Abstract
We have re-examined DNase I footprinting data for the binding of transcription factor IIIA (TFIIIA) to the 5S RNA gene, taking into account the protein-DNA contacts observed in the crystal structure of the DNase I/DNA complex (1, 2). This structure was not available when many of the original footprinting experiments on the TFIIIA/DNA complex were performed. In this way the pattern of DNase I cleavage can be interpreted to map out with greater precision the regions on the 5S DNA occupied by TFIIIA. Then, assuming the binding site for a zinc-finger may be the same as that found in the structure of the zinc-finger protein Zif268/DNA complex (3), and taking into account footprinting data for truncated forms of TFIIIA, the TFIIIA zinc-fingers were fitted within the permitted regions. On the basis of this, an alignment of the zinc-fingers of TFIIIA with its DNA binding site is proposed, which combines features of earlier models (4).
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg J. M. Zinc finger domains: hypotheses and current knowledge. Annu Rev Biophys Biophys Chem. 1990;19:405–421. doi: 10.1146/annurev.bb.19.060190.002201. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Christensen J. H., Hansen P. K., Lillelund O., Thøgersen H. C. Sequence-specific binding of the N-terminal three-finger fragment of Xenopus transcription factor IIIA to the internal control region of a 5S RNA gene. FEBS Lett. 1991 Apr 9;281(1-2):181–184. doi: 10.1016/0014-5793(91)80388-j. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Klug A. Mode of interaction of the zinc finger protein TFIIIA with a 5S RNA gene of Xenopus. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5528–5532. doi: 10.1073/pnas.87.14.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M., Rhodes D., Klug A. Location of the primary sites of micrococcal nuclease cleavage on the nucleosome core. J Mol Biol. 1983 Oct 25;170(2):423–446. doi: 10.1016/s0022-2836(83)80156-9. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Fairall L., Harrison S. D., Travers A. A., Rhodes D. Sequence-specific DNA binding by a two zinc-finger peptide from the Drosophila melanogaster Tramtrack protein. J Mol Biol. 1992 Jul 20;226(2):349–366. doi: 10.1016/0022-2836(92)90952-g. [DOI] [PubMed] [Google Scholar]
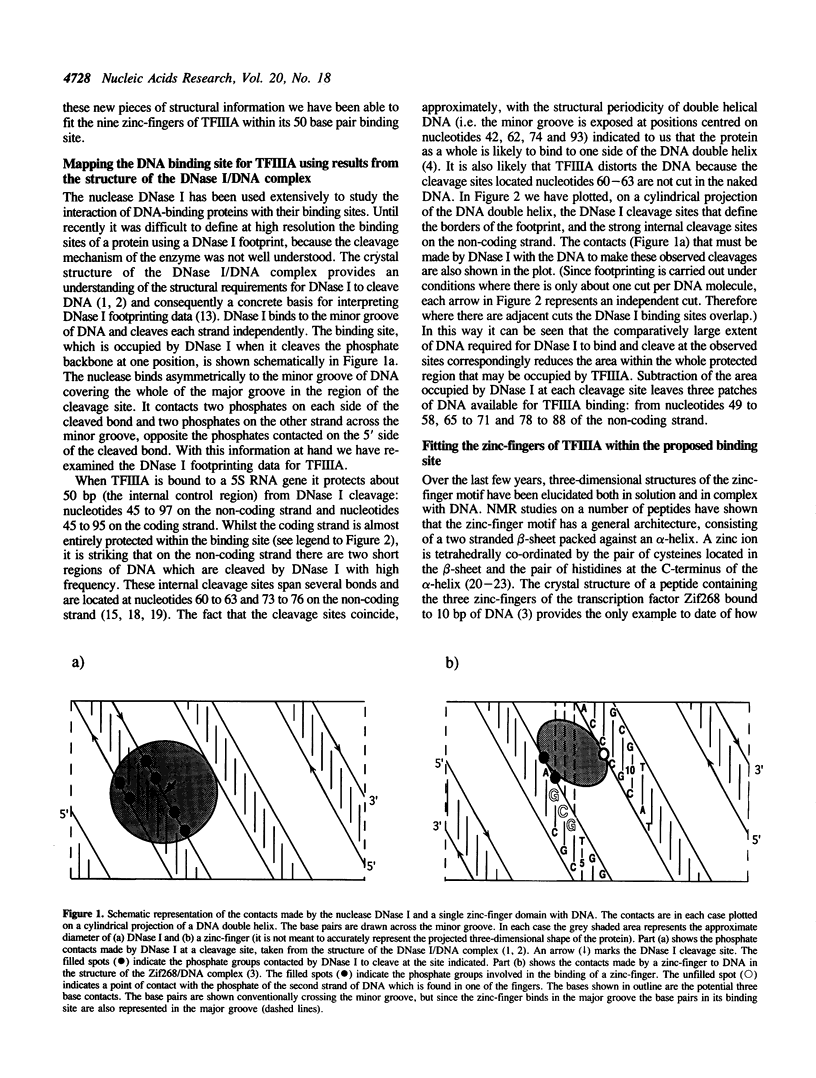
- Fairall L., Rhodes D., Klug A. Mapping of the sites of protection on a 5 S RNA gene by the Xenopus transcription factor IIIA. A model for the interaction. J Mol Biol. 1986 Dec 5;192(3):577–591. doi: 10.1016/0022-2836(86)90278-0. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Klevit R. E., Herriott J. R., Horvath S. J. Solution structure of a zinc finger domain of yeast ADR1. Proteins. 1990;7(3):215–226. doi: 10.1002/prot.340070303. [DOI] [PubMed] [Google Scholar]
- Kochoyan M., Havel T. F., Nguyen D. T., Dahl C. E., Keutmann H. T., Weiss M. A. Alternating zinc fingers in the human male associated protein ZFY: 2D NMR structure of an even finger and implications for "jumping-linker" DNA recognition. Biochemistry. 1991 Apr 9;30(14):3371–3386. doi: 10.1021/bi00228a004. [DOI] [PubMed] [Google Scholar]
- Lahm A., Suck D. DNase I-induced DNA conformation. 2 A structure of a DNase I-octamer complex. J Mol Biol. 1991 Dec 5;222(3):645–667. doi: 10.1016/0022-2836(91)90502-w. [DOI] [PubMed] [Google Scholar]
- Lee D. K., Evans R. K., Blanco J., Gottesfeld J., Johnson J. D. Contacts between 5 S DNA and Xenopus TFIIIA identified using 5-azido-2'-deoxyuridine-substituted DNA. J Biol Chem. 1991 Sep 5;266(25):16478–16484. [PubMed] [Google Scholar]
- Lee M. S., Gippert G. P., Soman K. V., Case D. A., Wright P. E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989 Aug 11;245(4918):635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- Liao X. B., Clemens K. R., Tennant L., Wright P. E., Gottesfeld J. M. Specific interaction of the first three zinc fingers of TFIIIA with the internal control region of the Xenopus 5 S RNA gene. J Mol Biol. 1992 Feb 20;223(4):857–871. doi: 10.1016/0022-2836(92)90248-i. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Nakaseko Y., Nasmyth K., Rhodes D. Zinc-finger motifs expressed in E. coli and folded in vitro direct specific binding to DNA. Nature. 1988 Mar 17;332(6161):284–286. doi: 10.1038/332284a0. [DOI] [PubMed] [Google Scholar]
- Nardelli J., Gibson T. J., Vesque C., Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991 Jan 10;349(6305):175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- Neuhaus D., Nakaseko Y., Nagai K., Klug A. Sequence-specific [1H]NMR resonance assignments and secondary structure identification for 1- and 2-zinc finger constructs from SW15. A hydrophobic core involving four invariant residues. FEBS Lett. 1990 Mar 26;262(2):179–184. doi: 10.1016/0014-5793(90)80184-k. [DOI] [PubMed] [Google Scholar]
- Omichinski J. G., Clore G. M., Appella E., Sakaguchi K., Gronenborn A. M. High-resolution three-dimensional structure of a single zinc finger from a human enhancer binding protein in solution. Biochemistry. 1990 Oct 9;29(40):9324–9334. doi: 10.1021/bi00492a004. [DOI] [PubMed] [Google Scholar]
- Omichinski J. G., Clore G. M., Robien M., Sakaguchi K., Appella E., Gronenborn A. M. High-resolution solution structure of the double Cys2His2 zinc finger from the human enhancer binding protein MBP-1. Biochemistry. 1992 Apr 28;31(16):3907–3917. doi: 10.1021/bi00131a004. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K., Appella E., Omichinski J. G., Clore G. M., Gronenborn A. M. Specific DNA binding to a major histocompatibility complex enhancer sequence by a synthetic 57-residue double zinc finger peptide from a human enhancer binding protein. J Biol Chem. 1991 Apr 15;266(11):7306–7311. [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Jackson I. J., Brown D. D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984 Jun;37(2):645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Thukral S. K., Eisen A., Young E. T. Two monomers of yeast transcription factor ADR1 bind a palindromic sequence symmetrically to activate ADH2 expression. Mol Cell Biol. 1991 Mar;11(3):1566–1577. doi: 10.1128/mcb.11.3.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana K. E., Churchill M. E., Tullius T. D., Brown D. D. Mapping functional regions of transcription factor TFIIIA. Mol Cell Biol. 1988 Apr;8(4):1684–1696. doi: 10.1128/mcb.8.4.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]


