Abstract
Forkhead box class O 3a (FOXO3a) is a transcription factor and tumor suppressor linked to longevity that determines cell fate through activating transcription of cell differentiation, survival, and apoptotic genes. Recruitment of the coactivator CBP/p300 is a crucial step in transcription, and we revealed that in addition to conserved region 3 (CR3) of FOXO3a, the C-terminal segment of CR2 (CR2C) binds CBP/p300 and contributes to transcriptional activity. CR2C and CR3 of FOXO3a interact with the KIX domain of CBP/p300 at both “MLL” and “c-Myb” binding sites simultaneously. A FOXO3a CR2C-CR3 peptide in complex with KIX exists in equilibrium between two equally populated conformational states, one of which has CR2C bound to the MLL site and CR3 bound to the c-Myb site, whereas in the other, CR2C and CR3 bind the c-Myb and MLL sites, respectively. This promiscuous interaction between FOXO3a and CBP/p300 is further supported by additional binding sites on CBP/p300, namely, the TAZ1 and TAZ2 domains. In functional studies, our structure-guided mutagenesis showed that both CR2C and CR3 are involved in the activation of certain endogenous FOXO3a target genes. Further, phosphorylation of S626, a known AMP-dependent protein kinase target in CR3, increased affinity for CBP/p300 and the phosphomimetic mutation enhanced transactivation of luciferase. These findings underscore the significance of promiscuous multivalent interactions and posttranslational modification in the recruitment of transcriptional coactivators, which may allow transcription factors to adapt to various gene-specific genomic and chromatin structures and respond to cell signals.
Keywords: NMR solution structure, intrinsic disorder, promiscuous binding
The forkhead box class O (FOXO) protein subfamily contains four members: FOXO1, FOXO3a, FOXO4, and FOXO6 (1, 2). FOXO3a is ubiquitously expressed in all tissues (2) and activates the transcription of many genes that regulate differentiation, cell cycle, DNA repair, stress resistance, metabolism, and apoptosis (1). FOXO3a is a 71-kDa intrinsically disordered protein (IDP) that contains only one structured domain, the 100-residue forkhead (FH) domain (3), which binds a consensus DNA sequence called forkhead response element (FRE). There are three additional conserved regions (CR1-CR3) across this subfamily (Fig. 1A), and it has been reported that CR3 is the transactivation domain (TAD), mediating the association with the coactivator CBP/p300 through binding to its KIX domain (4, 5). We previously discovered an intramolecular interaction between the FH and CR3 domains (3), and demonstrated that FH binding to FRE releases CR3, allowing it to bind KIX (6). The KIX domain has two distinct binding grooves that can simultaneously engage the TADs of c-Myb and MLL, thus designated the c-Myb and MLL sites (7). MLL and c-Myb bind KIX in a cooperative manner through their “ΦXXΦΦ” motifs, where “Φ” is a hydrophobic residue, and “X” is an arbitrary residue (7). A number of other transcription factors bearing this motif (e.g., CREB, c-Jun, p53) have also been found to interact with KIX (7–9) (Fig. 1B).
Fig. 1.
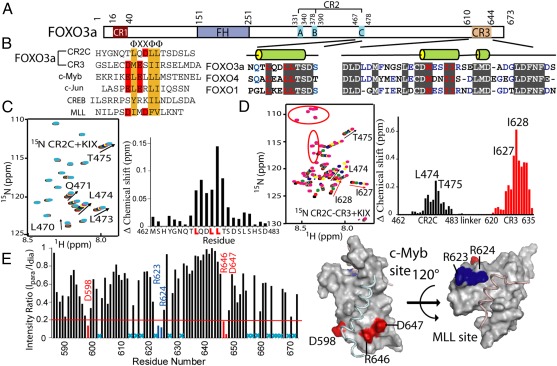
Characterization of interactions between FOXO3a and the CBP KIX domain. (A) Domain architecture of FOXO3a, indicating the FH domain and three conserved regions (CR1–3). Sequence alignments of CR2C and CR3 from FOXO proteins are shown with the predicted secondary structure. (B) Sequence alignment of KIX-interacting peptides. The hydrophobic residues of the “ΦXXΦΦ” motif are colored orange, and negatively charged residues are colored red. (C) NMR titration of 15N-labeled CR2C (0.2 mM) with KIX. (Left) HSQC of 15N-labeled CR2C (black) overlaid with spectra collected during titration with KIX: 0.1 (red), 0.35 (green), 0.85 (blue), 1.35 (yellow), 1.85 (pink), and 4.75-fold (cyan). Perturbed resonances are labeled. (Right) Normalized chemical shift change of each CR2C residue on addition of 4.75-fold excess KIX. The “LXXLL” motif is highlighted. (D) NMR titration of 15N-labeled CR2C-CR3 (0.16 mM) with KIX. (Left) HSQC of 15N-labeled CR2C-CR3 (black) overlaid with spectra collected during titration with KIX: 0.25 (red), 0.75 (green), 1.75 (blue), 3.25 (yellow), and 4.75-fold (pink). Perturbed resonances are labeled. Red circles indicate Gly and Ser residues in the engineered linker. (Right) Normalized chemical shift change of each CR2C-CR3 residue on addition of 4.75-fold excess KIX. CR2C and CR3 residues are plotted in black and red, respectively, and the most perturbed residues are labeled. (E) Probing CR2C-CR3 binding to KIX by PRE. (Left) Peak intensity ratio (Ipara /Idia) of each KIX residue in the presence of CR2C-CR3 peptide tagged with Mn2+ (para) vs. Ca2+ (dia). Cyan X's indicate residues for which this ratio cannot be determined because of overlapped peaks. Residues exhibiting large broadening (ratio <0.2, red line) are labeled. (Right) Broadened residues mapped on the ternary KIX–c-Myb–MLL complex (PDB ID code 2AGH). The c-Myb and MLL sites are indicated, and broadened residues in each site are colored red and blue, respectively. All structure figures were prepared using PyMol (The PyMOL Molecular Graphics System, Schrödinger, LLC).
FOXOs recruit CBP/p300 to the FRE, which, in turn, recruits general transcriptional machinery and also remodels chromatin through histone acetyltransferase activity (10). However, CBP/p300 also acetylates the FH domain, which impairs DNA binding and attenuates transcription (11). The CR2, which consists of three separate regions (A, B, and C) (Fig. 1A), may also play a role in the transactivation activity of FOXOs. Chromosomal translocations create MLL-FOXO fusion proteins composed of the DNA binding region of MLL and the C terminus of either FOXO3a (T228-G673) or FOXO4. In addition to CR3, immortalization of myeloid progenitors by this fusion requires the C-terminal segment of CR2 (CR2C) (5, 12), which contains a ΦXXΦΦ motif (Fig. 1B). Whether CR2C is involved in transactivation by native FOXOs is unknown; however, it has been shown that CR2C binds Sirt1 (13). The histone deacetylase activity of Sirt1 results in transcription silencing (14), but Sirt1 also deacetylates the FH domain, restoring transcriptional activity of FOXO3a; thus, the net effect of Sirt1 on transcription is cell type- and gene-dependent (15). As such, the role of CR2C is not fully understood. Moreover, the transactivation activity of FOXO3a is regulated by AMP-dependent protein kinase (AMPK) phosphorylation (16). Among six AMPK phosphorylation sites on FOXO3a, five are located in the C-terminal intrinsically disordered region (IDR), including S626 in CR3; however, the mechanism of phosphorylation-regulated transcription is elusive.
In this study, we found that in addition to CR3, CR2C interacts with the KIX domain and both regions bind KIX simultaneously. We determined the solution structures of KIX in complex with a fusion peptide of CR2C and CR3, which binds to the MLL and c-Myb sites in equilibrium with two distinct orientations. The promiscuity of these TADs is further evidenced by our discovery that they also bind the TAZ1 and TAZ2 domains of CBP/p300. The transactivation function of CR2C and CR3 was characterized by luciferase assays and examined in the context of full-length FOXO3a by analysis of target gene expression. Our studies provide insight into the mechanism by which CBP/p300 is recruited by FOXO3a through multiple promiscuous and dynamic interactions between two TADs and KIX, TAZ1 and TAZ2, as well as how AMPK phosphorylation enhances this process.
Results
CR2C of FOXO3a Contributes to KIX Domain Binding.
Characterization of oncogenic MLL-FOXO fusion proteins has implicated CR2C in transcription activation; however, little is known about the underlying mechanism or how CR2C and CR3 may cooperate in transactivation (5, 12). These regions are predicted to be intrinsically disordered, consistent with CD spectra of isolated CR2C (M462-D483) and CR3 (L620-A635) peptides (Fig. S1A), but may have some propensity to form short secondary structure elements (Fig. 1A). We identified an “LXXLL” motif in the CR2C region, and demonstrated a low-affinity interaction with KIX (G586-L672) using NMR titrations (Fig. 1C). The conserved Leu residues in the LXXLL motif exhibited the largest chemical shift changes, and we estimated the Kd to be >500 μM, whereas that of CR3 is about 240 μM (Table 1 and Table S1). Although CR2C binds weakly to KIX, when linked to CR3, the binding affinity of the fusion is higher than either peptide alone. A fusion peptide of CR2C and CR3 connected by a (GGGS)3 linker (designated as CR2C-CR3) bound to KIX with a Kd of 85 μM, similar to that of the native FOXO3a C terminus, in which CR2C is linked to CR3 via ∼150 unstructured amino acids (Table 1 and Table S1). However, the engineered peptide is substantially more stable, thus enabling structural characterization of the KIX complex.
Table 1.
Dissociation constants (Kd) for the interactions of FOXO3a transactivation peptides with CBP domains
| KIX | KIX MLLmut | KIX c-Mybmut | KIX doublemut | TAZ1 | TAZ2 | NCBD | |
| CR3Trp | 237 ± 40 | 302 ± 43 | 311 ± 65 | NA | 72 ± 8 | 68 ± 9 | NA |
| pCR3 | 157 ± 39 | 192 ± 34 | 164 ± 28 | NA | 23 ± 2 | 15 ± 1 | NA |
| CR2C-CR3 | 85 ± 18 | 258 ± 100 | 335 ± 86 | NBD | 71 ± 6 | 33 ± 3 | NBD |
Kd values (μM) were determined by ITC. NA, experiment was not performed, and the data are not available; NBD, no binding detected, indicating that any interaction is too weak to be detected by ITC.
CR2C-CR3 Interacts with KIX in Two Orientations.
Titration with CR2C perturbed peaks from KIX residues in both the c-Myb and MLL sites (Fig. S2A), indicating that like CR3 (6), CR2C also binds both sites. We titrated 15N-labeled CR2C-CR3 with unlabeled KIX and observed that both CR2C and CR3 bound KIX simultaneously, whereas the linker was not affected by KIX. The reverse titration demonstrated that both the c-Myb and MLL sites of KIX are engaged by CR2C-CR3 (Fig. 1D and Fig. S2B). Paramagnetic relaxation enhancement (PRE) experiments were conducted to investigate whether either of the two TADs in CR2C-CR3 binds preferentially to either site of KIX and to probe the binding orientation. Perturbation of 15N-labeled KIX resonances was examined in the presence of CR2C-CR3 bearing an EDTA tag on the lone Cys residue (C622) located in the N terminus of CR3 (Fig. S3A). The resonances most broadened in the presence of chelated Mn2+ vs. Ca2+ (peak intensity ratio: Ipara/Idia <0.2, where Ipara and Idia are peak intensities in the presence of the paramagnetic and diamagnetic ions, respectively) were from D598, R646, and D647 (c-Myb site) and from R623 and R624 (MLL site) (Fig. 1E), indicating that in the context of the fusion protein, CR3 still binds both sites. Taken together, these data demonstrate that CR2C-CR3 binds to KIX in two different orientations that exist in fast-exchange equilibrium on the NMR chemical shift time scale. In one orientation, CR2C binds the c-Myb site and CR3 occupies the MLL site (designated 2b3l), and in the other, CR2C binds the MLL site and CR3 occupies the c-Myb site (2l3b). Residues in the c-Myb and MLL sites were broadened to a similar extent (Fig. 1E), suggesting that the 2b3l and 2l3b conformers are similarly populated. This is further supported by the observation that CR3 has similar binding affinity for KIX constructs bearing mutations that disrupt the MLL site (I660A and L664A) or the c-Myb site (A654Q, Y650A, and Y658A), without affecting folding (Table 1 and Fig. S1 B and C).
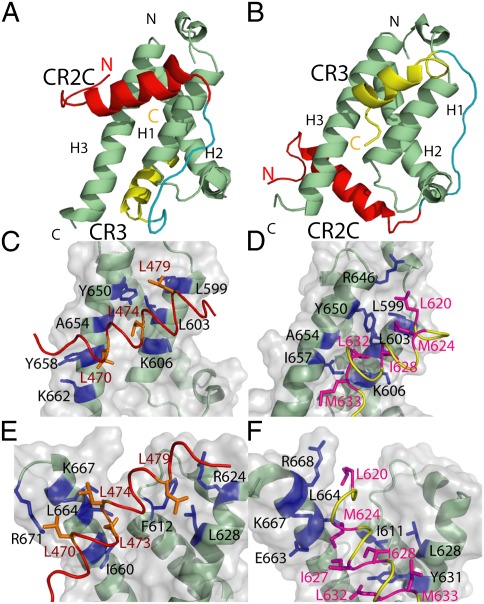
Solution Structures of KIX in Complex with CR2C-CR3.
Manually and automatically assigned NOEs from CR2C-CR3 in complex with WT KIX domain as well as CR2C or CR3 peptides bound to MLL-site or c-Myb–site KIX mutants (strategy described SI Methods and Fig. S4, structure statistics presented in Table S2) were used to determine structures of both the 2b3l and the 2l3b conformations (ribbon diagram in Fig. 2 A and B and 20-conformer ensemble in Fig. S5 A and B). CR2C and CR3 exhibit increased helicity on binding KIX (Fig. S3B) and adopt helical structures in both conformations. KIX adopts a conformation composed of three α-helices and two 310 helices, similar to that in the ternary complex with c-Myb and MLL (7). Consistent with the PRE results, C622 is found near R623 and R624 in the 2b3l conformation and near D598, R646, and D647 in the 2l3b conformation (Fig. S5 C and D). In the c-Myb site, CR2C and CR3 interact with a common set of residues, including L599, L603, K606, Y650, and A654, whereas CR2C forms additional interactions with Y658 and K662 and CR3 forms additional interactions with R646 and I657 (Fig. 2 C and D). Strikingly, CR2C and CR3 bind the MLL site in different orientations. CR2C extensively contacts a surface composed of F612, R624, L628, I660, L664, K667, and R671, whereas CR3 interacts mainly with I611, L628, Y631, E663, L664, K667, and R668 (Fig. 2 E and F). The binding surfaces of the peptides are dominated by hydrophobic residues. For CR2C, the ΦXXΦΦ motif leucines (L470, L473, and L474), together with L479, interact with KIX (Fig. 2 C and E), whereas the binding surface of CR3 is more extensive, involving the ΦXXΦΦ motif residues M624, I627, and I628 together with L620, L632, and M633 (Fig. 2 D and F).
Fig. 2.
Solution structures of KIX in complex with FOXO3a CR2C-CR3. CR2C-CR3 interacts with KIX in two distinct conformations. Lowest energy solution structure of KIX and CR2C-CR3 in the 2b3l conformation with CR2C bound to the c-Myb site and CR3 bound to the MLL site (A) and 2l3b with CR2C bound to the MLL site and CR3 bound to the c-Myb site (B). The backbones of KIX, CR2C, CR3, and the linker are shown in green, red, yellow, and cyan, respectively. Each structural component of KIX and the termini of KIX and CR2C-CR3 are labeled. CR2C in the c-Myb site (C), CR3 in the c-Myb site (D), CR2C in the MLL site (E), and CR3 in the MLL site (F). The backbones of KIX, CR2C, and CR3 are colored as above, and the surface of KIX is shown in gray. The side chains of CR2C and CR3 that interact with KIX are shown in orange and magenta, respectively, and the side chains of KIX that interact with CR2C or CR3 are shown in blue.
Structural Comparison with Other KIX-TAD Complexes.
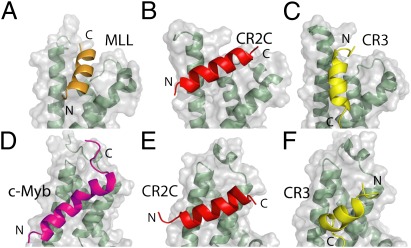
Wright and colleagues (7–9) have extensively characterized interactions between the KIX domain and a number of TAD peptides, revealing a surprising degree of plasticity. A comparison of our structures of KIX in complex with two FOXO3a TADs with the structure of KIX in complex with the TADs of c-Myb and MLL (7) further highlights the intriguing plasticity of molecular recognition by the KIX domain. CR2C and CR3 can each occupy both the c-Myb and MLL sites. CR2C binds to the MLL site in an orientation rotated ∼60° relative to the MLL peptide, whereas CR3 is rotated ∼180° (Fig. 3 A–C). A binding orientation similar to CR2C was recently observed in a complex of KIX with the HEB TAD (PDB ID code 2KWF). This illustrates the remarkable plasticity of the MLL site, which can accommodate these different peptides in various orientations with no substantial structural rearrangement. In the c-Myb site, CR2C binds in the same manner as c-Myb peptide; however, once again, CR3 binds in the opposite orientation (Fig. 3 D–F). The unique orientation of CR3 in both sites is supported by our PRE data (Fig. 1E) and may be determined, in part, by L620, which is not part of the ΦXXΦΦ motif but is involved in a network of interactions with both KIX sites (Fig. 2 D and F).
Fig. 3.
Structural comparison of KIX complexes with CR2C, CR3, c-Myb, and MLL. KIX in complex with MLL (PDB ID code 2AGH) (A), CR2C bound to the KIX MLL site (B), CR3 bound to the KIX MLL site (C), KIX in complex with c-Myb (PDB ID code 2AGH) (D), CR2C bound to the KIX c-Myb site (E), and CR3 bound to the KIX c-Myb site (F). The N and C termini are labeled for each peptide.
Full p300 Transactivation Activity of FOXO3a Requires both KIX Sites.
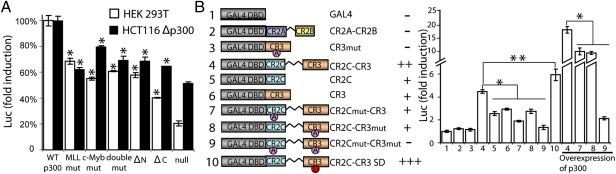
Using luciferase assays in HEK 293T cells, we found that a fusion protein composed of GAL4 DBD and CR2C-CR3 activated the expression of luciferase more than fourfold, and with exogenous co-overexpression of p300, this increased to 20-fold (Fig. S6A). To determine whether both the MLL- and c-Myb binding sites of KIX are involved in FOXO3a transactivation, we specifically disrupted each site in full-length p300 using mutations corresponding to those that abolished CR2C-CR3 binding to each site of the KIX domain. Mutation of either binding site in the KIX domain of p300 reduced luciferase expression; however, mutation of both sites did not further reduce the transactivation activity (Fig. 4A). These experiments were repeated in HCT116 cells lacking endogenous p300 to minimize background luciferase expression (Fig. S6A). Luciferase expression was lower in these cells because of lower transfection efficiency and transgene expression; however, the KIX mutations exhibited the same trend as in HEK 293T cells (Fig. 4A).
Fig. 4.
Role of FOXO3a CR2C and CR3 and two KIX binding sites in luciferase transactivation. (A) HEK 293T or p300-null HCT116 cells were transfected with vector encoding luciferase fused to five GAL4 binding sites and pCMX vector encoding GAL4 DBD fused to CR2C-CR3, as well as to vector encoding p300 [WT or p300 lacking TAZ1 and KIX domains (p300ΔN); lacking TAZ2 (p300ΔC); or bearing mutations in the KIX MLL site, c-Myb site, or both). Luciferase activity was normalized to the cells overexpressing WT-p300 for each cell line. Significant differences (determined by ANOVA) are indicated. *P < 0.01. (B) HEK 293T cells were transfected with luciferase vector (as above), and vectors encoding GAL4 DBD were fused to CR2A-CR2B or to WT or mutant CR2C, CR3, or CR2C-CR3 fusion as indicated (Left). The four CR2C-CR3 fusions were also assayed in the presence of overexpressed p300. Luciferase activity was normalized to cells overexpressing GAL4 DBD alone. *P < 0.01; **P < 0.05. All experiments were repeated three times, and the data are shown as mean ± SEM.
Other Domains of CBP/p300 Contribute to FOXO3a Binding.
KIX mutations that disrupt binding of CR2C-CR3 impaired but did not completely abolish p300-mediated transactivation activity (Fig. 4A). We speculated that CR2C or CR3 might interact with other TADs of CBP/p300 (Fig. S6C); thus, we tested the interaction between CR2C-CR3 and the TAZ1, TAZ2, and NCBD domains of CBP. Addition of TAZ1 (A340-R439) or TAZ2 (S1764-Q1855) induced large chemical shift perturbations in the 1H-15N HSQC spectra of 15N-labeled CR2C-CR3, whereas there were no significant perturbations detected on addition of NCBD (P2059-Q2117) (Fig. S6 D–F). Using isothermal titration calorimetry (ITC), we determined that CR2C-CR3 binds to TAZ1 with a Kd value of ∼70 μM and to TAZ2 with a Kd value of ∼30 μM; thus, TAZ2 shows slightly higher affinity than KIX (Table 1). To determine whether the TAZ domains play a role in FOXO3a transactivation in the cell, we performed luciferase assays in HEK 293T cells with p300 constructs lacking TAZ1 and KIX (p300ΔN, residues T1032-H2414) or the TAZ2 domain (p300ΔC, residues M1-Q1736). Luciferase transactivation by p300ΔN and p300ΔC was reduced by 42% and 60%, respectively, relative to full-length p300 (Fig. 4A), indicating that no single p300 domain is absolutely required for transactivation, rather multiple interactions between FOXO3a and p300 cooperating to mediate recruitment. This was also confirmed in the HCT116 cells with luciferase assays (Fig. 4A).
CR2C and CR3 Work in Concert to Achieve Full Transactivation Activity.
To dissect the role of each putative FOXO3a TAD, we assayed luciferase transactivation by means of a series of constructs, including GAL4 DBD fused to CR2C alone, CR3 alone, the CR2C-CR3 fusion, a CR2C mutant (L474A, designated CR2Cmut), a CR3 double mutant (I627A and I628A, designated CR3mut), and various combinations of WT and mutant CR2C and CR3 regions, as well as CR2A-CR2B (Fig. 4B). ITC results show that these mutations disrupted the binding of CR2C and CR3 to KIX, TAZ1, and TAZ2 (Table S1). The CR2C-CR3 exhibited strong transactivation activity, which was reduced by about half when either CR2C or CR3 was mutated or when CR2C or CR3 was expressed alone, and transactivation was essentially abolished by mutation of both CR2C and CR3 (Fig. 4B). Co-overexpression of p300 increased transactivation of luciferase, but the same trend was observed across this series of constructs (Fig. 4B). CR2A-CR2B exhibited no appreciable transactivation activity (Fig. 4B), suggesting that the functions of these conserved regions do not involve CBP/p300 recruitment.
AMPK Phosphorylation Enhances CBP/p300 Binding and Transactivation.
It has been shown that the net effect of AMPK phosphorylation enhances FOXO3a-dependent transcription; however, the contribution of individual phosphorylation sites has not been examined (16). An AMPK phosphorylation site (S626) was found within the ΦXXΦΦ motif of CR3; thus, we compared the binding of synthetic phosphorylated CR3 (pCR3) vs. unphosphorylated CR3 to KIX, TAZ1, and TAZ2 using ITC. Consistent with the reported role of AMPK, phosphorylation of S626 increased the affinity for KIX by ∼1.5-fold (Kd of 157 vs. 237 μM), increased the affinity for TAZ1 approximately threefold (Kd of 23 vs. 72 μM), and increased the affinity for TAZ2 approximately fourfold (Kd of 15 vs. 68 μM) (Table 1). Furthermore, the phosphomimetic S626D mutation modestly increased transactivation in the luciferase assays (Fig. 4B), suggesting that phosphorylation of this site increases FOXO3a-dependent transcription activation, probably by stabilizing interactions with CBP/p300. Titration of 15N-labeled KIX indicated that pCR3 binds both the c-Myb and MLL sites (Fig. S2C), and ITC demonstrated that S626 phosphorylation enhanced the affinity for both sites to a modest but significant extent (Table 1). In our structures, S626 is solvent-exposed and does not directly contact either the c-Myb or MLL site of KIX. However, S626 is surrounded by numerous positively charged residues in the MLL site [R623, R624 (∼10 Å), K667 (<8 Å), R668, R669, and R671], suggesting several potential interaction sites for phospho-S626 (Fig. S5E). Indeed, R623, R624, and R671 exhibit larger chemical shift perturbations when titrated with pCR3 vs. CR3 (Fig. S2C). In the c-Myb site, H602, H605, K606, H651, and Y650 are candidates for interaction with the phosphate group if the phosphorylated CR3 peptide binds with a subtle structural rearrangement relative to CR3 (Fig. S5F).
CR2C and CR3 both Act as TADs in Transcription of FOXO3a-Target Genes.
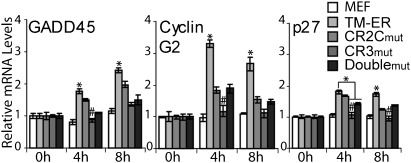
To probe the function of CR2C and CR3 in full-length FOXO3a, we established mouse embryonic fibroblast (MEF) cell lines each stably transformed with constructs encoding a fusion between the ligand binding domain of estrogen receptor (ER) and full-length FOXO3a, with three mutations (TM) that block Akt-dependent translocation (FOXO3aTM-ER) (17) or FOXO3aTM-ER bearing the CR2C and/or CR3 mutations. The mRNA levels of 10 known FOXO3a-target genes were examined by quantitative RT-PCR (qRT-PCR) after activation by 4-hydroxy tamoxifen (4-OHT), which drives FOXO3aTM-ER into the nucleus. Under our assay conditions, the cell cycle-regulating genes GADD45, Cyclin G2, and p27 were up-regulated significantly (>1.5-fold) by FOXO3aTM-ER on 4-OHT treatment (Fig. 5), but little effect was detected on PUMA, Bim, DDB1, and Pck2 (Fig. S7A). The mRNA of FasL, p21, and mG6PC could not be detected in this system. Both CR2C and CR3 were required for the full transcription activation function because mutation of either region impaired the up-regulation of GADD45, Cyclin G2, and p27 mRNAs. Mutation of CR3 nearly abolished transcriptional activation, whereas CR2C mutation was less disruptive; however, combined mutation of both regions surprisingly resulted in slightly higher gene expression than mutation of CR3 alone (Fig. 5). CR2C binds to KIX weakly, but its presence enhances the binding of the CR2C-CR3 fusion peptide (Table 1). However, CR2C also provides a binding site for Sirt1, which represses gene transcription through histone deacetylation (13, 14). Mutation of the CR2C LXXLL motif is expected to attenuate recruitment of Sirt1 as well as CBP/p300; thus, the modest reduction in transactivation caused by the CR2C mutation and the recovery associated with combining the CR2C mutation with CR3mut represent the net effect of disrupting interactions with both CBP/p300 and Sirt1.
Fig. 5.
Role of CR2C and CR3 in transactivation of endogenous target genes by full-length FOXO3a. MEF cells were stably transformed with FOXO3aTM-ER bearing WT or mutated CR2C and/or CR3 regions. After activation by 4-OHT treatment, the mRNA level of each FOXO3a target gene in each cell line was assayed by RT-PCR and normalized to the basal level determined before 4-OHT treatment. Cells expressing FOXO3aTM-ER had a higher level of the target gene mRNA than the cells expressing the CR2C/CR3 mutants (determined by ANOVA). *P < 0.05. #CR3mut cells exhibited lower mRNA level than CR2Cmut or doublemut (P < 0.05). All experiments were repeated three times, and the data are shown as mean ± SEM.
Discussion
IDPs play important roles in transcription, translation, and signal transduction (18). The C-terminal IDR of FOXO3a contains the CR2 and CR3 TADs. In the current study, we demonstrated that both CR2C and CR3 interact with the KIX domain of CBP/p300. CR2C-CR3 associates with KIX promiscuously, in that each TAD can bind either the c-Myb or MLL site, resulting in two stable conformations. The intrinsic disorder and promiscuous binding may confer some kinetic and thermodynamic advantages for coactivator recruitment. The concentration of CBP/p300 is limiting; thus, the rates of encounter and formation of transcription factor–coactivator complexes are important for gene transcription. The fly-casting model suggests that relative to structured proteins, long IDRs provide a greater capture radius for the binding partner (19). Indeed, comparison of luciferase transactivation by CR2C-CR3 constructs connected by the engineered (GGGS)3 linker (4.5-fold) vs. the ∼150-residue WT sequence (36-fold) revealed that the native linker was more active. A threefold longer linker composed of the same repeats (GGGS)9 conferred 6.2-fold transactivation, suggesting that a long flexible linker can promote transactivation (Fig. S6B). One drawback of IDPs is that their translational diffusion is slower than that of structured proteins (20); however, we previously showed that FOXO3a adopts a semicompact state in which FH associates with CR3 until this interaction is disrupted by recognition of the FRE in a gene promoter (6). Once FOXO3a is anchored to the promoter, the translational diffusion is no longer relevant. The low affinity of individual TAD/CBP interactions may be an advantage for rapid switching between on and off states, and some FOXO target gene promoters contain multiple copies of FRE, which contribute to the local enrichment of FOXO3a, offsetting the low binding affinity. Indeed, only genes with multiple FREs in their promoter sequences [3 in GADD45, 5 in Cyclin G2, and ≥2 in p27) were significantly activated by FOXO3a TM-ER in our qRT-PCR assay. In contrast, PUMA, Bim, and DDB1, each of which has a single FRE in its promoter (17, 21), showed little up-regulation, suggesting that cooperation of other transcription factors (e.g., p53) or cofactors may be required (Fig. S7B).
Some transcription factors (e.g., CREB) preferentially bind one domain of CBP/p300, whereas others (e.g., p53, EKLF) bind multiple CBP/p300 TADs (8, 22, 23). This multivalent binding can increase the binding affinity for CBP/p300, which would be further enhanced by the presence of tandem TADs (24). In addition to the KIX domain, we identified TAZ1 and TAZ2 as interacting partners for FOXO3a, which contribute to transactivation. Although the interaction between CR2C-CR3 and KIX is relatively weak (85 μM), additional interactions with the TAZ1/2 domains may promote the formation of productive transcription factor–coactivator complexes. The presence of long IDRs linking the FH domain, CR2C, and CR3 is invariant among mammalian isoforms of FOXO through evolution, suggesting that these flexible regions are functionally important, which is supported by our finding that a longer CR2C-CR3 linker enhanced transactivation. In addition to the kinetic advantage for CBP/p300 recruitment, the conformational plasticity of these long IDRs may enable the promiscuous multivalent binding of CR2C and CR3 to the KIX and TAZ domains. We speculate that this plastic mode of binding would favor productive encounters with CBP/p300 by offering adaptability to variables in gene and chromosome structure for proper positioning of the coactivator with respect to the general transcription machinery anchored to the TATA box. This plasticity would likewise facilitate cooperation with other transcription factors (Fig. S7B). A recent study of FOXO in oxidative stress showed that CBP/p300 becomes covalently attached to FOXO through a disulfide bond with a conserved Cys residue in CR3 (25). There is no Cys in KIX; thus, our finding that CR3 interacts with the Cys-rich TAZ1/2 domains suggests that they may be involved.
Many transcription factors are regulated by phosphorylation of their TADs, but the consequences are unpredictable. For instance, phosphorylation of the CREB KID domain can increase or decrease CBP/p300 binding and transcription, depending on the site (8, 26). The C-terminal IDR of FOXO3a contains five AMPK phosphorylation sites, including S626 in CR3, and AMPK phosphorylation has been reported to promote transcription of FOXO3a target genes (16). We found that phosphorylation of S626 increased the affinity of CR3 for both binding sites of KIX, as well as TAZ1 and TAZ2, and that a single phosphomimetic mutation (S626D) was sufficient to increase transactivation of the luciferase gene, thus providing a mechanism for the reported regulation of FOXO3a transactivation by AMPK. In the context of a multivalent complex, the increased affinities to each of four CBP/p300 binding sites for the phosphorylated form of CR3 may cooperatively lead to a much larger overall increase in affinity for the full-length protein. Furthermore, enhancement of the encounter rate by nonspecific electrostatic interactions may contribute to the increase in affinity. In addition, phosphorylation of the other four AMPK sites within the IDR may generate a “negative electrostatic field,” which has been suggested to promote complex formation through long-range electrostatic effects (18).
The FOXO3a and p53 tumor suppressors share certain functional similarities. Both transcription factors integrate many signaling pathways and regulate both cell cycle and apoptotic genes, some of which are shared targets (e.g., GADD45, p21cip1, PUMA). FOXO3a and p53 both contain two tandem intrinsically disordered TADs that interact with multiple domains of their shared coactivator CBP/p300 (22) (Table 1). The promiscuous interaction between CR2C-CR3 and KIX is similar to the model of Wright and colleagues (9) of the binding of the two TADs of p53 to KIX. However, each p53 TAD is capable of binding in two orientations to both the c-Myb site and the MLL site, potentially supporting a fully promiscuous ensemble composed of as many as eight binding modes. In contrast to p53, CR2C and CR3 of FOXO3a bind specifically in one direction at each site, which reduces the degree of promiscuity and makes it possible to determine high-resolution structures of two equally populated conformations. FOXO3a and p53 both act as integrators of cell signaling through mechanisms that involve posttranslational modifications of their IDRs. We found that phosphorylation of CR3 enhanced CBP/p300 binding and transactivation, similar to the effect of phosphorylation of several sites within the TADs of p53 (27).
Our studies present a very dynamic picture of FOXO3a-dependent transcription activation. The intramolecular interaction between the FH and CR3 domains blocks nonspecific interactions with DNA and keeps FOXO3a in a semicompact conformation, promoting translational diffusion, thus enhancing the rate of encounter with promoter regions (3, 6). After FH recognizes the FRE, CR3 is released (6); together, CR3 and CR2C interact promiscuously with many domains of CBP/p300, including both KIX sites, TAZ1 and TAZ2. CR2C-CR3 associates with KIX in equilibrium between two states, and we determined the solution structures illustrating molecular details of these two binding conformations, which provide more insight into the promiscuous interactions of intrinsically disordered transcription factors and coactivators. Such multivalent binding is likely relevant to a number of transcription factors, because several contain tandem TADs linked by IDRs (e.g., c-Myc, HIF-1α, c-Jun, Gcn4, p53, HEB; Fig. S7C). Structural and biochemical characterization of the newly discovered interaction between CR2C-CR3 and TAZ1 and TAZ2 is needed, and the interplay between FOXO3a, CBP/p300, and Sirt1, which contributes to determining cell fate, is worth further investigation.
Methods
All CBP domains and unphosphorylated FOXO3a peptides used in this study were expressed as recombinant GST fusion proteins in Escherichia coli and purified under native conditions, with the exception of His-tagged KIX, which was refolded after purification under denaturing conditions. NMR spectra and PRE data were collected and processed as described previously (3, 6). The 3D structures of KIX and the CR2C–CR3 complex were calculated as described previously (3). Calorimetric titrations were carried out on a Microcal VP-ITC titration calorimeter. Luciferase assays and qRT-PCR were conducted with cultured HEK 293T, HCT116 Δp300, and MEF cells. Complete methods are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank Dr. P. Wright for CBP domain plasmids, Dr. J. Zhang for luciferase assay vectors, Drs. K. Miyazono and T. Imamura for the WT and mutant p300 expression vectors, Dr. C. Caldas for p300-deficient HCT116 cells, and Dr. S. Smith for stimulating discussions. The Canadian Foundation for Innovation provided an 800-MHz NMR spectrometer. This work is supported by the Canadian Institutes of Health Research (M.I.). M.I. holds a Canada Research Chair in Cancer Structural Biology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The NMR structures, atomic coordinates, chemical shifts, and restraints have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2LQH and 2LQI) and BioMagResBank, http://www.bmrb.wisc.edu/ (BMRB ID codes 18314 and 18315).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119073109/-/DCSupplemental.
References
- 1.Burgering BM. A brief introduction to FOXOlogy. Oncogene. 2008;27:2258–2262. doi: 10.1038/onc.2008.29. [DOI] [PubMed] [Google Scholar]
- 2.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 3.Wang F, et al. Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53. J Mol Biol. 2008;384:590–603. doi: 10.1016/j.jmb.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 4.Nasrin N, et al. DAF-16 recruits the CREB-binding protein coactivator complex to the insulin-like growth factor binding protein 1 promoter in HepG2 cells. Proc Natl Acad Sci USA. 2000;97:10412–10417. doi: 10.1073/pnas.190326997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.So CW, Cleary ML. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol Cell Biol. 2002;22:6542–6552. doi: 10.1128/MCB.22.18.6542-6552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F, et al. Synergistic interplay between promoter recognition and CBP/p300 coactivator recruitment by FOXO3a. ACS Chem Biol. 2009;4:1017–1027. doi: 10.1021/cb900190u. [DOI] [PubMed] [Google Scholar]
- 7.De Guzman RN, Goto NK, Dyson HJ, Wright PE. Structural basis for cooperative transcription factor binding to the CBP coactivator. J Mol Biol. 2006;355:1005–1013. doi: 10.1016/j.jmb.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 8.Radhakrishnan I, et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 9.Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. Mapping the interactions of the p53 transactivation domain with the KIX domain of CBP. Biochemistry. 2009;48:2115–2124. doi: 10.1021/bi802055v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 11.Matsuzaki H, et al. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci USA. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.So CW, Cleary ML. Common mechanism for oncogenic activation of MLL by forkhead family proteins. Blood. 2003;101:633–639. doi: 10.1182/blood-2002-06-1785. [DOI] [PubMed] [Google Scholar]
- 13.Nakae J, et al. The LXXLL motif of murine forkhead transcription factor FoxO1 mediates Sirt1-dependent transcriptional activity. J Clin Invest. 2006;116:2473–2483. doi: 10.1172/JCI25518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brachmann CB, et al. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 15.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 16.Greer EL, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 17.You H, et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med. 2006;203:1657–1663. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2010;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- 19.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Liu Z. Kinetic advantage of intrinsically disordered proteins in coupled folding-binding process: A critical assessment of the “fly-casting” mechanism. J Mol Biol. 2009;393:1143–1159. doi: 10.1016/j.jmb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Sunters A, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795–49805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- 22.Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc Natl Acad Sci USA. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mas C, et al. Structural and functional characterization of an atypical activation domain in erythroid Kruppel-like factor (EKLF) Proc Natl Acad Sci USA. 2011;108:10484–10489. doi: 10.1073/pnas.1017029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dansen TB, et al. Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat Chem Biol. 2009;5:664–672. doi: 10.1038/nchembio.194. [DOI] [PubMed] [Google Scholar]
- 26.Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee CW, Ferreon JC, Ferreon AC, Arai M, Wright PE. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc Natl Acad Sci USA. 2010;107:19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.