Abstract
Signatures of balancing selection can highlight polymorphisms and functions that are important to the long-term fitness of a species. We performed a first genome-wide scan for balancing selection in a bacterial species, Staphylococcus aureus, which is a common cause of serious antimicrobial-resistant infections of humans. Using a sliding window approach, the genomes of 16 strains of S. aureus, including 5 new genome sequences presented here, and 1 outgroup strain of S. epidermidis were scanned for signatures of balancing selection. A total of 195 short windows were investigated based on their extreme values of both Tajima's D (>2.03) and π/K ratios (>0.12) relative to the rest of the genome. To test the unusualness of these windows, an Approximate Bayesian Computation framework was used to select a null demographic model that better accounted for the observed data than did the standard neutral model. A total of 186 windows were demonstrated to be unusual under the null model and, thus, represented candidate loci under balancing selection. These 186 candidate windows were located within 99 candidate genes that were spread across 62 different loci. Nearly all the signal (97.2%) was located within coding sequences; balancing selection on gene regulation apparently occurs through the targeting of global regulators such as agr and gra/aps. The agr locus had some of the strongest signatures of balancing selection, which provides new insight into the causes of diversity at this locus. The list of candidate genes included multiple virulence-associated genes and was significantly enriched for functions in amino acid and inorganic ion transport and metabolism and in defense mechanisms against innate immunity and antimicrobials, highlighting these particular functions as important to the fitness of this pathogen.
Keywords: balancing selection, population genomics, Approximate Bayesian Computation, bacterial evolution, Staphylococcus aureus
Introduction
The human genome has been comprehensively mined for signatures of positive selection for nearly a decade. Many of these studies have focused on identifying targets of short-term selective sweeps, where the population frequency of a favorable allele has increased and variation at linked sites has been eliminated (Akey et al. 2002; Nielsen et al. 2005; Sabeti et al. 2007). Fewer studies have focused on comprehensively identifying targets of long-term balancing selection, where multiple favorable alleles have been maintained at intermediate frequencies in the population, reaching neither fixation nor extinction, and variation at linked sites has been increased (Bubb et al. 2006; Andrés et al. 2009). Balancing selection can occur where a selective advantage is conferred by a heterozygous genotype, by the rarity of an allele (i.e., frequency-dependent selection), or where selective pressures fluctuate in space or time (Charlesworth 2006). Certain genes of the human immune system present strong signatures of balancing selection (Andrés et al. 2009), and the human leukocyte antigen (HLA) locus is one of the more widely studied examples (Hughes and Yeager 1998). Other examples include pathogen recognition molecules (Ferrer-Admetlla et al. 2008), inflammatory mediators (Wilson et al. 2006; Fumagalli, Pozzoli, et al. 2009), and effector molecules such as antimicrobial peptides (AMPs) (Cagliani et al. 2008; Hollox and Armour 2008). Furthermore, balancing selection may affect variation in human genes that encode pathogen receptors and in other genes that influence infectious disease susceptibility (Bamshad et al. 2002; Fumagalli, Cagliani, et al. 2009).
Signatures of balancing selection are also evident in the genomes of human pathogens. Some of these signatures may be a direct result of long-term host–pathogen interactions. This possibility is under active investigation for the malarial parasite, Plasmodium falciparum, where new vaccine candidates are being compiled based on the signatures left by host immunity (Tetteh et al. 2009; Nygaard et al. 2010; Ochola et al. 2010). No comprehensive genome-wide scans for balancing selection have been performed for a bacterial species. However, some examples of balancing selection in pathogenic bacteria have been developed. For instance, the outer surface protein C (ospC) of the causative agent of Lyme disease, Borrelia burgdorferi, has been suggested to be under balancing selection. Possible causes for the maintenance of ospC polymorphisms include the frequency-dependent selection of host immunity (Wang et al. 1999) or the heterogeneous environments that different vertebrate hosts present to the bacteria (Brisson and Dykhuizen 2004). Another possible example involves the maintenance of diversity in the O-antigen encoded by the rfb locus of Salmonella enterica. It has been suggested that this diversity is caused by the feeding preferences of host-specific amoeba (Wildschutte and Lawrence 2007). In both examples, no single allele confers a fitness advantage in all the different environments that the bacteria inhabit; thus, the selected polymorphisms from each environment become balanced in the population.
In this study, we performed a genome-wide scan for balancing selection in a premier human pathogen, the bacterium Staphylococcus aureus. Although S. aureus harmlessly colonizes the anterior nares of up to 30% of the human population (van Belkum et al. 2009), it is a common cause of infections that range in severity from relatively mild skin boils and self-limiting food poisoning to life-threatening osteomyelitis, pneumonia, and endocarditis (Boucher et al. 2010). By the year 2005, approximately 478,000 S. aureus–related hospitalizations were reported annually in the United States (Klein et al. 2007). Facilitating its success as a pathogen, S. aureus strains have acquired resistance to nearly all classes of antibiotics including the stalwarts of antistaphylococcal therapy, the beta-lactams and glycopeptides (Rehm and Tice 2010). Strains that are resistant to all beta-lactams, which are known as methicillin-resistant S. aureus (MRSA), have expanded their niche beyond the healthcare setting to include the community (DeLeo et al. 2010). We sought to determine whether balancing selection was detectable in S. aureus, whether it was more frequent in coding or noncoding DNA and whether overt virulence genes or other genes were the more frequent targets. The results provide new insights into the causes of genetic variation in S. aureus, and they highlight particular polymorphisms and functions of consequence to this pathogen.
Materials and Methods
Bacterial Strains
Staphylococcus aureus population genetic structure consists of well-defined groups of closely related clones, called clonal complexes (CCs), most of which nest within two predominant subspecies groups (Feil et al. 2003; Robinson et al. 2005). Two CC10 strains (D139 and H19) and three CC30 strains (E1410, M809, and WW2703/97) were sequenced for comparative genomics studies. These five genome sequences were combined with 11 other publicly available sequences (fig. 1) to capture both intra- and inter-CC genetic variations. In addition, we selected an outgroup sequence from S. epidermidis strain RP62a (Gill et al. 2005). Strains were grown overnight on blood agar plates or in tryptic soy broth at 37 °C. Strains were stored long term at −80 °C in 15% glycerol/tryptic soy broth (v/v) stocks. Bacterial genomic DNA was extracted using commercial kits (Qiagen and Invitrogen) as per the manufacturer's protocols.
FIG. 1.
Staphylococcus aureus genome phylogeny and details of the strains and sequences used in this study. Numbers within circles indicate the clonal complex of the strains. Numbers within squares indicate the subspecies group of the strains. The phylogeny was constructed using RAxML v7.0 (Stamatakis 2006) using a GTR + Γ + I model of nucleotide substitution on the aligned genome sequences. Nonparametric bootstrapping with 1,000 replicates revealed that all nodes had 100% support except the node marked with an asterisk, which had 97% support. Sequence type (ST) is based on multilocus sequence typing data.
Genome Sequencing, Assembly, and Annotation
High-quality draft genome sequences were determined for the five S. aureus strains using 454 FLX pyrosequencing (Roche). DNA fragment libraries were constructed and paired-end reads of up to 3 kb were obtained according to the manufacturer's recommendations. Genome assembly was performed using the program Newbler. Reads were assembled into contigs and scaffolds using the runAssembly script with the parameters –ar –rip –g. RNAmmer was used to scan the assembly for at least one 16S ribosomal RNA (rRNA) sequence of an appropriate class, and the best match to a collection of high-quality reference 16S sequences from the Ribosomal Database Project (RDP) was identified using the RDP k-mer matching method. As a final step, contaminating sequences were removed by querying assemblies via BLAST against several databases, including a mitochondrial database, UniVecCore, and the NCBI nonredundant (NR) database.
Genome assemblies were annotated using both ab initio–based and evidence-based approaches. GeneMark (Borodovsky et al. 1995), Glimmer3 (Delcher et al. 1999), Zcurveb (Guo et al. 2003), and MetaGene (Noguchi et al. 2006) were used to predict ab initio gene models. Open reading frames were constructed from BLASTX hits against the NCBI NR protein database, as part of the evidence-based approach; BLAST hits were required to have an e value better than 1 × 10−10 to be used as BLAST evidence. Annotations from highly curated reference genomes were transferred to genome assemblies where available, in order to improve automated annotation. Hmmer searches were used to identify Pfam and TIGRfam domains; Pfam/TIGRfam domains were located based on six-frame translations of the genomic sequence using a Pfam/TIGRfam library. RNAmmer (Lagesen et al. 2007) was used to identify rRNAs, tRNAScan-SE (Lowe and Eddy 1997) was used to identify tRNA encoding sequences and RFAM (Griffiths-Jones et al. 2005) was used to detect other common RNA features. Putative gene loci were determined by the combination of ab initio predictions, transferred reference gene models, and models generated from BLASTX hits. The most-likely nonconflicting gene models were selected for each locus, based on the best evidence available. Gene models with evident discrepancies were manually reviewed.
Genome Alignment and Scan
The genome sequences of 16 S. aureus strains (fig. 1) and a single S. epidermidis outgroup strain were aligned using the progressiveMauve algorithm of Mauve v2.3.1 (Darling et al. 2010) with default parameters. Locally collinear blocks (LCBs), which are alignment blocks that contain potential positional homologies, were only included in subsequent analyses if they contained aligned sequence for all 17 strains. The included LCBs were concatenated based on their order in the finished reference genome of S. aureus strain N315. All gapped positions were excluded, including entire LCBs that represented accessory genomic regions of S. aureus and regions unique to either S. aureus or S. epidermidis.
Tajima's D was used to measure the allele frequency spectrum (Tajima 1989) and the ratio of intraspecific polymorphism (π) to interspecific divergence (K) was used to measure genetic variation within and between species (Hudson et al. 1987). D compares two estimators of the population-scaled mutation rate; D > 0 is expected to result from balancing selection as well as certain demographic processes such as population contractions and subdivisions. High π/K ratios reflect high levels of intraspecific polymorphism, as expected with balancing selection, while controlling for loci that have high mutation rates. These two measures reflect different aspects of genetic variation and together constitute a powerful 2D scan for balancing selection (Innan 2006; Andrés et al. 2009; Ochola et al. 2010).
The simulations of Nordborg and Innan (2003) indicate that the appropriate window size for detecting balancing selection with Tajima's D is determined by the ratio of the population-scaled mutation (θ) and recombination (ρ) rates. Too large a window will dilute the signal and too small a window will not include enough signal to reach statistical significance. At a θ/ρ of 10/1, which is similar to that estimated from our observed data using ClonalFrame v1.2 software (Didelot and Falush 2007) (per site: θ = 0.008, ρ = 0.0007), 200 bp windows are optimal and 100 bp windows are second best (Nordborg and Innan 2003). Our scan for balancing selection was carried out using 200 bp windows and repeated using 100 bp windows. The simulations of Nordborg and Innan (2003) did not use a gene conversion model of recombination as used here for bacteria, so the truly optimal window sizes for bacteria may be smaller than what we have used here (see Andolfatto and Nordborg 1998). D and π/K were calculated for consecutive nonoverlapping windows to simplify the summaries of the number of targets. Windows lacking single nucleotide polymorphisms (SNPs) among the S. aureus genomes were excluded because Tajima's D is undefined for such windows. Windows in the top 5% of the empirical distributions of both D and π/K were retained for further study.
Approximate Bayesian Computation
Although loci may have extreme values of D and π/K, it does not necessarily follow that they have been influenced by balancing selection. The different processes of natural selection and demography can leave similar patterns of genetic variation at a given locus (Tajima 1989). Neutrality tests are often performed to eliminate false positive signatures of selection, but the null model for these tests should attempt to account for confounders. An adequate demographic model provides a conservative null model for identifying loci with unusual patterns of genetic variation that may have resulted from selection (Andrés et al. 2009). Unfortunately, the demographic history of S. aureus is unexplored, and the bias introduced by our nonrandom sampling procedure has not been characterized. To address these challenges, an Approximate Bayesian Computation (ABC) framework (Beaumont et al. 2002) was used to select a null demographic model.
Under the standard neutral model (SNM), which assumes a constant-sized population, Tajima's D is expected to be zero (Tajima 1989). However, Tajima's D was positive when calculated from full-length S. aureus genomes (D = 1.27) and when averaged across 200 bp windows (D = 0.73); also, the empirical distribution of D across windows was shifted in the positive direction (fig. 2A). These observations suggested that the SNM may not adequately describe the observed data. Recent population contraction and population subdivision are both expected to result in positive values of D (Ingvarsson 2004; Thornton and Andolfatto 2006). Preliminary investigations indicated that simple population contraction models could potentially mimic both the observed D and π/K, whereas a simple model of population subdivision, including two subpopulations with no migration, could potentially mimic the observed D but not π/K due to the high π produced in the simulations. Viable population subdivision models would likely require much more complicated parameter-rich modeling. Thus, we focused further study on three models: the SNM, a recent population contraction model (PCM), and a recent bottleneck model (BNM). For both the PCM and the BNM, the positive values of Tajima's D would represent the retention of lineages that were prevalent before the contraction. Because of the nonrandom sampling of genomes in this study, we do not consider these models to reflect an exact demographic history of S. aureus; rather, these models provide potentially conservative alternatives to the SNM.
FIG. 2.
Empirical distributions of Tajima's D and π/K. (A) Bar graph shows the distribution of D for the 7,475 windows that contained SNPs among the S. aureus genomes. Line graph shows the averages from 1,000 simulations of the PCM using the parameter estimates obtained from the ABC analyses. (B) Distribution of π/K labeled as in panel A. (C) Relationship between D and π/K. Shaded bars in panels A and B and shaded diamonds in panel C represent the 195 windows from the top 5% of the empirical distributions of both measures of balancing selection.
We assumed that an ancestral population diverged and eventually produced outgroup (S. epidermidis) and ingroup (S. aureus) populations. The time of divergence, Td, was a parameter to be estimated for all three models. Following Innan (2006) and Putnam et al. (2007), Td can be roughly estimated in units of N generations as (K/π − 1)/2. From the observed data, the rough estimate of Td was 13.8–14.2 depending on whether full-length genomes or 200 bp window averages were used. Given these reference points, the prior distribution of Td was set to be uniform between 3 and 30. For the PCM and BNM, we further assumed that the ingroup population experienced an instantaneous population contraction. The reduction in population size, Nb, relative to the constant ancestral and outgroup population sizes, N = 1, was a parameter to be estimated for the PCM and BNM. The prior distribution of Nb was set to be uniform on the log10 scale between −4 and 0, with the lower limit eliminating most genetic variation and the upper limit allowing for no change in population size. For the PCM, the time of contraction was fixed at 0.005 coalescent units. For the BNM, we further assumed that the ingroup population experienced an instantaneous recovery back to the original population size. The strength of a bottleneck is the ratio between Nb and the duration time of the bottleneck (Ingvarsson 2008). Preliminary investigations indicated that multiple combinations of Nb and duration time appeared to have similar impacts on genetic variation, and we could not simultaneously estimate both parameters with the available summary statistics. We therefore fixed the duration time at 0.005 coalescent units and estimated the time of recovery, Tr. The prior distribution of Tr was set to be uniform on the log10 scale between −4 and 0, with the lower limit representing a very recent recovery and the upper limit being close to the estimated coalescent time of the most recent common ancestor of the sampled S. aureus genomes.
ClonalFrame v1.2 software (Didelot and Falush 2007) was used with default settings on the aligned S. aureus genomes to estimate mutation and recombination parameters. The program was run twice, and convergence of the Markov chains from the two runs was verified for all parameters. The estimated mutation and recombination parameters were averaged across the two runs and used as fixed parameters in the ABC analyses. Although ClonalFrame may underestimate bacterial recombination rates by a factor of two (Didelot et al. 2010), an additional simulation done with twice the recombination rate estimated here showed no large impact on the PCM's D and π/K distributions (data not shown).
Our ABC simulation pipeline used ms software (Hudson 2002) to simulate the coalescent tree with recombination, Seq-Gen v1.3.2 software (Rambaut and Grassly 1997) to sim-ulate sequences on the tree, and VariScan v2.0.2 software (Vilella et al. 2005) to calculate summary statistics from the sequences. Recombination was modeled as a gene conversion process by ms, which resulted in multiple trees that were passed to Seq-Gen as multiple partitions. Sequences were simulated, and interspecific divergence (K) was summarized, under a Jukes-Cantor model of nucleotide substitution. We wrote several C programs to tie ms, Seq-Gen, and VariScan into a loop and to evaluate output. Although preliminary investigations and various checks were done in an embarrassingly parallel fashion on Macintosh computers, the simulations involving 500,000 and 100,000 runs were done using resources at the Mississippi Center for Supercomputing Research.
For all three models, we simulated 500,000 data sets conditioned on the number of sequences, sequence length, S. aureus mutation and recombination parameters, and demographic parameters sampled from their priors. The simulated data sets were then summarized with five statistics that were informative for specific parameters: interspecific divergence (K), Watterson's θ, Fu and Li's D*, haplotype diversity, and number of haplotypes (Hudson et al. 1987; Fu and Li 1993; Pritchard et al. 1999). In theory, sufficient summary statistics are required for ABC analyses but, in practice, summary statistics are chosen to be strongly correlated with the parameters and weakly correlated with each other (Lopes and Beaumont 2010). Across simulations, K was strongly correlated with Td (Spearman's r > 0.99) regardless of whether it was calculated from full-length genomes or averaged across windows of the genomes. However, haplotype diversity and number of haplotypes, respectively, were uninformative for full-length genomes but provided important information for Nb (r = 0.57 and 0.46) and Tr (r = 0.49 and 0.62) when averaged across windows of the genomes. Preliminary investigations showed that these correlations were similar across a variety of window sizes (500 bp, 1 kb, 2 kb, 4 kb), so 1 kb window averages were used to summarize the data.
A standardized Euclidean distance, δ, was used to measure the resemblance between the summaries of simulated and observed data. For each simulation, δ was calculated as
 |
where, for the ith summary statistic, S is the statistic for the observed data, S* is the statistic for the simulated data, and SD(S*) is the standard deviation over all simulations. The 500,000 simulations were ranked by their δ's, and the parameter values from the top 500 simulations (0.001 acceptance rate) were selected as the approximate posterior distributions. The posteriors were smoothed with an Epanechnikov kernel and the mode and the 95% highest posterior density intervals were calculated, using the Locfit v1.5-6 module (Loader 1996) of the R v2.13.1 software package. The ratios of the acceptance rates of different equally weighted models over a range of δ's represent approximate Bayes factors (Leuenberger and Wegmann 2010), which were interpreted here according to Jeffreys (1998) to select a null model.
To construct null distributions of D and π/K at 200 bp and 100 bp window sizes, we simulated 100,000 data sets under the null model using the posterior modes as point estimates for the model parameters. Separate simulations were done to produce the results based on 200 bp and 100 bp windows. The “unusualness” of each observed window was measured as the proportion of simulated data sets that produced a window with both a D and a π/K as great or greater than the observed window.
Gene Assignment, Alignment, and Analyses of Putative Gene Functions
Windows showing evidence of balancing selection based on the above analyses were assigned to genes by BLASTN of the windows against the genome sequence of S. aureus strain N315. Gene sequences were then aligned using the ClustalW algorithm, implemented in MegAlign v7.1 (Lasergene). Gene sequences of unequal length were aligned based on amino acid sequences and back translated to nucleotide sequences. All alignments were manually inspected. Putative gene functions were examined through use of the Clusters of Orthologous Genes (COGs) database (Tatusov et al. 1997). Odds ratios were used to compare the proportions of COG functional categories among the list of candidate genes with the proportions in the remainder of the examined genome, using InStat v3.1 (GraphPad Software).
Results
Loci with Extreme Values of D and π/K
A progressiveMauve alignment of the genome sequences of 16 S. aureus strains and a single S. epidermidis outgroup strain produced 58 LCBs with sequence from all strains. After excluding gapped positions, the total alignment length was 1,601,384 bp, which represented an average of 57% coverage of these S. aureus genomes. The alignment revealed 42,673 SNPs among the S. aureus genomes and 408,076 SNPs that distinguished S. aureus from S. epidermidis. The alignment was scanned with 200 bp consecutive nonoverlapping windows. SNPs among the S. aureus genomes were present in 7,475 of the 8,007 windows. Tajima's D calculated from these windows ranged from −2.21 to 3.05, and the distribution was shifted in the positive direction, whereas π/K ranged from 0.001 to 0.49 (fig. 2A and B). Across these windows, a significant positive correlation was observed between D and π/K (Spearman's r = 0.492, P < 0.0001) (fig. 2C). This correlation reflected agreement between the two measures of balancing selection, even though it was much weaker than that found previously for P. falciparum genes (Ochola et al. 2010). A total of 195 windows were in the top 5% of the empirical distributions of both D and π/K. All of these 195 windows had D > 2.03 and π/K > 0.12.
Selection of a Null Model
The SNM and two potentially conservative alternatives, a recent PCM and a recent BNM, were examined using an ABC framework. The resemblance between the summaries of simulated and observed data, δ, provides an objective measure to compare models. Better models produce more simulations with lower δ's. The lowest δ's were 0.084 for the PCM, 0.094 for the BNM, and 0.327 for the SNM. The 500th δ's, which corresponded to the commonly used acceptance rate of 0.001 (Bertorelle et al. 2010), were 0.398 for the PCM, 0.437 for the BNM, and 1.288 for the SNM. Approximate Bayes factors were calculated as the ratios of the number of simulations produced by different models over a range of δ's (Leuenberger and Wegmann 2010). We examined δ's between 0.4 and 1.0, which corresponded to acceptance rates under the PCM of ∼0.001 to 0.023 and demonstrated the stability of the Bayes factors over these values (fig. 3). The Bayes factors comparing the PCM with the BNM were <2 over these δ's, which indicated that the evidence was slightly in favor of the PCM but barely worth mentioning (Jeffreys 1998). In contrast, the Bayes factors comparing the PCM and the BNM with the SNM were >100 over these δ's, which indicated that the evidence was decisive (Jeffreys 1998) in favor of the PCM and the BNM over the SNM.
FIG. 3.
Approximate Bayes factors to compare different models over a range of δ's. Diamonds indicate PCM versus SNM. Triangles indicate BNM versus SNM. Squares indicate PCM versus BNM.
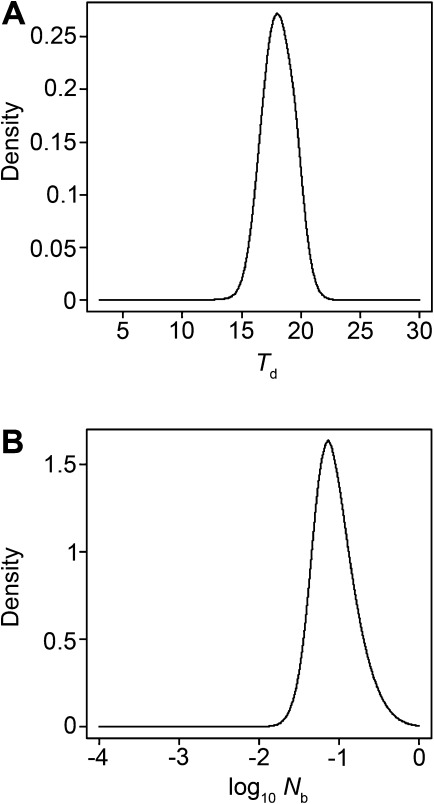
The PCM was selected as the null model because it was slightly favored over the BNM, it was simpler than the BNM (i.e., one less parameter), and it had well-estimated parameters. Moreover, the Tr parameter of the BNM was poorly estimated (data not shown). The Td and Nb values associated with the top 500 simulations of the PCM provided the approximate posterior distributions of these parameters (fig. 4). Both of these parameters were sampled from uninformative flat priors, but their posteriors clearly showed the signal that was captured by the ABC analyses. The point estimates and 95% credibility intervals were 18.0 (15.5, 20.8) for Td and 0.073 (0.029, 0.321) for Nb.
FIG. 4.
Approximate posterior distributions for the Td and Nb parameters under the PCM. Posteriors were smoothed with an Epanechnikov kernel as described in the text.
Simulations done with the parameter estimates can indicate various strengths and weaknesses of the selected null model (Thornton and Andolfatto 2006). Results for Tajima's D and π/K averaged over 1,000 simulations of the PCM are shown in figure 2A and B, respectively. Similar to the observed data, Tajima's D from simulated genomes was positive when calculated from full-length sequences (D = 0.51) and when averaged across 200 bp windows (D = 0.35), and the distribution was shifted in the positive direction (fig. 2A). However, the PCM produced an apparent excess of windows with negative D compared with the observed data. The distribution of π/K was also similar to the observed data, but the PCM produced an apparent excess of windows with low π/K (fig. 2B). The PCM appeared to provide a better fit to the observed data at the right tails of the distributions of D and π/K, where loci under balancing selection should occur.
Candidate Loci under Balancing Selection
The unusualness of the 195 observed windows with extreme values of D and π/K was tested under the PCM. Nine of these windows were discarded as false positives because D and π/K values at least as high as theirs occurred in 5% or more of the simulated data sets. The remaining 186 windows were considered to be unusual under the PCM and formed our list of candidate windows under balancing selection (supplementary table 1, Supplementary Material online). Most of the candidate windows were very unusual under the null model. For example, the 186 windows were unusual at P < 0.05, but 156 windows remained unusual at P < 0.025, and 110 windows remained unusual at P < 0.01. We note that the power of this test comes from considering D and π/K jointly. If we had relied on separate tests of D and π/K, only four windows would have been unusual for both measures of balancing selection at P < 0.05. The separate tests are less powerful because of the high values of D that are produced under the PCM; for example, 85% of the simulations produced windows with D > 2.0 and 49% of the simulations produced windows with D > 2.5.
The sequence alignment was reexamined to determine whether artifacts could have contributed to the signatures in the candidate windows. No significant difference in the mean or median number of gaps was observed for the 186 candidate windows in comparison to all other windows, indicating no specific problems with the quality of the candidate windows' sequence. Two candidate windows brought together genes that flanked large gaps of 3.9 and 12.1 kb; however, in both cases, two and three other candidate windows, respectively, were found in one of the flanking genes, indicating genuine signatures. All windows included only one sequence from each of the 17 strains, and synteny in S. aureus was confirmed for the regions that flanked each of the candidate windows, indicating no problems with the inclusion of paralogs or with wrongly assigned orthologs.
The 186 candidate windows were located within 99 candidate genes. This list of candidate genes was somewhat robust to the window size used to do the genome scan. When the scan was repeated using 100 bp windows, 23 candidate genes were found to be unique to the 200 bp analysis and 12 other genes were found to be unique to the 100 bp analysis (supplementary table 2, Supplementary Material online), whereas the remaining 76 candidate genes were present in both lists. The number of candidate windows located within candidate genes ranged from 1 to 9. A total of 97.2% of the candidate windows' sequence was located within coding DNA. In total, the candidate windows covered 2.3% of the examined sequence and 5.7% of the examined genes. These results were consistent with other results from simulations and from human studies that indicate that balancing selection is a rarely detected form of positive selection (Nordborg and Innan 2003; Bubb et al. 2006; Andrés et al. 2009).
Evidence for Clustering of Candidate Loci
Nonsynonymous mutations were lacking in 36 of the 186 candidate windows (supplementary table 1, Supplementary Material online). Although these windows could be false positives or they could be the result of unexpectedly strong selection on codon preference or mRNA structure/stability, they could also be the result of physical linkage (i.e., hitchhikers) to the actual targets of balancing selection. Indeed, both the mean and the median distance between these 36 candidate windows and the remaining 150 candidate windows were significantly different (closer) than the distance between 1,000 random windows and the 150 candidate windows (P < 0.0001). Furthermore, 47 of the 99 candidate genes occurred in 18 clusters with two or more adjacent genes under balancing selection. If candidate genes separated by single noncandidate genes were counted in the clusters, then 59 of the 99 candidate genes occurred in 22 clusters (supplementary table 3, Supplementary Material online). This evidence for physical linkage and clustering of the targets of balancing selection in S. aureus contrasts with the narrow targets found in humans (Andrés et al. 2009).
Putative Functions of Candidate Loci
The functions of the 99 candidate genes were diverse. However, three COG functional categories, which included amino acid transport and metabolism, inorganic ion transport and metabolism, and defense mechanisms, were significantly enriched (i.e., overrepresented) among the candidate genes in comparison to the remainder of the examined genome (table 1). When the COG functional categories were reexamined using the gene list based on 100 bp windows, only the defense category was significantly enriched among the genes under balancing selection. Further investigation revealed that some candidate genes not assigned defense functions based on their COG categories have experimental evidence supporting a defense function (discussed below). Thus, S. aureus genes involved in defense against innate immunity and antimicrobials have been especially important to the fitness of the species.
Table 1.
Analyses of COG Functional Categories for Enrichment and Depletion among the 99 Candidate Genes under Balancing Selection.
| COG Category | Functions | Odds Ratioa |
| C | Energy production and conversion | 1.00 (0.40, 2.53) |
| D | Cell cycle control | 0.48 (0.03, 8.12) |
| E | Amino acid transport and metabolism | 1.95 (1.11, 3.42) |
| F | Nucleotide transport and metabolism | 0.92 (0.28, 3.01) |
| G | Carbohydrate transport and metabolism | 1.34 (0.60, 2.98) |
| H | Coenzyme transport and metabolism | 0.64 (0.20, 2.06) |
| I | Lipid transport and metabolism | 1.20 (0.36, 3.92) |
| J | Translation, ribosomal structure, and biogenesis | 0.37 (0.11, 1.18) |
| K | Transcription | 0.43 (0.13, 1.38) |
| L | Replication, recombination, and repair | 0.36 (0.09, 1.49) |
| M | Cell wall/membrane/envelope biogenesis | 0.24 (0.03, 1.78) |
| N | Cell motility | 1.64 (0.09, 29.98) |
| O | Posttranslational modification, protein turnover, and chaperones | 0.59 (0.14, 2.45) |
| P | Inorganic ion transport and metabolism | 2.81 (1.53, 5.14) |
| Q | Secondary metabolism biosynthesis, transport, and catabolism | 0.87 (0.12, 6.52) |
| R | General function prediction only | 1.00 (0.54, 1.88) |
| S | Function unknown | 0.54 (0.22, 1.36) |
| T | Signal transduction mechanisms | 2.00 (0.77, 5.15) |
| U | Intracellular trafficking, secretion, and vesicular transport | 0.38 (0.02, 6.32) |
| V | Defense mechanisms | 6.84 (2.41, 19.43) |
95% confidence intervals in parentheses. Underlined indicates significant enrichment.
A Locus with Classical Characteristics of Balancing Selection
The accessory gene regulator (agr) locus encodes a two-component signal transduction system that is involved in quorum sensing and in regulating the expression of many genes, including most known virulence genes of S. aureus (for a review, see Novick and Geisinger 2008). Four distinct agr allelic groups, and one hybrid group, have been identified within this species (Ji et al. 1997; Jarraud et al. 2000; Robinson et al. 2005). Some of these groups have been shown to inhibit the regulatory activities of each other (Ji et al. 1997) and some of these groups appear to be associated with characteristics such as glycopeptide resistance and production of certain toxins (Jarraud et al. 2000; Sakoulas et al. 2002; Verdier et al. 2004).
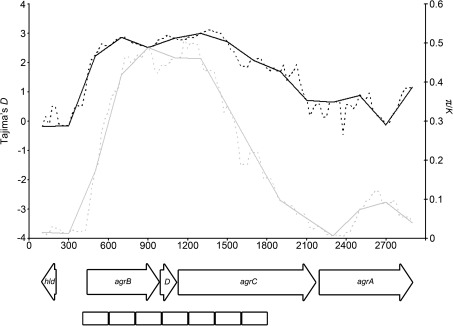
Windows from the agr locus ranked among the top candidates under balancing selection, indicating strong signatures at this locus. For example, three of the top ten most extreme Tajima's D and five of the top ten most extreme π/K were from windows that belonged to the agr locus. One window from the agr locus was also among the four windows, mentioned above, that were unusual when D and π/K were considered separately. The strongest signatures of balancing selection at the agr locus were located within the known hypervariable region that spans the 3′ end of agrB, all of agrD, and the 5′ end of agrC (fig. 5). Based on these results, we propose that the agr locus may serve as a positive control for future studies of balancing selection in S. aureus.
FIG. 5.
Sliding window analysis of Tajima's D and π/K at the agr locus. Windows are 200 bp, x axis indicates the midpoints of the windows. Black lines show D, gray lines show π/K. Solid lines show nonoverlapping windows, dashed lines show windows with 25 bp steps. Arrows represent the positions of open reading frames, boxes represent the seven candidate windows under balancing selection at this locus.
Discussion
Balancing selection is expected to leave signatures in the allele frequency spectrum and in the ratio of intraspecific polymorphism to interspecific divergence, which is amplified and made more specific when considered together (Innan 2006; Andrés et al. 2009; Ochola et al. 2010). Demography can influence genetic variation in ways similar to that of natural selection, but demography's influence is genome wide, whereas selection's influence is localized. A conservative approach to identify the targets of balancing selection is to locate extreme genetic signatures and then test their unusualness under a demographic model that attempts to account for confounders. ABC provides a flexible framework for selecting a null model (Bertorelle et al. 2010), even when nothing is known about the demographic history of the species of interest. Using these procedures, we localized the peak signatures of balancing selection in the S. aureus genome to 186 candidate windows within 99 candidate genes that were spread across 62 different loci.
When performing a genome-wide scan for balancing selection, it is helpful to have a locus known to be under balancing selection to serve as a positive control. In human studies, the HLA locus serves as a positive control for balancing selection (Garrigan and Hedrick 2003), whereas in P. falciparum studies, the ama1 gene has served as a positive control (Tetteh et al. 2009). In this study, the agr locus had some of the strongest signatures of balancing selection and was proposed to be a positive control for future studies. Besides the potential functional differences of agr allelic groups within S. aureus, noted above, other staphylococcal species are known to have an agr locus with different allelic groups than those of S. aureus (Dufour et al. 2002), suggesting that agr polymorphism is an ancient characteristic (Wright et al. 2005). It is tempting to speculate that agr may be under balancing selection in other staphylococcal species. Polymorphisms that traverse species boundaries can provide evidence for long-term balancing selection, though this can be difficult to detect (Wiuf et al. 2004).
Balancing selection in S. aureus affects substantially more coding sequence than noncoding sequence. Rather than targeting multiple noncoding regulatory sequences, balancing selection apparently impacts gene regulation through the targeting of global regulators such as the agr and glycopeptide resistance–associated/AMP-sensing (gra/aps) (Cui et al. 2005; Li et al. 2007) loci. A number of candidate genes have links to virulence, either directly through damaging host products (aur) or indirectly through metabolism (aroA, oppF) or resistance to innate immunity and antimicrobials (aur, fmtC/mprF, norA, snoABCDE, vraG) or they would be suspected of a link to virulence through their iron-scavenging function (sstAC) or cell wall attachment (sasC). (Prokesova et al. 1991; Mei et al. 1997; Morrissey et al. 2000; Kristian et al. 2003; Sieprawska-Lupa et al. 2004; Bayer et al. 2006; Buzzola et al. 2006; Mwangi et al. 2007; Howden et al. 2008; Sass and Bierbaum 2009; Schroeder et al. 2009). Thus, balancing selection may have played some role in the development and/or maintenance of virulence in this species. Of interest, genes involved in amino acid and inorganic ion transport and metabolism, and defense mechanisms, were enriched among the list of candidate genes under balancing selection. The enrichment of the defense category was robust to the window size used to detect the signatures of balancing selection. We therefore focus further discussion on this category of candidate genes.
Two candidate genes that belonged to the defense category were vraF and vraG. These genes encode a putative ATP-binding cassette transporter and are regulated by the adjacent upstream global regulatory system, graRS/apsRS (Meehl et al. 2007), which were also among our list of candidate genes. vraG contained nine separate candidate windows, the most of any candidate gene (supplementary table 3, Supplementary Material online). Thus, this locus has particularly strong signatures of balancing selection. Mutations in vraG and graS/apsS have been implicated in the evolution of vancomycin resistance within patients infected by S. aureus (Mwangi et al. 2007; Howden et al. 2008). Mutations in graS/apsS can also confer resistance to cationic AMPs (Sass and Bierbaum 2009), which are products of the innate immune system of humans and other organisms.
Interestingly, several other candidate genes have been shown to be involved in defense against innate immunity even though their COG categories do not indicate that function. For example, the aur metalloprotease is able to degrade the cationic AMP, LL-37 (Sieprawska-Lupa et al. 2004). The fmtC/mprF gene product transfers L-lysine residues to phosphatidylglycerol located on the outer leaflet of the cell membrane, which helps to repulse cationic AMPs (Kristian et al. 2003). Another candidate gene, lysP, which encodes a putative lysine permease, might impact the available pool of lysine, though its role in cationic AMP resistance has not been examined. The sno locus, which included five separate candidate genes, is involved in resistance to the cationic AMP, thrombin-induced platelet microbicidal protein 1 (Bayer et al. 2006). Thus, genes involved in glycopeptide and cationic AMP resistance are well represented among our list of candidate genes under balancing selection. Further study of the candidate genes and residues may offer new insights into resistance mechanisms and may guide the development of therapeutics based on these types of molecules.
Both aur and norA have been noted in previous studies to be subdivided into two main allelic groups within S. aureus (Sabat et al. 2000; Noguchi et al. 2004). No distinct biological functions or fitness advantages have been attributed to these allelic groups in either gene that might explain their maintenance. Sabat et al. (2008) concluded that the aur gene was influenced strongly by purifying selection due to the sparse number of nonsynonymous mutations and that the gene had undergone recombination. The possibility of negative purifying selection at some sites of this gene and positive balancing selection at other sites is not incompatible. For example, purifying selection could eliminate polymorphisms that cause a deviation from the predicted fitness optima represented by the two allelic groups.
This study had limitations. First, the sample consisted of a relatively small number of genomes that do not represent a random sample of known S. aureus diversity. Tajima's D is a frequency-based statistic, so values calculated from smaller sample sizes will be less accurate than those from larger sample sizes. We also did not consider polymorphisms within S. epidermidis, which would be expected to have some impact on K. However, multiple strains from five of the major S. aureus CCs and both subspecies groups were included in this sample, and 186 candidate windows were sufficiently unusual under a conservative null model to suggest that processes other than demography or sampling bias had produced their signatures. Since 30% or less of the human population carries S. aureus in their anterior nares (van Belkum et al. 2009), whereas skin colonization by the closely related species, S. epidermidis, is ubiquitous (Otto 2009), a population contraction demographic model for S. aureus is biologically plausible. Even so, this null model may not be adequate for subsequent studies that assemble a random sample of S. aureus genomes. The “contraction” detected here may simply reflect the diversity missed through nonrandom sampling. Further study of the demographic history of S. aureus using a random sample of genomes could provide a valuable null model for identifying unusual loci. Second, we expect to have missed some targets of balancing selection. We have focused this study on the core genome and have ignored the accessory genome (e.g., plasmids, phage, mobile genetic elements). The presence or absence of accessory genes has been suggested to represent a balanced polymorphism in a pathogenic bacteria of plants (Araki et al. 2006). The use of outgroups that are more closely related to S. aureus, such as the divergent CC75 strain of S. aureus that was recently sequenced (Holt et al. 2011), might allow more of the S. aureus genome to be probed. Finally, although this study performed a scan for balancing selection due to sampling and modeling limitations related to population subdivision, we cannot discount the possibility that some of the signatures identified here might have alternative selective causes such as positive diversifying selection acting differently in the well-defined CCs. Further study is needed to address these limitations.
Balancing selection affects genetic variation in both the S. aureus genome and the immune system of its human host. Multiple genes that provide defense against innate immunity and antimicrobials show robust evidence of balancing selection in this pathogen. Other candidate genes under balancing selection may reflect the need of S. aureus to adapt to different environments. Further study can reveal the precise mechanisms that maintain these balanced polymorphisms and the extent to which they reflect long-term host–pathogen interactions.
Supplementary Material
Supplementary tables 1–3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We are grateful to Davida Smyth and Agnes Wong for technical assistance, to Maria Giovanni (NIAID), Bruce Birren, and the Broad Sequencing Platform (Broad Institute) for their support and assistance with the genome sequencing, to Brian Hopkins and the Mississippi Center for Supercomputing Research for the allocation of computing time, disk space, and technical support, and to the associate editor and three anonymous reviewers for their insightful critiques. The sequencing project was funded in part by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, under contract number HHSN27220090018C. This work was supported by grant number GM080602 from the National Institute of General Medical Sciences, National Institutes of Health to D.A.R.
References
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P, Nordborg M. The effect of gene conversion on intralocus associations. Genetics. 1998;148:1397–1399. doi: 10.1093/genetics/148.3.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés AM, Hubisz MJ, Indap A, et al. (12 co-authors) Targets of balancing selection in the human genome. Mol Biol Evol. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H, Tian D, Goss EM, Jakob K, Halldorsdottir SS, Kreitman M, Bergelson J. Presence/absence polymorphism for alternative pathogenicity islands in Pseudomonas viridiflava, a pathogen of Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:5887–5892. doi: 10.1073/pnas.0601431103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Takeuchi F, Kuroda M, et al. (14 co-authors) Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Mummidi S, Gonzalez E, et al. (11 co-authors) A strong signature of balancing selection in the 5′ cis-regulatory region of CCR5. Proc Natl Acad Sci U S A. 2002;99:10539–10544. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer AS, McNamara P, Yeaman MR, Lucindo N, Jones T, Cheung AL, Sahl HG, Proctor RA. Transposon disruption of the complex I NADH oxidoreductase gene (snoD) in Staphylococcus aureus is associated with reduced susceptibility to the microbicidal activity of thrombin-induced platelet microbicidal protein 1. J Bacteriol. 2006;188:211–222. doi: 10.1128/JB.188.1.211-222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertorelle G, Benazzo A, Mona S. ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol Ecol. 2010;19:2609–2625. doi: 10.1111/j.1365-294X.2010.04690.x. [DOI] [PubMed] [Google Scholar]
- Borodovsky MJ, McIninch JD, Koonin EV, Rudd KE, Medigue C, Danchin A. Detection of new genes in a bacterial genome using Markov models for three gene classes. Nucleic Acids Res. 1995;23:3554–3562. doi: 10.1093/nar/23.17.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H, Miller LG, Razonable RR. Serious infections caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2010;51(Suppl 2):S183–S197. doi: 10.1086/653519. [DOI] [PubMed] [Google Scholar]
- Brisson D, Dykhuizen DE. ospC Diversity in Borrelia burgdorferi. Genetics. 2004;168:713–722. doi: 10.1534/genetics.104.028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb KL, Bovee D, Buckley D, et al. (12 co-authors) Scan of human genome reveals no new loci under ancient balancing selection. Genetics. 2006;173:2165–2177. doi: 10.1534/genetics.106.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzola FR, Barbagelata MS, Caccuri RL, Sordelli DO. Attenuation and persistence of and ability to induce protective immunity to a Staphylococcus aureus aroA mutant in mice. Infect Immun. 2006;74:3498–3506. doi: 10.1128/IAI.01507-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R, Fumagalli M, Riva S, Pozzoli U, Comi GP, Menozzi G, Bresolin N, Sironi M. The signatures of long-standing balancing selection at the human defensin β-1 promoter. Genome Biol. 2008;9:R143. doi: 10.1186/gb-2008-9-9-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Lian J, Neoh H, Reyes E, Hiramatsu K. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2005;49:3404–3413. doi: 10.1128/AAC.49.8.3404-3413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzburg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated methicillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Lawson D, Darling A, Falush D. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics. 2010;186:1435–1449. doi: 10.1534/genetics.110.120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep BA, Gill SR, Chang RF, et al. (12 co-authors) Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Dufour P, Jarraud S, Vandenesch F, Greenland T, Novick RP, Bes M, Etienne J, Lina G. High genetic variability of the agr locus in Staphylococcus species. J Bacteriol. 2002;184:1180–1186. doi: 10.1128/jb.184.4.1180-1186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Cooper JE, Grundmann H, et al. (12 co-authors) How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–3316. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Bosch E, Sikora M, et al. (11 co-authors) Balancing selection is the main force shaping the evolution of innate immunity genes. J Immunol. 2008;181:1315–1322. doi: 10.4049/jimmunol.181.2.1315. [DOI] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Cagliani R, Pozzoli U, Riva S, Comi GP, Menozzi G, Bresolin N, Sironi M. Widespread balancing selection and pathogen-driven selection at blood group antigen genes. Genome Res. 2009;19:199–212. doi: 10.1101/gr.082768.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Pozzoli U, Cagliani R, Comi GP, Riva S, Clerici M, Bresolin N, Sironi M. Parasites represent a major selective force for interleukin genes and shape the genetic predisposition to autoimmune conditions. J Exp Med. 2009;206:1395–1408. doi: 10.1084/jem.20082779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Hedrick PW. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution. 2003;57:1707–1722. doi: 10.1111/j.0014-3820.2003.tb00580.x. [DOI] [PubMed] [Google Scholar]
- Gill SR, Fouts DE, Archer GL, et al. (29 co-authors) Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy AF, Worrell V, Orvis J, Roe BA, Dyer DW, Iandolo JJ. The Staphylococcus aureus NCTC 8325 genome. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-positive pathogens. 2nd ed. Washington (DC): ASM Press; 2006. pp. 381–412. [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FB, Ou HY, Zhang CT. ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res. 2003;31:1780–1789. doi: 10.1093/nar/gkg254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Feil EJ, Lindsay JA, et al. (45 co-authors) Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox EJ, Armour JAL. Directional and balancing selection in human beta-defensins. BMC Evol Biol. 2008;8:113. doi: 10.1186/1471-2148-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt DC, Holden MT, Tong SY, et al. (11 co-authors) A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol. 2011;3:881–895. doi: 10.1093/gbe/evr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden BP, Stinear TP, Allen DL, Johnson PD, Ward PB, Davies JK. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:3755–3762. doi: 10.1128/AAC.01613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Yeager M. Natural selection at major histocompatibility loci of vertebrates. Annu Rev Genet. 1998;32:415–435. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK. Population subdivision and the Hudson-Kreitman-Aguade test: testing for deviations from the neutral model in organelle genomes. Genet Res. 2004;83:31–39. doi: 10.1017/s0016672303006529. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK. Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics. 2008;180:329–340. doi: 10.1534/genetics.108.090431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H. Modified Hudson-Kreitman-Aguade test and two-dimensional evaluation of neutrality tests. Genetics. 2006;173:1725–1733. doi: 10.1534/genetics.106.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarraud S, Lyon GJ, Figueiredo AM, Gérard L, Vandenesch F, Etienne J, Muir TW, Novick RP. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol. 2000;182:6517–6522. doi: 10.1128/jb.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys H. Theory of probability. 3rd ed. Oxford: Oxford University Press; 1998. [Google Scholar]
- Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg Infect Dis. 2007;13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristian SA, Dürr M, van Strijp JA, Neumeister B, Peschel A. MprF-mediated lysinylation of phospholipids in Staphylococcus aureus leads to protection against oxygen-independent neutrophil killing. Infect Immun. 2003;71:546–549. doi: 10.1128/IAI.71.1.546-549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Ohta T, Uchiyama I, et al. (37 co-authors) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger C, Wegmann D. Bayesian computation and model selection without likelihoods. Genetics. 2010;184:243–252. doi: 10.1534/genetics.109.109058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol. 2007;66:1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- Loader C. Local likelihood density estimation. Ann Stat. 1996;24:1602–1618. [Google Scholar]
- Lopes JS, Beaumont MA. ABC: a useful Bayesian tool for the analysis of population data. Infect Genet Evol. 2010;10:826–833. doi: 10.1016/j.meegid.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehl M, Herbert S, Gotz F, Cheung A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:2679–2689. doi: 10.1128/AAC.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J-M, Nourbakhsh F, Ford CW, Holden DW. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- Morrissey JA, Cockayne A, Hill PJ, Williams P. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect Immun. 2000;68:6281–6288. doi: 10.1128/iai.68.11.6281-6288.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi MM, Wu SW, Zhou Y, et al. (11 co-authors) Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A. 2007;104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi N, Okada H, Narui K, Sasatsu M. Comparison of the nucleotide sequence and expression of norA genes and microbial susceptibility in 21 strains of Staphylococcus aureus. Microb Drug Resist. 2004;10:197–203. doi: 10.1089/mdr.2004.10.197. [DOI] [PubMed] [Google Scholar]
- Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Innan H. The genealogy of sequences containing multiple sites subject to strong selection in a subdivided population. Genetics. 2003;163:1201–1213. doi: 10.1093/genetics/163.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- Nygaard S, Braunstein A, Malsen G, et al. (12 co-authors) Long- and short-term selective forces on malaria parasite genomes. PLoS Genet. 2010;6:e1001099. doi: 10.1371/journal.pgen.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochola LI, Tetteh KK, Stewart LB, Riitho V, Marsh K, Conway DJ. Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Mol Biol Evol. 2010;27:2344–2351. doi: 10.1093/molbev/msq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Staphylococcus epidermidis—the “accidental” pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Seielstad MT, Perez-Lezaun A, Feldman MW. Population growth of human Y-chromosomes: a study of Y chromosome microsatellites. Mol Biol Evol. 1999;16:1791–1798. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- Prokesova L, Porwit-Bóbr Z, Baran K, Potempa J, Pospisil M, John C. Effect of metalloproteinase from Staphylococcus aureus on in vitro stimulation of human lymphocytes. Immunol Lett. 1991;27:225–230. doi: 10.1016/0165-2478(91)90156-5. [DOI] [PubMed] [Google Scholar]
- Putnam AS, Scriber JM, Andolfatto P. Discordant divergence times among Z-chromosome regions between two ecologically distinct swallowtail butterfly species. Evolution. 2007;61:912–927. doi: 10.1111/j.1558-5646.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Grassly NC. Seq-Gen: an application for the Monte-Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- Rehm SJ, Tice A. Staphylococcus aureus: methicillin-susceptible S. aureus to methicillin-resistant S. aureus and vancomycin-resistant S. aureus. Clin Infect Dis. 2010;51(Suppl 2):S176–S182. doi: 10.1086/653518. [DOI] [PubMed] [Google Scholar]
- Robinson DA, Monk AB, Cooper JE, Feil EJ, Enright MC. Evolutionary genetics of the accessory gene regulator (agr) locus in Staphylococcus aureus. J Bacteriol. 2005;187:8312–8321. doi: 10.1128/JB.187.24.8312-8321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabat AJ, Kosowska K, Poulsen K, Kasprowicz A, Sekowska A, van den Burg B, Travis J, Potempa J. Two allelic forms of the aureolysin gene (aur) within Staphylococcus aureus. Infect Immun. 2000;68:973–976. doi: 10.1128/iai.68.2.973-976.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabat AJ, Wladyka B, Kosowska-Shick K, Grundmann H, van Dijl JM, Kowal J, Appelbaum PC, Dubin A, Hryniewicz W. Polymorphism, genetic exchange and intragenic recombination of the aureolysin gene among Staphylococcus aureus strains. BMC Microbiol. 2008;8:129. doi: 10.1186/1471-2180-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, et al. (264 co-authors) Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoulas G, Eliopoulos GM, Moellering RC, Jr., Wennersten C, Venkataraman L, Novick RP, Gold HS. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother. 2002;46:1492–1502. doi: 10.1128/AAC.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass P, Bierbaum G. Native graS mutation supports the susceptibility of Staphylococcus aureus strain SG511 to antimicrobial peptides. Int J Med Microbiol. 2009;299:313–322. doi: 10.1016/j.ijmm.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Schroeder K, Jularic M, Horsburgh SM, et al. (11 co-authors) Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS One. 2009;4:e7567. doi: 10.1371/journal.pone.0007567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieprawska-Lupa M, Mydel P, Krawczyk K, et al. (15 co-authors) Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob Agents Chemother. 2004;48:4673–4679. doi: 10.1128/AAC.48.12.4673-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tetteh KK, Stewart LB, Ochola LI, Amambua-Ngwa A, Thomas AW, Marsh K, Weedall GD, Conway DJ. Prospective identification of malaria parasite genes under balancing selection. PLoS One. 2009;4:e5568. doi: 10.1371/journal.pone.0005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K, Andolfatto P. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics. 2006;172:1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A, Melles DC, Nouwen J, van Leeuwen WB, van Wamel W, Vos MC, Wertheim HFL, Verbrugh HA. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol. 2009;9:32–47. doi: 10.1016/j.meegid.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Verdier I, Reverdy ME, Etienne J, Lina G, Bes M, Vandenesch F. Staphylococcus aureus isolates with reduced susceptibility to glycopeptides belong to accessory gene regulator group I or II. Antimicrob Agents Chemother. 2004;48:1024–1027. doi: 10.1128/AAC.48.3.1024-1027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, Blanco-Garcia A, Hutter S, Rozas J. VariScan: analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics. 2005;21:2791–2793. doi: 10.1093/bioinformatics/bti403. [DOI] [PubMed] [Google Scholar]
- Wang IN, Dykhuizen DE, Qiu W, Dunn JJ, Bosler EM, Luft BJ. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics. 1999;151:15–30. doi: 10.1093/genetics/151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildschutte H, Lawrence JG. Differential Salmonella survival against communities of intestinal amoebae. Microbiology. 2007;41:10095–10104. doi: 10.1099/mic.0.2006/003616-0. [DOI] [PubMed] [Google Scholar]
- Wilson JN, Rockett K, Keating B, Jallow M, Pinder M, Sisay-Joof F, Newport M, Kwiatkowski D. A hallmark of balancing selection is present at the promoter region of interleukin 10. Genes Immun. 2006;7:680–683. doi: 10.1038/sj.gene.6364336. [DOI] [PubMed] [Google Scholar]
- Wiuf C, Zhao K, Innan H, Nordborg M. The probability and chromosomal extent of trans-specific polymorphism. Genetics. 2004;168:2363–2372. doi: 10.1534/genetics.104.029488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JS, Traber KE, Corrigan R, Benson SA, Musser JM, Novick RP. The agr radiation: an early event in the evolution of staphylococci. J Bacteriol. 2005;187:5585–5594. doi: 10.1128/JB.187.16.5585-5594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.