (SA) or E. coli (EC) and discrimination of MSSA (MS) from MRSA (MR) by direct detection of mecA transcripts in spiked blood. The natural logs of the observed counts for each of four species-specific transcripts were averaged to generate the organism ID score. +, spiked urine; −, healthy control urine. Each point represents a different isolate. (B) Discrimination of ciprofloxacin-sensitive (S) from ciprofloxacin-resistant (R) E. coli in spiked blood or urine using expression signatures (expressed as SPD). (C) Detection of species-specific transcripts in clinical urine specimens. + indicates the presence (>105/mL); – indicates the absence (<105/mL) of the indicated species. Each point represents a different urine specimen. (D) Discrimination of CIPS from CIPR E. coli in validated E. coli-positive (>105/mL) clinical urine specimens using expression signatures (expressed as SPD). Dashed lines in A and C indicate three SDs from mean of control, nonorganism samples. Dashed lines in B and D indicate three SDs from the mean of resistant samples.
Finally, we applied this method directly to clinical specimens by testing 34 urine specimens collected from patients suspected of having urinary tract infections based on a positive urine analysis. A small aliquot of urine was pulsed with ciprofloxacin for 30 min, lysed, and assayed in parallel with an unexposed aliquot, using a single probe set that included both identification and drug-susceptibility probes. For comparison, standard culture-based diagnostic tools were used for organism identification and drug-susceptibility testing [growth on selective/differential media, testing of isolates using API 20E strips (bioMerieux, Inc.), and microtiter plate-based MIC determinations; see SI Appendix, Tables S8 and S9]. We were able to identify K. pneumoniae and P. aeruginosa in all specimens infected with these organisms at levels greater than the standard clinical cutoff (>105 organisms per milliliter of urine); these scores for these culture-positive specimens differed from the scores of the culture-negative specimens by >12 SDs (Fig. 5C). Results for E. coli were less clear because background counts for the E. coli probes generally were higher than for the P. aeruginosa and K. pneumoniae probes. This background also was observed with nonclinical materials and blank controls across multiple probe lots, suggesting that the background signal is inherent to the NanoString assay and could represent low levels of nucleic acid contamination in reagents. Nevertheless, all 17 E. coli-positive specimens scored higher than the 17 E. coli-negative specimens, with 13 of the E. coli-positive specimens scoring more than three SDs higher than the mean of the negative specimens. Finally, using the data from the spiked urine samples (Fig. 5B) as a training data set to define the ciprofloxacin sensitivity signature in urine, we evaluated the 13 unknown E. coli-infected urine specimens for ciprofloxacin sensitivity (Fig. 5D and SI Appendix, Table S9). Samples were scored as sensitive if the SPD score fell more than three SDs from the mean SPD of the resistant samples used in the training set. Using this cutoff, all 13 samples were characterized correctly (100% accuracy, P = 0.00037).
Discussion
The current wealth of genomic and transcriptomic data available should promote the development of substantially improved diagnostics for infectious diseases. Our work suggests that an RNA-based diagnostic can capitalize on this information by taking advantage of both static genome sequences and dynamic transcriptomic responses. Although additional testing will be required to determine the full applicability of the approach, direct RNA detection has the potential to identify most classes of pathogens and to allow simultaneous phenotypic determination of antibiotic susceptibility within a single assay. An RNA-centered diagnostic has two important advantages. First, the natural abundance of single-stranded mRNA allows its direct detection via hybridization, without purification or amplification. First, the natural abundance of single-stranded mRNA allows its direct detection via hybridization, without purification, thus eliminating the challenges of enzymatic amplification. More critically, antibiotic-elicited RNA signatures are a rapid indicator of antibiotic susceptibility; because these signatures are among the first cellular changes that occur upon drug exposure, they should allow detection of resistance long before the traditional phenotype of growth inhibition can be observed. Moreover, we have demonstrated that transcriptional profiles reflect physiologic phenotypes regardless of the underlying genotypic basis for resistance. RNA detection thus has the potential to accelerate, unify, and simplify diagnosis, particularly with regards to determining antibiotic susceptibilities. Recently, RNA signatures have been recognized as an important tool in guiding clinical practice in cancer (30). Our work extends this application by measuring dynamic changes in RNA signatures upon antibiotic exposure rather than static baseline transcriptional profiles.
Although this paper provides a proof of principle, considerable additional work will be required to yield a clinic-ready, RNA-based diagnostic for infectious diseases. First, probe-set design will benefit from the availability of more sequenced genomes and additional transcriptomic data. Second, it will be necessary to develop a fully automated, integrated system for sample processing and detection, which will drive the speed, sensitivity, and detection limit of the system [the current assay detects 40–90 bacteria grown in culture; see SI Appendix, Fig. S6]. Engineering also can facilitate the development of systems suited to resource-poor settings. Third, it will be necessary to benchmark the RNA signatures with conventional assays for organism identification and drug-susceptibility determination and to run trials to determine statistically meaningful sensitivities and specificities for the method. As an important step, the work described here demonstrates the feasibility of this approach and raises the possibility that an RNA-based diagnostic could meet the critical need for more rapid diagnosis of infectious disease.
Materials and Methods
For expanded details of methods, see SI Appendix.
Selection of Organism-Identification Probes.
To select nCounter probes for differential detection of Gram-negative organisms, we compared all publically available sequenced genomes for relevant organisms (SI Appendix, Table S10). We identified genes conserved within each species by selecting coding sequences (CDS) having at least 50% identity over at least 70% of the CDS length for all sequenced genomes for pathogenic strains of that species available at the time the probe sets were generated. We broke the CDS into overlapping 50-mers and retained only those 50-mers with 100% identity within a species and having no greater than 50% identity to a CDS in any other species in the study. Available published expression data in Gene Expression Omnibus were reviewed, and genes with good expression under most conditions were selected. Full details of this process are described in SI Appendix.
Bacterial and Fungal Culture.
All Gram-negative bacterial clinical isolates were obtained from the Brigham and Women's Hospital microbiology laboratory, and clinical M. tuberculosis isolates were obtained from the Massachusetts Supranational Tuberculosis Reference Laboratory. Bacterial or yeast cultures were grown to an OD600 of ∼1 in LB medium. For mixing experiments, equal numbers of bacteria as determined by OD600 were combined before lysis for nCounter analysis. For spiked urine experiments, E. coli was grown overnight at 37 °C in sterile urine, diluted 1:20, and grown to OD600 of 0.1–0.3 in fresh urine before harvest or antibiotic exposure, as described below. Mycobacterium isolates were grown in Middlebrook 7H9 medium (BD Diagnostics) supplemented with OADC (BD Diagnostics) and 0.2% glycerol.
Viral and Plasmodium Infections.
HeLa cells (2 × 106), 293T cells (2 × 105), and human peripheral blood monocytes (5 × 105) were infected with HSV-1 strain KOS and HSV-2 strain 186 Syn+, influenza A PR8, or HIV-1 NL-ADA, respectively, at the noted MOIs. Primary red blood cells (5 × 109) were infected with P. falciparum strain 3D7 until they reached the noted levels of parasitemia. At the indicated times, the cells were washed once with PBS and harvested as described below.
Antibiotic Exposure.
Cultures of E. coli or P. aeruginosa were grown to an OD600 of ∼1 in LB. Cultures then were divided into two samples, one of which was treated with antibiotic (E. coli: ciprofloxacin 4–8 μg/mL or 300 ng/mL or ampicillin 500 μg/mL for 10 min, gentamicin 64 μg/mL for 30 min; P. aeruginosa: ciprofloxacin 16 μg/mL for 30 min). Cultures of S. aureus or Enterococcus were grown to an OD600 of ∼1 in LB. Cultures then were exposed to cloxacillin (25 μg/mL) or vancomycin (128 μg/mL), respectively, for 30 min. Cultures of M. tuberculosis were grown to midlog phase and then were normalized to OD600 of 0.2. Two milliliters of each culture were treated with no antibiotic or with one of the following (final concentration): isoniazid 0.2–1.0 μg/mL for 3 h; streptomycin 5 μg/mL for 6 h, or ciprofloxacin 5 μg/mL for 3 h. Clinical samples, including spiked blood, spiked urine, and discarded patient urines, were exposed to 4 μg/mL ciprofloxacin for 30 min. All bacterial cultures and clinical materials were maintained at 37 °C during drug exposure.
Sample Processing.
Gram-negative cultures were lysed directly in RLT buffer (Qiagen). In addition, Gram-positive cultures were disrupted mechanically with bead beating. For spiked blood samples, 1 × 107 bacteria were added per milliliter of blood. Samples then were added to PAXgene blood RNA tubes (PreAnalytix GmbH) and were processed according to the manufacturer's protocol through the first centrifugation. Then supernatant was aspirated, pellets were resuspended in RLT buffer, and lysates were used directly in hybridizations. For spiked urine and clinical specimens, urine was added directly to RLT buffer. Mycobacterial cultures were centrifuged and then were resuspended in TRIzol (Gibco) with or without mechanical disruption by bead beating, and the initial aqueous phase was collected for analysis. Viral and parasite RNA were prepared similarly using TRIzol and chloroform. For all lysates, 3–5 μL were used directly in hybridizations according to standard nCounter protocols. For organism identification, raw scores were normalized to internal hybridization controls. To determine antibiotic susceptibility, raw counts were normalized to the mean of the middle 50% of all probes for a sample, and fold induction for each gene was determined by comparing antibiotic-treated with untreated samples.
Processing of Clinical Urine Specimens.
Discarded, de-identified urine specimens positive for nitrites and leukocytes were obtained from the Brigham and Women's Hospital clinical laboratory. The time between collection and processing ranged from 3–48 h. Laboratory sample processing was minimal, consisting of a 30-min exposure of a collected urine specimen to antibiotic followed by rapid lysis in RLT buffer. nCounter transcript detection was performed according to the manufacturer's recommendation for analysis of mammalian genomes (18-h hybridization followed by a 5-h analysis on an automated instrument). Microbial identification and antibiotic susceptibility were determined independently using standard laboratory protocols. For full details see SI Appendix.
Supplementary Material
Acknowledgments
We thank Nathan Hasely for helpful discussions regarding analysis of the data, Marci DeGrace and Nir Hacohen (Massachusetts General Hospital) for the gift of the influenza samples, and Monica Diez and Subra Suresh (Massachusetts Institute of Technology) for the gift of the Plasmodium falciparum samples. This work was funded by National Institutes of Health (NIH) Grants U54 AI057159 (to D.T.H.), and AI 063106 (to D.M.K.). A.K.B. was supported by a Parker B. Francis Foundation Fellowship and NIH Grant K08 AI080944. S.A.S. was supported by a Helen Hay Whitney Fellowship. K.F.B. was supported by National Research Service Award Fellowship AI 081477.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119540109/-/DCSupplemental.
References
- 1.Park MM, Davis AL, Schluger NW, Cohen H, Rom WN. Outcome of MDR-TB patients, 1983-1993. Prolonged survival with appropriate therapy. Am J Respir Crit Care Med. 1996;153:317–324. doi: 10.1164/ajrccm.153.1.8542137. [DOI] [PubMed] [Google Scholar]
- 2.Paul M, et al. Systematic review and meta-analysis of the efficacy of appropriate empiric antibiotic therapy for sepsis. Antimicrob Agents Chemother. 2010;54:4851–4863. doi: 10.1128/AAC.00627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ince J, McNally A. Development of rapid, automated diagnostics for infectious disease: Advances and challenges. Expert Rev Med Devices. 2009;6:641–651. doi: 10.1586/erd.09.46. [DOI] [PubMed] [Google Scholar]
- 4.Weile J, Knabbe C. Current applications and future trends of molecular diagnostics in clinical bacteriology. Anal Bioanal Chem. 2009;394:731–742. doi: 10.1007/s00216-009-2779-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham J, Perkins M. Diagnostics for Tuberculosis: Global Demand and Market Potential. Geneva: WHO; 2006. [Google Scholar]
- 6.Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. 2004;4:337–348. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Versalovic J, Lupski JR. Molecular detection and genotyping of pathogens: More accurate and rapid answers. Trends Microbiol. 2002;10(10, Suppl):S15–S21. doi: 10.1016/s0966-842x(02)02438-1. [DOI] [PubMed] [Google Scholar]
- 8.Muldrew KL. Molecular diagnostics of infectious diseases. Curr Opin Pediatr. 2009;21:102–111. doi: 10.1097/MOP.0b013e328320d87e. [DOI] [PubMed] [Google Scholar]
- 9.Mahony JB. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21:716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stamper PD, Cai M, Howard T, Speser S, Carroll KC. Clinical validation of the molecular BD GeneOhm StaphSR assay for direct detection of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in positive blood cultures. J Clin Microbiol. 2007;45:2191–2196. doi: 10.1128/JCM.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll KC. Rapid diagnostics for methicillin-resistant Staphylococcus aureus: Current status. Mol Diagn Ther. 2008;12:15–24. doi: 10.1007/BF03256265. [DOI] [PubMed] [Google Scholar]
- 12.Boehme CC, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palomino JC. Molecular detection, identification and drug resistance detection in Mycobacterium tuberculosis. FEMS Immunol Med Microbiol. 2009;56:103–111. doi: 10.1111/j.1574-695X.2009.00555.x. [DOI] [PubMed] [Google Scholar]
- 14.Sangurdekar DP, Srienc F, Khodursky AB. A classification based framework for quantitative description of large-scale microarray data. Genome Biol. 2006;7:R32. doi: 10.1186/gb-2006-7-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 16.Wolk DM, et al. Multicenter evaluation of the Cepheid Xpert methicillin-resistant Staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J Clin Microbiol. 2009;47:758–764. doi: 10.1128/JCM.01714-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farley JE, et al. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay to culture by use of BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures from an at-risk community population. J Clin Microbiol. 2008;46:743–746. doi: 10.1128/JCM.02071-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamper PD, Cai M, Lema C, Eskey K, Carroll KC. Comparison of the BD GeneOhm VanR assay to culture for identification of vancomycin-resistant enterococci in rectal and stool specimens. J Clin Microbiol. 2007;45:3360–3365. doi: 10.1128/JCM.01458-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kehl KS, Havens P, Behnke CE, Acheson DW. Evaluation of the premier EHEC assay for detection of Shiga toxin-producing Escherichia coli. J Clin Microbiol. 1997;35:2051–2054. doi: 10.1128/jcm.35.8.2051-2054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brazas MD, Hancock REW. Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2005;49:3222–3227. doi: 10.1128/AAC.49.8.3222-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson GG, Moreau-Marquis S, Stanton BA, O'Toole GA. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun. 2008;76:1423–1433. doi: 10.1128/IAI.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson SV, Berg CM. Inhibition of amino acid transport in Escherichia coli by some beta-lactam antibiotics. Antimicrob Agents Chemother. 1977;11:968–973. doi: 10.1128/aac.11.6.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanoulle X, et al. Structural analysis of Escherichia coli OpgG, a protein required for the biosynthesis of osmoregulated periplasmic glucans. J Mol Biol. 2004;342:195–205. doi: 10.1016/j.jmb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Minion J, Leung E, Menzies D, Pai M. Microscopic-observation drug susceptibility and thin layer agar assays for the detection of drug resistant tuberculosis: A systematic review and meta-analysis. Lancet Infect Dis. 2010;10:688–698. doi: 10.1016/S1473-3099(10)70165-1. [DOI] [PubMed] [Google Scholar]
- 25.Boshoff HIM, et al. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: Novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 26.Fu LM, Shinnick TM. Understanding the action of INH on a highly INH-resistant Mycobacterium tuberculosis strain using Genechips. Tuberculosis (Edinb) 2007;87:63–70. doi: 10.1016/j.tube.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz J. Mechanisms of resistance to quinolones: Target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee A, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 30.Zoon CK, et al. Current molecular diagnostics of breast cancer and the potential incorporation of microRNA. Expert Rev Mol Diagn. 2009;9:455–467. doi: 10.1586/erm.09.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.