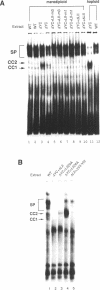
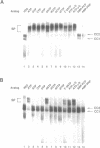
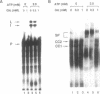
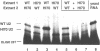
Abstract
The in vitro spliceosome assembly pathway is conserved between yeast and mammals as U1 and U2 snRNPs associate with the pre-mRNA prior to U5 and U4/U6 snRNPs. In yeast, U1 snRNP-pre-mRNA complexes are the first splicing complexes visualized on native gels, and association with U1 snRNP apparently commits pre-mRNA to the spliceosome assembly pathway. The current study addresses U2 snRNP addition to commitment complexes. We show that commitment complex formation is relatively slow and does not require ATP, whereas U2 snRNP adds to the U1 snRNP complexes in a reaction that is relatively fast and requires ATP or hydrolyzable ATP analogs. In vitro spliceosome assembly was assayed in extracts derived from strains containing several U1 sRNA mutations. The results were consistent with a critical role for U1 snRNP in early complex formation. A mutation that disrupts the base-pairing between the 5' end of U1 snRNA and the 5' splice site allows some U2 snRNP addition to bypass the ATP requirement, suggesting that ATP may be used to destabilize certain U1 snRNP:pre-mRNA interactions to allow subsequent U2 snRNP addition.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Legrain P., Rosbash M. The yeast PRP6 gene encodes a U4/U6 small nuclear ribonucleoprotein particle (snRNP) protein, and the PRP9 gene encodes a protein required for U2 snRNP binding. Mol Cell Biol. 1990 Dec;10(12):6417–6425. doi: 10.1128/mcb.10.12.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M., Jr, Igel A. H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990 Dec;4(12A):2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- Barabino S. M., Blencowe B. J., Ryder U., Sproat B. S., Lamond A. I. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990 Oct 19;63(2):293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Brody E., Abelson J. The "spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985 May 24;228(4702):963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987 Nov;1(9):1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Dalbadie-McFarland G., Abelson J. PRP5: a helicase-like protein required for mRNA splicing in yeast. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4236–4240. doi: 10.1073/pnas.87.11.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985 Aug;42(1):345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987 Jun 19;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Kretzner L., Rymond B. C., Rosbash M. S. cerevisiae U1 RNA is large and has limited primary sequence homology to metazoan U1 snRNA. Cell. 1987 Aug 14;50(4):593–602. doi: 10.1016/0092-8674(87)90032-8. [DOI] [PubMed] [Google Scholar]
- Lamm G. M., Blencowe B. J., Sproat B. S., Iribarren A. M., Ryder U., Lamond A. I. Antisense probes containing 2-aminoadenosine allow efficient depletion of U5 snRNP from HeLa splicing extracts. Nucleic Acids Res. 1991 Jun 25;19(12):3193–3198. doi: 10.1093/nar/19.12.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P., Seraphin B., Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988 Sep;8(9):3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. L., Kretzner L., Seraphin B., Rosbash M. Universally conserved and yeast-specific U1 snRNA sequences are important but not essential for U1 snRNP function. Genes Dev. 1990 Oct;4(10):1766–1774. doi: 10.1101/gad.4.10.1766. [DOI] [PubMed] [Google Scholar]
- Lin R. J., Newman A. J., Cheng S. C., Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985 Nov 25;260(27):14780–14792. [PubMed] [Google Scholar]
- Michaud S., Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991 Dec;5(12B):2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992 Feb 21;68(4):743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991 Apr 5;65(1):115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rosbash M. Specific small nuclear RNAs are associated with yeast spliceosomes. Cell. 1986 Jun 20;45(6):869–877. doi: 10.1016/0092-8674(86)90561-1. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rymond B. C., Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. 1986 Nov 27-Dec 3Nature. 324(6095):341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- Pruzan R., Furneaux H., Lassota P., Hong G. Y., Hurwitz J. Assemblage of the prespliceosome complex with separated fractions isolated from HeLa cells. J Biol Chem. 1990 Feb 15;265(5):2804–2813. [PubMed] [Google Scholar]
- Reed R. Protein composition of mammalian spliceosomes assembled in vitro. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8031–8035. doi: 10.1073/pnas.87.20.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990 Mar;10(3):1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988 Nov 18;242(4881):1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Specific and stable intron-factor interactions are established early during in vitro pre-mRNA splicing. Cell. 1985 Nov;43(1):131–142. doi: 10.1016/0092-8674(85)90018-2. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Rosbash M. A chemical modification/interference study of yeast pre-mRNA spliceosome assembly and splicing. Genes Dev. 1988 Apr;2(4):428–439. doi: 10.1101/gad.2.4.428. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Rosbash M. Differential nuclease sensitivity identifies tight contacts between yeast pre-mRNA and spliceosomes. EMBO J. 1986 Dec 20;5(13):3517–3523. doi: 10.1002/j.1460-2075.1986.tb04677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymond B. C., Torrey D. D., Rosbash M. A novel role for the 3' region of introns in pre-mRNA splicing of Saccharomyces cerevisiae. Genes Dev. 1987 May;1(3):238–246. doi: 10.1101/gad.1.3.238. [DOI] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Abovich N., Rosbash M. Genetic depletion indicates a late role for U5 snRNP during in vitro spliceosome assembly. Nucleic Acids Res. 1991 Jul 25;19(14):3857–3860. doi: 10.1093/nar/19.14.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA-pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991 May;10(5):1209–1216. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatei K., Kimura K., Ohshima Y. New methods to investigate ATP requirement for pre-mRNA splicing: inhibition by hexokinase/glucose or an ATP-binding site blocker. J Biochem. 1989 Sep;106(3):372–375. doi: 10.1093/oxfordjournals.jbchem.a122860. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]