Abstract
Although human bitter taste perception is hypothesized to be a dietary adaptation, little is known about genetic signatures of selection and patterns of bitter taste perception variability in ethnically diverse populations with different diets, particularly from Africa. To better understand the genetic basis and evolutionary history of bitter taste sensitivity, we sequenced a 2,975 bp region encompassing TAS2R38, a bitter taste receptor gene, in 611 Africans from 57 populations in West Central and East Africa with diverse subsistence patterns, as well as in a comparative sample of 132 non-Africans. We also examined the association between genetic variability at this locus and threshold levels of phenylthiocarbamide (PTC) bitterness in 463 Africans from the above populations to determine how variation influences bitter taste perception. Here, we report striking patterns of variation at TAS2R38, including a significant excess of novel rare nonsynonymous polymorphisms that recently arose only in Africa, high frequencies of haplotypes in Africa associated with intermediate bitter taste sensitivity, a remarkably similar frequency of common haplotypes across genetically and culturally distinct Africans, and an ancient coalescence time of common variation in global populations. Additionally, several of the rare nonsynonymous substitutions significantly modified levels of PTC bitter taste sensitivity in diverse Africans. While ancient balancing selection likely maintained common haplotype variation across global populations, we suggest that recent selection pressures may have also resulted in the unusually high level of rare nonsynonymous variants in Africa, implying a complex model of selection at the TAS2R38 locus in African populations. Furthermore, the distribution of common haplotypes in Africa is not correlated with diet, raising the possibility that common variation may be under selection due to their role in nondietary biological processes. In addition, our data indicate that novel rare mutations contribute to the phenotypic variance of PTC sensitivity, illustrating the influence of rare variation on a common trait, as well as the relatively recent evolution of functionally diverse alleles at this locus.
Keywords: adaptive evolution, genotype–phenotype association, African genetic diversity, excess of rare mutations
Introduction
The ability to taste the bitter antithyroid compound phenylthiocarbamide (PTC) is a highly variable trait in humans, and a large fraction of the phenotypic variance in PTC perception has been attributed to genetic variation at TAS2R38, a bitter taste receptor gene, located on chromosome 7 (Drayna 2005). To date, studies have identified five polymorphisms at this gene, three of which cause amino acid substitutions (at positions 49, 262, and 296 in the TAS2R38 receptor protein) that form two common amino acid haplotypes in non-Africans: a dominant PAV “taster” haplotype and a recessive AVI “nontaster” haplotype which confer PTC bitter taste sensitivity and insensitivity, respectively (Kim et al. 2004; Campbell and Tishkoff 2008; Tepper 2008).
Haplotype variation at TAS2R38 is also associated with the ability to taste naturally occurring bitter substances and dietary preference. For example, in a study of bitter taste perception of food, PAV homozygotes rated vegetables containing glucosinolate (a natural bitter tasting compound present in cruciferous vegetables, such as broccoli, watercress, and kale) as 60% more bitter than AVI homozygotes, whereas PAV/AVI heterozygotes assigned intermediate scores of bitterness to most glucosinolate-generating vegetables (Sandell and Breslin 2006). Furthermore, an analysis of food selection in an Italian population showed that AVI/AVI nontaster homozygotes consumed more cruciferous vegetables than individuals carrying a single copy of the PAV taster haplotype (Sacerdote et al. 2007; Garcia-Bailo et al. 2009). Thus, genetic variation at this locus has an effect on human bitter taste perception, resulting in a range of responses to dietary sources of bitter compounds (Kim et al. 2004; Sandell and Breslin 2006; El-Sohemy et al. 2007; Garcia-Bailo et al. 2009; Peyrot des Gachons et al. 2009). Recent data also showed an association between TAS2R38 haplotype variation and overeating (Dotson et al. 2010). These studies suggest that genetic variation at TAS2R38 may have important implications for food acceptance, food intake, and long-term health (Drewnowski and Rock 1995; Garcia-Bailo et al. 2009).
Although PTC bitter taste perception is a classical polymorphic trait, it has never previously been studied in a large set of ethnically diverse populations with different diets. A prior genetic analysis of TAS2R38 in Africans (consisting of 31 individuals predominantly from Cameroon) and non-Africans (consisting of 134 individuals) detected signatures of balancing selection, including intermediate frequencies of the PAV taster and AVI nontaster haplotypes, and a low level of genetic differentiation among global populations (Wooding et al. 2004). The authors further suggest that patterns of diversity at this locus may be selectively advantageous as a means to detect and avoid bitter toxins in food (Wooding et al. 2004). However, the small number of Africans included in this prior study did not preclude the possibility that ethnically diverse Africans with different diets and high levels of genetic substructure may show distinct patterns of variability and signatures of selection. Furthermore, there have not previously been studies of PTC taste perception variability and genotype–phenotype associations in indigenous African populations.
Because of the high levels of genetic substructure in Africa (Tishkoff et al. 2009) and the evidence for local adaptation in response to diet (Tishkoff et al. 2007), we hypothesized that ethnically diverse African populations practicing diverse modes of subsistence (such as hunting/gathering, pastoralism, agriculture, and agro-pastoralism) may have different frequencies of genetic variants at TAS2R38 due to selection pressure resulting from highly diverse diets. In order to test this hypothesis, we examined nucleotide sequence variation within the single coding exon of the TAS2R38 gene and additional flanking regions in 611 Africans from 57 populations and in 132 geographically diverse non-Africans. We also examined genotype–phenotype associations in 463 Africans from the above populations to determine how variation in Africa influences bitter taste perception. Here, we report striking signatures of selection at TAS2R38, including a significant excess of novel rare nonsynonymous polymorphisms in African populations, high frequencies of haplotypes in Africa associated with intermediate taste sensitivity, remarkably similar common haplotype frequencies in divergent African populations, and an ancient coalescence time of variation (>2.1 My) in globally diverse populations. Furthermore, several of the rare polymorphisms significantly modified the ability to detect PTC bitterness in Africans. This study, the largest to date on African genetic diversity at the TAS2R38 locus, serves as a model system for understanding the influence of natural selection, as well as common and rare amino acid variation, on normal variable traits in humans.
Materials and Methods
Population Samples
The African samples in this study consisted of 611 individuals originating from 57 populations in West Central Africa (Cameroon) and East Africa (Kenya and Tanzania) that practice diverse modes of subsistence (such as hunting–gathering, pastoralism, and agriculture) and a comparative set of 132 non-Africans from the Middle East, Europe, South Asia, East Asia, and the Americas (supplementary table S1, Supplementary Material online). African populations that shared genetic similarity as well as cultural and/or linguistic properties (e.g., Pygmy hunter–gatherers, Khoesan-speaking hunter–gatherers, and Niger–Kordofanian speakers) were pooled for analysis in the present study based on a recent analysis of population structure in Africa (Tishkoff et al. 2009).
Institutional Review Board approval for this project was obtained from the University of Maryland at College Park and the University of Pennsylvania, and written informed consent was obtained from all African participants. Research/ethics approval and permits were obtained from the following institutions prior to sample collection: The Commission for Science and Technology and the National Institute for Medical Research in Dar es Salaam, Tanzania; the Kenya Medical Research Institute in Nairobi, Kenya; the Ministry of Health and National Committee of Ethics, Cameroon.
Phenotype Determination
Levels of PTC bitter taste sensitivity were measured for 463 individuals from the above sampled populations originating from Cameroon and Kenya using a modification of the classic recognition threshold method described by Harris and Kalmus (1951). Specifically, the test was administered to subjects using serial dilutions of PTC in vials labeled 13, 11, 9, 7, 5, 3, and 1, with vial 13 containing the most dilute solution of PTC and vial 1 containing the most concentrated solution. Raw PTC score represents the point in the successive dilution taste test at which individuals were able to detect the bitter taste of PTC. Raw PTC scores were adjusted for age and sex using the equation outlined in supplementary materials and methods(Supplementary Material online).
DNA Sequencing
A 2,975 bp region, encompassing the TAS2R38 gene (1,002 bp) and additional noncoding regions flanking either side of the gene (1,973 bp), was amplified by polymerase chain reaction (supplementary fig. S1; primers are listed in supplementary materials and methods, Supplementary Material online). All nucleotide sequence data were generated using the 3730xl automated sequencer (Applied Biosystems).
Haplotype Analysis
The program PHASE v.2.1, which implements a Bayesian statistical method, was used (Stephens et al. 2001) to reconstruct multisite haplotypes from sequence data for subsequent analyses. Haplotype names are assigned based on the amino acids present at positions 49, 262, and 296 in the TAS2R38 receptor protein (e.g., PAV has proline–alanine–valine present at positions 49, 262, and 296, respectively). The genealogical relationships among haplotypes were constructed using the median-joining algorithm implemented in the Network 4.5 program (Bandelt et al. 1999). The resulting topology was the tree with the minimum number of changes among all possible maximum parsimony trees from a given data set.
Tests of Genotype–Phenotype Association
We used least-squares linear regression and the nonparametric Kruskal-Wallis method to test for significant genotype–phenotype associations (for both raw PTC scores and PTC scores adjusted for age and sex) (supplementary materials and methods, Supplementary Material online). Using a linear regression approach, we also measured the within-locus dominance effect of the PAV haplotype on PTC response in our sample. Dominance is defined as the deviation of the heterozygote phenotype value from the mean value of the two homozygote phenotype values. If d = 0, there is no dominance and the locus is said to show additive effects; d is positive if the PAV haplotype is dominant and negative if the non-PAV haplotype (e.g., AAV, AAI, or AVI) is dominant (supplementary materials and methods, Supplementary Material online) These analyses were performed using the SPSS statistical package.
Functional Rare Variants
The effect of rare nonsynonymous amino acid substitutions on PTC response was assessed using a General Linear Model Analysis of Variance (GLM ANOVA) adjusting for age and sex in 463 Africans. Specifically, we compared mean raw PTC score of two groups: 1) individuals homozygous for two common haplotypes without rare amino acid substitutions (defined only by amino acid variation at positions 49, 262, and 296) and 2) individuals who were either homozygous or heterozygous for haplotypes containing low-frequency nonsynonymous amino acid substitutions, referred to as subhaplotypes, within each genotypic class (e.g., PAV/PAV and AAI/AVI represent genotypic classes). Statistical significance was determined after applying a Bonferroni correction for multiple comparisons in the GLM ANOVA analysis. A nonparametric Mann–Whitney test was also performed to confirm statistically significant results using the GLM ANOVA. The above analyses were completed using SPSS (supplementary materials and methods, Supplementary Material online).
To determine possible functional effects of amino acid changes, we used the Polyphen-2 algorithm (http://genetics.bwh.harvard.edu/pph2/) to predict which nonsynonymous substitutions change protein function. PolyPhen-2 infers whether an amino acid change is “benign,” “possibly damaging,” or “probably damaging” based on evolutionary and structural information (Ramensky et al. 2002).
Age Estimates of Mutations
The GENETREE program (Griffiths 2007) was used to estimate the expected ages of mutations and the expected time since the most recent common ancestor (TMRCA) for the coding region polymorphisms. This method is based on the coalescent process and assumes an infinite alleles model and no recombination. Haplotypes and sites that were not compatible with the infinite-sites model (which were six singleton haplotypes) were removed from the analysis. Each simulation produced an ordering of coalescent and mutation events. The age of mutations and the TMRCA of the gene tree were also estimated for each run (Griffiths 2007). The expected age was estimated from the weighted average of the simulated ages over different independent runs (supplementary materials and methods, Supplementary Material online).
Population Differentiation
Estimates of FST were computed as the mean number of differences between different pairs of sequences within the same subpopulation (West Central Africans, East Africans, and non-African were defined as subpopulations), and the mean number of differences between pairs of sequences randomly sampled from different subpopulations using the program DNAsp 5.10 (Hudson et al. 1992; Librado and Rozas 2009). FST was also calculated for the three common single nucleotide polymorphisms (SNPs) associated with PTC sensitivity using the program FSTAT 2.9.3.2 (Goudet 2002). These FST estimates were then compared to genome-wide estimates of FST for HapMap Phase III SNP data in the SNP@Evolution database (Cheng et al. 2009).
Tests of Neutrality
A McDonald–Kreitman test, which compares the ratio of nonsynonymous and synonymous mutations within and between species, was performed in African and non-African populations (McDonald and Kreitman 1991). We used three individuals belonging to Pan troglodytes (the common chimpanzee) for the between-species comparison downloaded from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) and Ensembl (http://uswest.ensembl.org) databases. Tajima's D (DT) was also calculated to test for a departure from neutrality based on the comparison of θW (the number of segregating sites) and θπ (pairwise nucleotide divergence) in African and non-African populations (McDonald and Kreitman 1991). Under neutrality, π and θW are expected to be equal assuming constant population size (Tishkoff and Verrelli 2003). In addition, a sliding window analysis of DT and θπ was performed to identify regions within the 2,975 bp sequence that depart from neutral expectation. Sliding window length varied between 50 and 100 sites, and the step size was 25. All tests of neutrality and the statistical significance of these tests were computed under the assumption of constant population size in DNAsp 5.10 (Librado and Rozas 2009).
Coalescent Simulations of Population Demography
Using the ms program (Hudson 1990), statistical significance of observed DT values was also determined by simulating a range of demographic scenarios of population growth (supplementary table S7, Supplementary Material online). In Africans, we simulated a 10-, 15-, 20-, and 40-fold increase in population size (from an initial population size of 10,000 individuals) starting at 70 kya until the present. In non-Africans, we simulated several demographic models involving a population bottleneck (from an initial population size of 10,000 individuals and decreasing to 3,000 individuals) at 60,000 years ago, corresponding to the approximate time modern humans migrated out of Africa, followed by a 10-, 20-, and 40-fold increase in population size starting at 50 kya until the present (supplementary table S13, Supplementary Material online).
Results
Patterns of Nucleotide Diversity
Nucleotide diversity statistics indicate higher levels of pairwise differences per nucleotide between sequences (π) in the coding region (1.57 × 10−3, 1.62 × 10−3, and 1.56 × 10−3 in Cameroonian, East African, and non-African populations) relative to the noncoding flanking regions in all populations (supplementary table S5, Supplementary Material online). A total of 61 SNPs were identified in the coding and flanking regions in global populations (supplementary table S2, Supplementary Material online). Furthermore, African populations had the largest total number of SNPs (54, supplementary table S3, Supplementary Material online) compared with non-Africans (13). While prior studies identified no more than five SNPs within the coding region of TAS2R38, we uncovered 21 SNPs, producing 21 haplotypes predominantly in African populations (supplementary fig. S2, Supplementary Material online). Of these 21 SNPs, 19 were nonsynonymous polymorphisms that caused amino acid substitutions (supplementary table S4, Supplementary Material online). The majority of these nonsynonymous SNPs were specific to African populations, and occurred at very low frequency (12 nonsynonymous SNPs at <1% frequency in the pooled African data set). Only three nonsynonymous polymorphisms previously associated with PTC taste sensitivity (i.e., nucleotides G145C, T785C, and A886G at amino acid positions 49, 262, and 296, respectively) were observed at moderate to intermediate frequency in our worldwide samples (31.1–47.6%). In addition, these common nonsynonymous SNPs consistently occurred at high frequency in diverse African populations regardless of geographic origin or mode of subsistence (supplementary fig. S3, Supplementary Material online).
Haplotype Variation
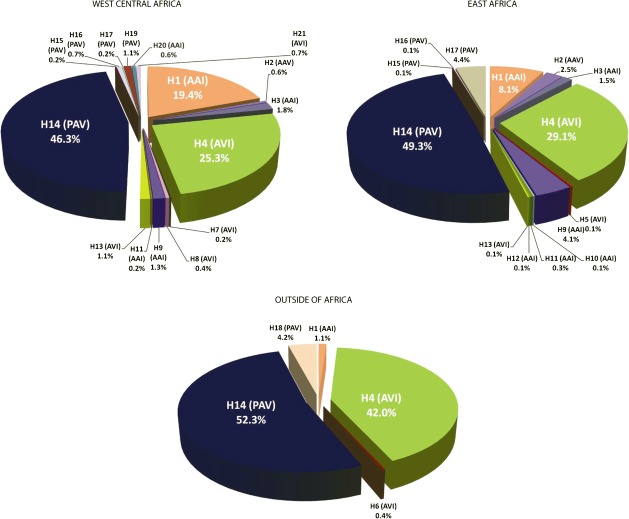
Amino acid haplotype frequencies for African and non-African populations are shown in figure 1. Non-Africans have two main haplotypes, the taster PAV and nontaster AVI haplotypes, which together account for 99% of sampled chromosomes. In contrast, among Africans, in addition to the high-frequency PAV and AVI haplotypes, we also found the “intermediate” AAI haplotype at moderate to high frequency (between 14% and 23%) and the “intermediate” AAV haplotype at lower frequency (∼1% and 2.5% in West Central and East Africa, respectively) (fig. 1). A low-frequency AAI haplotype (1%) was also found in a few individuals from the Middle East and Sardinia, regions with low levels of recent gene flow from Africa (Ehret 2002) (supplementary table S6C, Supplementary Material online). We also identified a number of novel PAV, AVI, and AAI subhaplotypes, defined by rare polymorphic sites (most of which are nonsynonymous) in addition to the three common functional SNPs, primarily in African populations (supplementary table S6A–C, Supplementary Material online; fig. 1). These subhaplotypes constitute 8.5%, 11.5%, and 4.5% of total haplotype variation in Cameroonian, East African, and non-African populations, respectively.
FIG. 1.
Haplotype and subhaplotype frequencies in African and non-African populations. PAV, AVI, AAI, and AAV haplotypes are listed within or in close proximity to each section of the pie charts. The alphanumeric designations outside of parentheses correspond to the list of haplotypes in supplementary table S4 and supplementary table S6 (Supplementary Material online). The common amino acid haplotypes associated with PTC sensitivity are as follows: H14(PAV), H4(AVI), H1(AAI), and H2(AAV). The other haplotypes, referred to as subhaplotypes, are defined by rare variants present on common haplotype backgrounds.
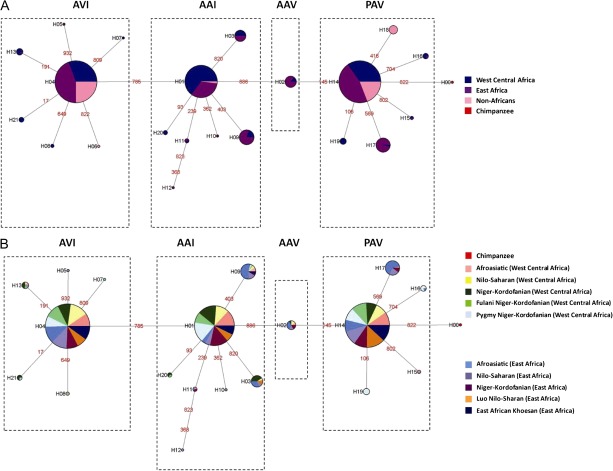
To examine the genealogical relationships among amino acid haplotypes, median-joining networks were constructed (Bandelt et al. 1999) (fig. 2A). These genealogies indicate that haplotype diversity grouped into four distinct clades, with the PAV and AVI haplotype clades separated by a number of mutational steps. Furthermore, the AAI and AAV clades were located intermediately between the divergent PAV and AVI haplotype clades. Notably, common haplotypes were observed at similar frequencies in diverse African populations that practice different modes of subsistence or live in different environments (fig. 2B; supplementary fig. S3 and tables S6A and S6B, Supplementary Material online), indicating that common haplotype variation is not correlated with geographic region or diet. The low-frequency amino acid subhaplotypes in Africa also showed a “star-like” pattern of variation, typically radiating from a common PAV, AAI, or AVI haplotype by single mutational steps. The absence of reticulations among haplotypes in these networks suggests that little recombination has occurred in the coding exon of TAS2R38. Pairwise linkage disequilibrium plots also showed strong allelic associations between the three common functional SNPs within the coding region of this gene (supplementary fig. S4, Supplementary Material online).
FIG. 2.
Networks of inferred haplotype relationships based on global and African data. Genealogical relationships of haplotypes were constructed using a maximum parsimony approach for (A) global populations and (B) West Central Africa and East Africa. Circles represent amino acid haplotypes, and the size of the circles is proportional to the number of chromosomes with a given haplotype. Colors within each haplotype represent the proportion of individuals within a population or geographic region with that given haplotype. The lines and numbers are the mutational changes between haplotypes.
Genotype–Phenotype Association
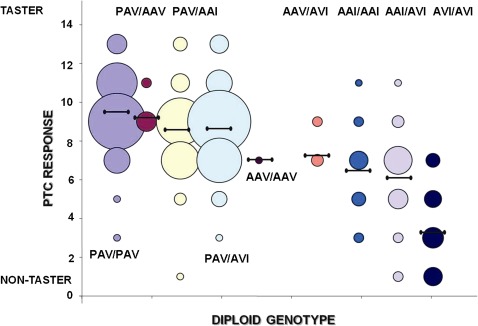
Recognition threshold levels for tasting PTC bitterness were measured for 463 individuals from linguistically and culturally diverse populations originating from Cameroon and Kenya. A plot of phenotype scores for the diploid haplotype genotypes (diplotypes) defined by the three common variants at positions 49, 262, and 296 is shown in figure 3. Our results indicate that PAV taster homozygotes possess a greater sensitivity to PTC compared with AVI nontaster homozygotes (mean of 9.51 ± 1.66 and 3.36 ± 2.11, respectively; fig. 3) consistent with prior studies (Bufe et al. 2005; Kim and Drayna 2005). Furthermore, there are a number of diplotypes, such as AAI/AVI, AAV/AAV, AAI/AAI, and AAV/AVI, which are associated with intermediate PTC sensitivity in Africans (mean of 6.32 ± 2.06 for pooled genotypes; fig. 3). Overall, the distribution of PTC bitter taste phenotypes is unimodal in African populations in contrast to previous studies that described PTC taste sensitivity as a bimodal trait in non-African populations (supplementary fig. S5, Supplementary Material online).
FIG. 3.
A plot of the range of raw phenotype scores (PTC response) for each genotypic class in 463 African individuals. The y axis is PTC response, which is the PTC score of individuals tested in the field. PTC response ranges from 13 to 1, with score 13 representing high PTC sensitivity and score 1 representing low PTC sensitivity. Genotypes are listed in close proximity to each column of bubbles, and the size of bubbles is proportional to the number of individuals with a given PTC score. Horizontal lines indicate the mean PTC score for each genotypic class.
We used both linear regression and the nonparametric Kruskal–Wallis method to test for correlation between diplotype and bitter taste phenotypes in 463 individuals. We observed a significant association (P < 0.001 for both methods), indicating that diplotype at TAS2R38 is a strong predictor of PTC response in Africans (supplementary tables S7 and S8, Supplementary Material online). However, the regression analyses indicate that genetic variability at this locus explained ∼35% of the variance in PTC sensitivity, suggesting that other genetic, epigenetic, and/or environmental factors likely contribute to PTC bitter taste perception in African populations. Based on a linear regression model incorporating additive (a) and dominance (d) parameters and PTC phenotypes adjusted for age and sex, we also estimated the dominance effect of the PAV haplotype in Africans. We found that the positive dominance coefficient (d = 1.105) was significantly different (P < 0.001) from the null hypothesis of d = 0, indicating that PAV is dominant (relative to non-PAV haplotypes) with PAV heterozygotes having a larger mean phenotypic value than expected under a model of additivity (supplementary table S9, Supplementary Material online).
Functional Rare Variants
To assess whether or not rare amino acid variants also influence PTC phenotypic variance, we compared mean PTC score in two groups: 1) individuals homozygous for two common haplotypes (defined by amino acid variants at positions 49, 262, and 296) and 2) individuals who were either homozygous or heterozygous for a low-frequency nonsynonymous subhaplotype within each genotypic class in figure 3 using a GLM ANOVA adjusting for age and sex (table 1). This analysis indicated that the difference in mean PTC score between groups with and without rare nonsynonymous variants in the AAI/AVI genotypic class was statistically significant (P < 0.001) after Bonferroni correction (the group with rare subhaplotypes had a lower mean score), suggesting an effect of rare amino acid haplotypes on PTC bitter taste sensitivity. This between-group difference in PTC score within the AAI/AVI genotype class was also significant using the nonparametric Mann–Whitney test (table 1). For other genotype classes, particularly those without a dominant PAV haplotype, individuals with rare nonsynonymous subhaplotypes had a lower mean PTC score compared with individuals with two common haplotypes, although the difference in mean score was not statistically significant (table 1).
Table 1.
Phenotypic Effects of Rare Nonsynonymous Variants in African Populations.
| Genotype A | Genotype B | Mean Difference (Genotype A − Genotype B) | Standard Error | Significance (Exact P Value) |
| AVI/AVI | At least one AVI subhaplotype | 0.90 | 1.77 | 0.614 |
| N = 25 | N = 3 | |||
| Mean PTC score = 3.350 | Mean PTC score = 2.447 | |||
| AAI/AVI | At least one AAI or AVI subhaplotype | 0.000 (P < 0.001) *** | ||
| N = 28 | N = 7 | 3.33 | 0.82 | Mann–Whitney (P = 0.005) ** |
| Mean PTC score = 6.788 | Mean PTC score = 3.459 | |||
| AAI/AAI | At least one AAI subhaplotype | 1.97 | 1.31 | 0.162 |
| N = 7 | N = 9 | |||
| Mean PTC score = 7.607 | Mean PTC score = 5.640 | |||
| AAV/AVI | At least one AVI subhaplotype | N/A | N/A | N/A |
| N = 1 | N = 0 | |||
| AAV/AAV | At least one AAV subhaplotype | N/A | N/A | N/A |
| N = 4 | N = 0 | |||
| PAV/AVI | At least one AVI or PAV subhaplotype | 0.22 | 0.49 | 0.659 |
| N = 134 | N = 18 | |||
| Mean PTC score = 8.572 | Mean PTC score = 8.356 | |||
| PAV/AAI | At least one PAV or AAI subhaplotype | 0.17 | 0.46 | 0.517 |
| N = 62 | N = 31 | |||
| Mean PTC score = 8.430 | Mean PTC score = 8.260 | |||
| PAV/AAV | At least one PAV subhaplotype | N/A | N/A | N/A |
| N = 9 | N = 0 | |||
| PAV/PAV | At least one PAV subhaplotype | 0.580 | 0.41 | 0.207 |
| N = 106 | N = 19 | |||
| Mean PTC Score = 9.580 | Mean PTC score = 9.000 |
Note.—In the GLM ANOVA adjusting for age and sex in 463 African individuals, raw PTC response was entered as the dependent variable; fixed variables consisted of two comparative groups: 1) individuals homozygous for two common haplotypes within a given genotypic class and 2) individuals who were either homozygous or heterozygous for a subhaplotype containing rare nonsynonymous variants within the same genotypic class. In this model, age and sex were included as covariates. A Bonferroni correction for multiple comparisons was applied, and only significant results are shown as underlined. Significant P values, P < 0.01 and P < 0.001, for the ANOVA and Mann–Whitney tests are indicated by ** and ***, respectively. N/A means the ANOVA could not be applied to the data.
We next examined the predicted functional effects of nonsynonymous substitutions using the Polyphen-2 algorithm, which infers whether or not amino acid changes are “benign,” “possibly damaging,” or “probably damaging.” We identified five low-frequency AAI or AVI subhaplotypes (H9–H13 in supplementary table S4, Supplementary Material online) that have amino acid changes predicted to be “possibly damaging” or “probably damaging” (at amino acid positions 64, 80, 121, and 135; corresponding nucleotides are 191, 239, 362, and 403, respectively, in supplementary table S4, Supplementary Material online). Two of these subhaplotypes (H9 and H13, which contain “damaging” amino acids at positions 135 and 64, respectively) comprised 17% of total haplotypes in the AAI/AVI genotypic class, likely contributing to the significant difference in mean PTC score between groups with and without rare nonsynonymous variation. The AVI and AAI subhaplotypes (H9–H13 in supplementary table S4, Supplementary Material online) also exist in a heterozygous state with the common PAV haplotype but do not have a significant influence on PTC taste sensitivity (table 1), due to the dominance effect of the PAV haplotype. The four “damaging” substitutions at amino acid positions 64, 80, 121, and 135 are located in the second transmembrane, second extracellular, and second cytoplasmic domains of the TAS2R38 protein (supplementary table S10, Supplementary Material online), pointing to regions of the bitter taste receptor that may affect function other than those containing the common amino acid variants. These “damaging” substitutions were found both in West Central and East African populations.
Age Estimates of TAS2R38 Variants
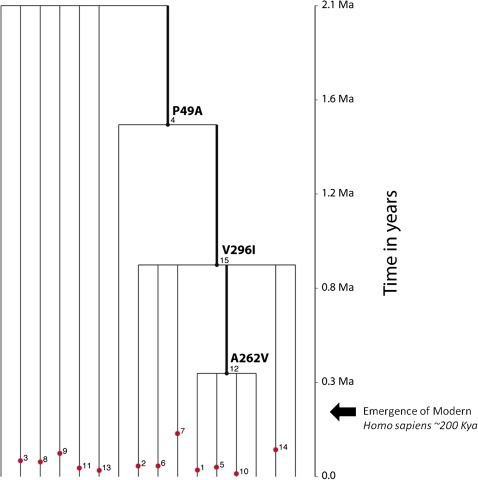
A gene tree was constructed for the global data set, and the TMRCA and ages of nucleotide polymorphisms were estimated using the program GENETREE (Griffiths 2007). This method yielded a mean TMRCA of 2.1 My, with a standard deviation of ±454,546 years (fig. 4; supplementary table S11, Supplementary Material online), which is among one of the oldest age estimates for autosomal loci (Blum and Jakobsson 2011). Our GENETREE analysis also indicated that the common variants associated with PTC bitter taste sensitivity are quite ancient, predating the origin of anatomically modern humans ∼200 kya. Specifically, the estimated mean ages of polymorphisms corresponding to amino acid sites 49, 262, and 296 were 1.3 million ± 242,211, 336,000 ± 89,845, and 1.0 million ± 267,268 years old, respectively. The lower frequency variants, including those that are associated with decreased PTC sensitivity, appear to be much younger in age, occurring within the last 200,000 years (supplementary table S11, Supplementary Material online).
FIG. 4.
Inferred coalescent gene tree of TAS2R38 nucleotide variation. Common mutations are represented by black dots and rare mutations are indicated by red dots. The ages of polymorphisms are mean estimates of TMRCA with standard deviations listed in supplementary table S11 (Supplementary Material online). Key mutations defining amino acid haplotype clades are numbered. The scale on the right represents time in years (Ma = millions of years ago) and the arrow represents the point at which modern humans evolved (kya = thousands of years ago). Mutation numbers (in small print) assigned by the GENETREE program are given within the tree and correspond to the mutation numbers in supplementary table S11 (Supplementary Material online).
Population Differentiation
Genetic differentiation at TAS2R38 among global populations was estimated based on FST analysis. The global FST value was estimated to be 0.012, a value much lower than previous estimates of FST in global populations, which typically range from 0.10 to 0.16 (Campbell and Tishkoff 2008), suggesting a low level of genetic differentiation between geographically diverse populations. The global FST values for common SNPs at amino acid positions 49, 262, and 296 were 0.004, 0.019, and 0.006, respectively, which represent FST outliers, falling in the low end (within the third percentile) of the empirical genome-wide distribution of FST values derived from HapMap Phase III SNP data. These unusually low FST values are consistent with a scenario of balancing selection maintaining similar allele frequencies across populations.
Tests of Neutrality
To test whether or not patterns of variation observed at TAS2R38 fit a neutral model of evolution, the McDonald–Kreitman test was used to compare the ratio of nonsynonymous and synonymous mutations within and between species (McDonald and Kreitman 1991). Our results showed a statistically significant (P < 0.05 for East African and pooled African populations) or borderline significant (P = 0.062 in Cameroon) excess of nonsynonymous substitutions in Africa (supplementary table S12, Supplementary Material online). The ratio of nonsynonymous to synonymous substitutions in non-Africans, however, did not differ significantly from neutral expectation (P = 0.56) (supplementary table S12, Supplementary Material online).
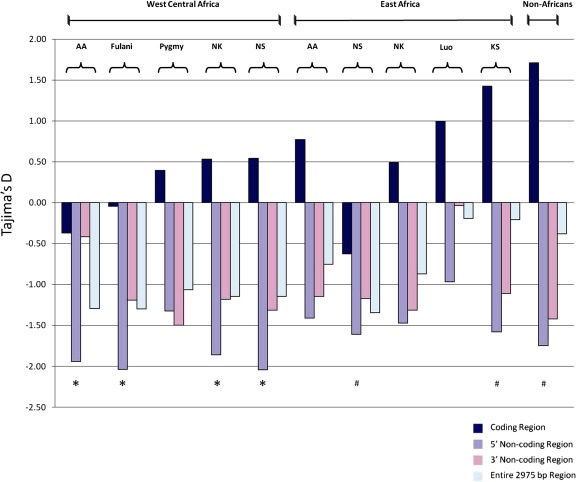
Estimates of DT were also calculated for 1) the coding exon of TAS2R38, 2) the 5′ and 3′ flanking noncoding regions, individually, and 3) the entire 2,975 bp region (supplementary table S5, Supplementary Material online). Our data revealed a strikingly different pattern of variation in the noncoding and coding regions of the gene (fig. 5). Specifically, the 5′ noncoding region showed consistently negative DT values that were statistically significant (P < 0.05) or marginally significant (0.05 < P < 0.1) in both African and non-African populations (fig. 5; for more detailed information on tested populations see supplementary table S5, Supplementary Material online). DT was also negative in the 3′ noncoding region but did not reach statistical significance (fig. 5; supplementary table S5, Supplementary Material online). In contrast, DT values were positive in 7 out of 10 African populations and in non-Africans, and DT was consistently more positive in the coding region relative to the noncoding flanking regions in all populations, particularly for the Hadza/Sandawe hunter–gatherers (DT = 1.42) and non-Africans (DT = 1.71) (fig. 5; supplementary table S5, Supplementary Material online), although these values did not significantly deviate from neutral expectation under the assumption of constant population size (fig. 5). However, because ancestral humans have increased in population size within the last 100,000 years (Marth et al. 2004; Voight et al. 2005), which can lead to negative DT values, departure from neutral expectation in the coding region was also tested under different demographic scenarios of population growth using coalescent simulations (Hudson et al. 1992) (supplementary table S13, Supplementary Material online). We found that observed DT values in the coding region were higher than expected, differing significantly from neutrality (P < 0.025) under scenarios of moderate population growth in both African and non-African populations (supplementary table S13, Supplementary Material online).
FIG. 5.
Tajima's D statistic for coding and noncoding regions in Africans and non-Africans. African populations that share genetic and linguistic similarity were grouped together as follows: Niger–Kordofanian speakers (NK), Nilo-Saharan speakers (NS), Afroasiatic speakers (AA), and Khoesan speakers (KS). Some genetically distinct populations within language families were classified as separate groups (e.g., Fulani Niger–Kordofanian speakers, Pygmy Niger–Kordofanian speakers, and Luo Nilo-Saharan speakers) based on a recent genetic study (Tishkoff et al. 2009). Statistically significant (P < 0.05) DT values are denoted by (*) and marginally significant (0.05 < P < 0.1) DT values are denoted by (#).
To further explore patterns of variation at TAS2R38, we also performed a sliding window analysis of DT and π across the entire 2,975 bp region that showed positive peaks in DT, corresponding to SNPs encoding substitutions at positions 49, 262, and 296 in the coding exon in all populations. These positive peaks were marginally significant at positions 49, 262, and 296 in non-Africans (0.05< P < 0.1) and were marginally significant (0.05 < P < 0.1) or statistically significant (P < 0.05) at positions 49 and 296 in African populations (supplementary fig. S6, Supplementary Material online), indicating deviations from neutrality at common variable sites associated with PTC sensitivity in diverse populations.
Discussion
Genotype–Phenotype Associations
Africans have higher levels of genetic and phenotypic diversity at the TAS2R38 locus relative to non-Africans. In particular, in Africa, we observed high frequencies of the PAV and AVI haplotypes common in non-Africans. However, in Africa, we also observed common AAV and AAI haplotypes, which are rare outside of Africa, as well as the presence of rare subhaplotypes, which have not previously been identified in human populations. Furthermore, our genotype–phenotype analysis of 463 Africans uncovered a relatively large number of individuals with intermediate PTC sensitivity (associated with AAV and AAI haplotypes) rarely observed outside of Africa, in addition to individuals with high and low PTC sensitivity, resulting in a unimodal distribution of bitter taste phenotypes. In contrast, previous studies of bitter taste perception have typically reported a bimodal distribution of PTC phenotypes in non-Africans (Kim et al. 2003, 2004; Prodi et al. 2004; Wooding et al. 2004). Thus, modern African populations have a wider range of bitter taste sensitivity than is observed outside of Africa. We also identified rare AAI subhaplotypes that were associated with a significant decrease in PTC sensitivity in Africa, indicating that functional rare variants present on common haplotype backgrounds can modify the genotype–phenotype relationship at the TAS2R38 locus, and contribute to the phenotypic variance in bitter taste sensitivity. Thus, the genetic architecture underlying bitter taste sensitivity is more complex than previously known.
Selection Maintaining Common Variation in Global Populations
Although we hypothesized that diverse African populations might show different patterns of variation at TAS2R38 due to local adaptation to diet, this is not what we found. Instead, we observed remarkably similar common haplotype frequencies in diverse African populations with divergent diets, a low level of genetic differentiation among global populations, and an ancient TMRCA of genetic variation at TAS2R38, which are indicative of long-term balancing selection (Tishkoff and Verrelli 2003). Furthermore, these genetic patterns are very different from what we have observed using neutral loci genotyped in many of the same African populations, which show high levels of substructure (Tishkoff et al. 2009). Overall, our results are in agreement with a prior genetic study (Wooding et al. 2004) that detected signatures of balancing selection at this locus predominantly in non-African populations. However, while this previous analysis (Wooding et al. 2004) showed that two main haplotypes occur at high frequency in worldwide populations due to balancing selection, our data indicate that several haplotypes are maintained at relatively high frequency by selection across diverse African populations. Our data further suggest that common amino acid variants at positions 49, 262, and 296 are the likely targets of balancing selection. In particular, our sliding window analyses showed strong deviations from neutrality at sites 49 and 296 in diverse populations, consistent with a scenario of balancing selection. Although prior in vitro studies have indicated that amino acid substitutions at sites 49 and 262 have a greater effect on PTC sensitivity than site 296 (Bufe et al. 2005; Miguet et al. 2006), our sliding window results suggest that variation at position 296 is functionally important in vivo. This finding is further supported by a recent analysis of bitter taste perception that showed that variation at position 296 is associated with increased sensitivity to the bitter compound 6-n-propylthiouracil which is chemically similar to PTC (Mennella et al. 2011). The fact that we observe few recombinant haplotypes, even in Africa, suggests that the combination of particular amino acids at multiple sites is crucial for function.
Wooding et al. (2006) demonstrated that the ability to taste the antithyroid compound PTC is an ancestral trait in both humans and chimpanzees and that variants associated with decreased sensitivity evolved independently in these species after their divergence ∼5–6 Ma. The TMRCA of nucleotide polymorphisms, assuming neutrality, at sites 49, 262, and 296 in our data set (∼1.3 million ± 242,211, 336,000 ± 89,845, and 1.0 million ± 267,268 years old, respectively) suggests that common variants influencing PTC sensitivity arose within the Homo lineage prior to the emergence of anatomically modern humans. The widespread occurrence of common haplotypes in globally diverse populations also indicates that common variation at TAS2R38 predates the evolution of modern Homo sapiens. Although a recent study inferred that the PAV haplotype is fixed in South African Khoesan speakers (SAK) based primarily on genotype information from common variants at positions 262 and 296 (Schuster et al. 2010), our sequence analysis of Pygmy populations and the East African Khoesan-speaking Hadza and Sandawe, which share ancient common ancestry with SAK (Tishkoff et al. 2007, 2009), showed the presence of PAV, AAI, and AVI common haplotypes, confirming the existence of genetic variability among highly divergent hunter–gatherer populations. The ancient age of common nucleotide variation is further supported by the recent finding of the P49A variant in a Neanderthal individual (Lalueza-Fox et al. 2009), implying that this polymorphism was present in the common ancestor of modern humans and Neandertals (Green et al. 2010). The deep coalescence time (TMRCA) for the origin of variation at TAS2R38 inferred in the current study is among the most ancient reported for autosomal loci (Kreitman and Di Rienzo 2004; Blum and Jakobsson 2011), which is consistent with long-term balancing selection maintaining common variation at this locus.
The persistence of common haplotypes over time and their widespread distribution across globally diverse populations suggest that these variants are functionally important. Indeed, the dominant PAV taster haplotype is hypothesized to have played a role in detecting potentially toxic substances, particularly antithyroid compounds, during hominid evolution. However, the selective force maintaining common AAV, AAI, and AVI haplotypes for extraordinarily long periods of time remains unclear. Although both AAV and AAI are associated with intermediate bitter taste sensitivity, the AAI haplotype is more common in Africa than AAV. Intriguingly, the AAV haplotype may represent a “stepping stone” to other more advantageous haplotype variation, such as AAI and AVI. We suggest that common PAV, AAI, and AVI haplotype variation may be maintained at high frequencies in response to selective pressures unrelated to diet. Indeed, recent studies have shown that bitter taste receptors are expressed in a variety of cell types in the human gastrointestinal tract (Rozengurt and Sternini 2007) and lungs (Shah et al. 2009; Deshpande et al. 2010), where they influence insulin and glucose levels (Dotson et al. 2008), eliminate harmful inhaled substances (Shah et al. 2009), and stimulate the relaxation of airways for improved breathing (Deshpande et al. 2010). These studies demonstrate that bitter taste loci have a number of different functions and raise the possibility that common variants at TAS2R38 may be under selection due to their physiological roles in human health beyond oral gustatory function. Though we cannot conclusively distinguish the selective forces maintaining common variation at TAS2R38, it is clear that genetic variation at this locus is diverse and has been functionally important long before modern Homo sapiens existed.
Excess of Rare Nonsynonymous Variation in Africa
We observed an excess of low-frequency polymorphisms in the 5′ and 3′ noncoding regions of TAS2R38, leading to negative DT values. This skew toward rare variants is consistent with a scenario of either positive directional selection or weak purifying selection (Campbell and Tishkoff 2008). We also observed a significant excess of rare nonsynonymous polymorphisms in the coding region, which contributed to slightly lower DT values in African relative to non-African populations. Although studies have shown that weak purifying selection (Bustamante et al. 2005; Campbell and Tishkoff 2008) or a relaxation of functional constraint (Harding et al. 2000) can lead to an excess of rare polymorphisms, these evolutionary scenarios cannot explain the unusually high level of rare nonsynonymous variants in Africa because we would expect a higher proportion of variants to be synonymous (given that a third of the possible mutational sites in the coding exon are expected to result in synonymous substitutions). Additionally, the population bottleneck event during the geographic expansion of modern humans from Africa within the last 100,000 years (Campbell and Tishkoff 2010) may have resulted in a loss of haplotype variation and an even more positive DT value in the coding region in non-Africans.
Our findings suggest the possibility that the excess of rare amino acid variation in Africa may result from recent selective pressures favoring an increase in the number of nonsynonymous mutations (relative to synonymous variation) on common haplotypes, which have been maintained by balancing selection. We hypothesize that the high levels of rare nonsynonymous substitutions may be selectively advantageous in nondietary physiological processes or that there has been recent selection for diminished taste sensitivity, as has been observed in studies of olfaction (Niimura and Nei 2003; Gimelbrant et al. 2004). Indeed, we observed a general trend of lower PTC sensitivity among individuals with rare amino acid substitutions compared with individuals homozygous for common haplotypes. In particular, several rare nonsynonymous variants, inferred as damaging, on AAI and AVI haplotypes were associated with a significant decrease in PTC bitter taste sensitivity. However, it is also possible that the evolution of these nonsynonymous mutations may represent an alternative strategy for generating diverse receptors within the human species that participate in a number of biological activities. We suggest that a complex model of selection, involving ancient balancing and recent diversifying selection, has maintained common and rare nonsynonymous variation, respectively, in the coding exon of TAS2R38 in Africa, implying a different selective history of rare variants at this locus in Africans compared with non-African populations. In addition, different types of selection may have operated in the noncoding regions compared with the coding exon of TAS2R38 in all populations which could have been facilitated by recombination between these genomic regions (supplementary fig. S4, Supplementary Material online).
To date, the genetic basis of complex traits, including disease susceptibility, is still poorly understood. It has been argued that alleles underlying complex diseases are relatively common, having a large effect on complex traits (the common disease/common variants hypothesis). Alternatively, it has also been suggested that common traits are influenced by rare susceptibility alleles at many loci each with small effect. Although recent studies have shown that rare alleles indeed contribute to medically relevant traits, such as lipoprotein or sterol levels (Cohen et al. 2004, 2006), our genetic analysis of PTC bitter taste sensitivity demonstrates that common and rare variants together can have a significant effect on normal phenotypic variation. In the future, studies of genotype–phenotype associations at other TAS2R bitter taste loci in diverse populations, including a wide range of ethnically distinct Africans, will help further clarify the contribution of both common and rare variation to bitter taste perception as well as improve our understanding of a trait that has been important in the evolution of the modern human lineage.
Supplementary Material
Supplementary materials and methods, figures S1–S6, and tables S1–S13 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Fathya Abdo, Eva Aluvalla, Ángel Carracedo, Graham Coop, Clara Elbers, Joseph Jarvis, Daniel Kariuki, Wen-Ya Ko, Joseph Lachance, Charla Lambert, Mingyao Li, Trini Miguel, Lilian Alando Nyindodo, Laura Scheinfeldt, Mark Shriver, Shamil Sunyaev, and Stephen Wooding for sample collection, valuable discussions, and/or statistical advice. We also thank the many Africans who generously donated their DNA and time so that we could learn more about the genetic basis and evolutionary history of bitter taste sensitivity in Africa. This study was funded by the US National Science Foundation (NSF) grant BCS-0552486, US NSF grant BCS-0827436, US National Institutes of Health (NIH) grant R01GM076637, US NIH Director's Pioneer Award Program DP1-OD-006445, and a David and Lucile Packard Career Award to S.A.T and a US NIH grant RO1 DC02995 to P.A.S.B.
References
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Blum MG, Jakobsson M. Deep divergences of human gene trees and models of human origins. Mol Biol Evol. 2011;28:889–898. doi: 10.1093/molbev/msq265. [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, et al. (14 co-authors). 2005. Natural selection on protein-coding genes in the human genome. Nature. 43:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA. The evolution of human genetic and phenotypic variation in Africa. Curr Biol. 2010;20:R166–R173. doi: 10.1016/j.cub.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Chen W, Richards E, Deng L, Zeng C. SNP@Evolution: a hierarchical database of positive selection on the human genome. BMC Evol Biol. 2009;9:221. doi: 10.1186/1471-2148-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande DA, Wang WC, McIlmoyle EL, Robinett KS, Schillinger RM, An SS, Sham JS, Liggett SB. Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction. Nat Med. 2010;16:1299–1304. doi: 10.1038/nm.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson CD, Shaw HL, Mitchell BD, Munger SD, Steinle NI. Variation in the gene TAS2R38 is associated with the eating behavior disinhibition in Old Order Amish women. Appetite. 2010;54:93–99. doi: 10.1016/j.appet.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson CD, Zhang L, Xu H, et al. (14 co-authors) Bitter taste receptors influence glucose homeostasis. PLoS One. 2008;3:e3974. doi: 10.1371/journal.pone.0003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayna D. Human taste genetics. Annu Rev Genomics Hum Genet. 2005;6:217–235. doi: 10.1146/annurev.genom.6.080604.162340. [DOI] [PubMed] [Google Scholar]
- Drewnowski A, Rock CL. The influence of genetic taste markers on food acceptance. Am J Clin Nutr. 1995;62:506–511. doi: 10.1093/ajcn/62.3.506. [DOI] [PubMed] [Google Scholar]
- Ehret C. The civilizations of Africa: a history to 1800. Charlottesville (VA): University of Virginia Press; 2002. [Google Scholar]
- El-Sohemy A, Stewart L, Khataan N, Fontaine-Bisson B, Kwong P, Ozsungur S, Cornelis MC. Nutrigenomics of taste—impact on food preferences and food production. Forum Nutr. 2007;60:176–182. doi: 10.1159/000107194. [DOI] [PubMed] [Google Scholar]
- Garcia-Bailo B, Toguri C, Eny KM, El-Sohemy A. Genetic variation in taste and its influence on food selection. OMICS. 2009;13:69–80. doi: 10.1089/omi.2008.0031. [DOI] [PubMed] [Google Scholar]
- Gimelbrant AA, Skaletsky H, Chess A. Selective pressures on the olfactory receptor repertoire since the human–chimpanzee divergence. Proc Natl Acad Sci U S A. 2004;101:9019–9022. doi: 10.1073/pnas.0401566101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. Fstat 2.9.3.2. [updated 2005 Aug 23; cited 2011 Jun 22] 2002 Available from: http://www2.unil.ch/popgen/softwares/fstat.htm. [Google Scholar]
- Green RE, Krause J, Briggs AW, et al. (56 co-authors) A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RC. Genetree v. 9.0. 2007. [cited 2011 Jun 22]. Available from: http://www.stats.ox.ac.uk/∼griff/software.html. [Google Scholar]
- Harding RM, Healy E, Ray AJ, et al. (11 co-authors) Evidence for variable selective pressures at MC1R. Am J Hum Genet. 2000;66:1351–1361. doi: 10.1086/302863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H, Kalmus H. The distribution of taste thresholds for phenylthiourea of 384 sib pairs. Ann Eugen. 1951;16:226–230. doi: 10.1111/j.1469-1809.1951.tb02475.x. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Gene genealogies and the coalescent process. Oxford: Oxford University Press; 1990. [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim UK, Breslin PA, Reed D, Drayna D. Genetics of human taste perception. J Dent Res. 2004;83:448–453. doi: 10.1177/154405910408300603. [DOI] [PubMed] [Google Scholar]
- Kim UK, Drayna D. Genetics of individual differences in bitter taste perception: lessons from the PTC gene. Clin Genet. 2005;67:275–280. doi: 10.1111/j.1399-0004.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Kreitman M, Di Rienzo A. Balancing claims for balancing selection. Trends Genet. 2004;20:300–304. doi: 10.1016/j.tig.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Lalueza-Fox C, Gigli E, de la Rasilla M, Fortea J, Rosas A. Bitter taste perception in Neanderthals through the analysis of the TAS2R38 gene. Biol Lett. 2009;5:809–811. doi: 10.1098/rsbl.2009.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Marth GT, Czabarka E, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Mennella JA, Pepino MY, Duke FF, Reed DR. Psychophysical dissection of genotype effects on human bitter perception. Chem Senses. 2011;36:161–167. doi: 10.1093/chemse/bjq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguet L, Zhang Z, Grigorov MG. Computational studies of ligand-receptor interactions in bitter taste receptors. J Recept Signal Transduct Res. 2006;26:611–630. doi: 10.1080/10799890600928210. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci U S A. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrot des Gachons C, Beauchamp GK, Breslin PA. The genetics of bitterness and pungency detection and its impact on phytonutrient evaluation. Ann N Y Acad Sci. 2009;1170:140–144. doi: 10.1111/j.1749-6632.2009.04594.x. [DOI] [PubMed] [Google Scholar]
- Prodi DA, Drayna D, Forabosco P, Palmas MA, Maestrale GB, Piras D, Pirastu M, Angius A. Bitter taste study in a sardinian genetic isolate supports the association of phenylthiocarbamide sensitivity to the TAS2R38 bitter receptor gene. Chem Senses. 2004;29:697–702. doi: 10.1093/chemse/bjh074. [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozengurt E, Sternini C. Taste receptor signaling in the mammalian gut. Curr Opin Pharmacol. 2007;7:557–562. doi: 10.1016/j.coph.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdote C, Guarrera S, Smith GD, et al. (13 co-authors) Lactase persistence and bitter taste response: instrumental variables and mendelian randomization in epidemiologic studies of dietary factors and cancer risk. Am J Epidemiol. 2007;166:576–581. doi: 10.1093/aje/kwm113. [DOI] [PubMed] [Google Scholar]
- Sandell MA, Breslin PA. Variability in a taste-receptor gene determines whether we taste toxins in food. Curr Biol. 2006;16:R792–R794. doi: 10.1016/j.cub.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Schuster SC, Miller W, Ratan A, et al. (48 co-authors) Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper BJ. Nutritional implications of genetic taste variation: the role of PROP sensitivity and other taste phenotypes. Annu Rev Nutr. 2008;28:367–388. doi: 10.1146/annurev.nutr.28.061807.155458. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, et al. (25 co-authors) The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Ranciaro A, et al. (19 co-authors) Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, Bufe B, Grassi C, Howard MT, Stone AC, Vazquez M, Dunn DM, Meyerhof W, Weiss RB, Bamshad MJ. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature. 2006;440:930–934. doi: 10.1038/nature04655. [DOI] [PubMed] [Google Scholar]
- Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.