Abstract
Nanopore analysis is an emerging single-molecule strategy for non-optical and high-throughput DNA sequencing, the principle of which is based on identification of each constituent nucleobase by measuring trans-membrane ionic current blockade or transverse tunnelling current as it moves through the pore. A crucial issue for nanopore sequencing is the fact that DNA translocates a nanopore too fast for addressing sequence with a single base resolution. Here we report that a transverse electric field can be used to slow down the translocation. We find 400-fold decrease in the DNA translocation speed by adding a transverse field of 10 mV/nm in a gold-electrode-embedded silicon dioxide channel. The retarded flow allowed us to map the local folding pattern in individual DNA from trans-pore ionic current profiles. This field dragging approach may provide a new way to control the polynucleotide translocation kinetics.
Nanopore sequencing is an emerging non-optical technology for high-throughput real-time single-molecule sequencing1,2,3. The basic idea is to identify each nucleobase by the size (or the electronic structure) through detecting a trans-pore ionic current blockade4,5,6,7,8 (or transverse tunnelling current3,9,10,11,12,13,14) during DNA translocation through a pore. A longstanding challenge for nanopore sequencing has been to slow down the flow speed of polynucleotides in the pore so as to achieve single-base spatial resolution6,7,8,9,10,11,12,13,14,15,16,17. While retarding of translocation and concomitant Ångstrom precision has recently been achieved in bioengineered nanopores by using a polymerase-DNA complex6, the system instability and limited pore-size selectivity of biological systems remains to be a critical issue for practical applications1. On the other hand, solid-state nanopores can serve as a robust and configurable single-molecule sensing platform. We present herein an electric-field dragging approach for retarding DNA translocation in a solid-state nanopore. We find that a transverse electric field of 10 mV/nm in an electrode-embedded silicon dioxide nanochannel slows down the biopolymer translocation velocity by more than two orders of magnitude. In addition, we observe field-induced unfolding of DNA. The results presented in this study suggest the usefulness of transverse field for providing essential conditions for “sequencing by tunnelling”: slow translocation of unfolded DNA through an electrode gap.
Results
We measured the ionic current flowing through an electrode embedded nanochannel sensor fabricated on a SiO2/Si wafer (Fig. 1a,b; Supplementary Fig. S and S21). Space between the nanoelectrodes defines the size of the fluidic channel, which is 200 nm × 50 nm × 60 nm (length (L) × width (w) × height (h)) (Fig. 1c). These electrodes were also used to exert a transverse field on biomolecules that electrophoretically passed through the channel. A patterned polydimethylsiloxane (PDMS) block was adhered on top of the nanostructure after treating the surface with oxygen plasma (Supplementary Fig. S3). For calibration of our fluidics device, 0.1 M KCl solution buffered with (HCl 10 mM, EDTA 1 mM) was injected, and ionic current across the fluidic channel Iion was measured under the bias voltage Vlong from −0.5 V to 0.5 V using two Ag/AgCl electrodes (Fig. 1d, blue plots)). The linear Iion−Vlong characteristics give ionic conductance of 20.7 nS, which agrees with a theoretical estimation e(μKnK+μClnCl)hw/L = 22.4 nS (dotted line in Fig. 1d), where μK(Cl) is the mobility of K+ (Cl−) ions and nK is the ion concentration. This suggests no leakage of fluid from the nanochannel. We also performed I–V measurements between the embedded Au nanoelectrodes (Fig. 1d, red plots). The transverse current Isens was smaller than 2 pA (below the resolution of our measurement system) over the range of transverse voltage Vtrans measured (see also Fig. S4). Two factors are presumably responsible for the low ionic conductance: the small Au surface area exposed to the solution, approximately 0.04 μm2, and the slow charge transfer kinetics at the Au electrode-solution interfaces18.
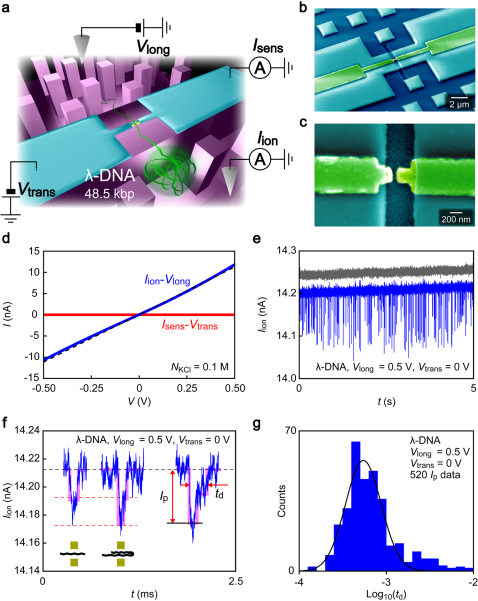
Figure 1. Electrical detection of DNA translocation in an electrode-embedded nanochannel sensor.
(a) Schematic illustration of an electrode-embedded nanochannel used for single-molecule detection of DNA translocation. 48.5 kbp λ-DNA dissolved in a KCl solution is electrophoretically drawn through the electrode channel. Meanwhile, the trans-channel ionic current blockade Iion and the transverse current Isens were measured simultaneously. (b) A false-colored scanning electron micrograph of the nanochannel sensor. A pair of nanoelectrodes (yellow) is used to apply a transverse field. Micropillar arrays at both sides of the electrode channel were used as a spacer for avoiding roof collapse upon channel sealing with a PDMS block. (c) A magnified view of the nanochannel/electrode structure. The electrode gap of size 50 nm × 60 nm × 200 nm (width × height × length) defines the fluidic channel dimension. (d) Ionic conductance of the electrode channel. The trans-channel ion current Iion measured in 0.1 M KCl solution increases nearly linearly with the driving voltage Vlong (blue plots), from which we obtained the channel conductance of 20 nS. The dotted line is the theoretical drift current flowing through the 50 nm channel. (e) A Iion trace acquired at a sampling rate of 1 MHz in a 10 nM λ-DNA solution under a driving voltage of Vlong = 0.5 V with no transverse field Vtrans (blue curve). Downward spike-like signals signify current blockade by polynucleotides passing through the electrode channel. Such characteristic current spikes were not observed in the control experiments conducted for the salt solution with no DNA added (gray curve). (f) A close view of a current spike, which is characterized by the magnitude of current blockade Ip and the translocation duration td. The black and red dotted lines denote the base level current and the characteristic blockade current of n × 14 pA (n = 1, 2) that correspond to translocation of (n - 1)-folded DNA, respectively. The purple lines are guide to the eyes for the current steps. (g) The distribution of td shown in logarithmic scale. The solid line is a Gaussian peak fitted to the histogram.
Detection of a single-molecule translocation event was examined by recording Iion in 0.1 M KCl solution containing 10 nM λ-DNA (48.5 kbp) under Vlong = 0.5 V at a sampling rate of 1 MHz with no transverse field. Spike-like current drops were observed that were indicative of a partial ion exclusion in the nanochannel upon DNA translocation (Fig. 1e)1,2,3,4. This was confirmed by the control measurements in a solution containing no DNA molecules in which we found only featureless Iion - t curves (Fig. 1e, gray plots). The frequency for observing current spikes, or equivalently the DNA capture rate, was about 20 Hz. Ionic current enhancement by the counteracting contribution of the counter charges of DNA molecules to the ion exclusion effect19 was not observed here. This is attributable to a substantial reduction of effective charge densities on DNA when the nanochannel size is considerably large compared to the polymer diameter15,19,20,21. Iion blockages are characterized by the amount of current blockage Ip and the spike width td (Fig. 1f). Ip reflects a volume of molecules that reside in the channel3,22. A rough estimation of the ionic current reduction by a volume exclusion for a stretched double-stranded DNA of 2 nm diameter in the channel yields Ip = 0.014 nA3. This information can be used to assess the molecular folding states as DNA with (n – 1) folds would produce a blockade current of n × 0.014 nA (Fig. 1f)3,22. We also deduce the average translocation speed from the td distribution that represents DNA residence time in the nanochannel (Fig. 1g). Typical td is about 0.5 ms, from which we obtain 97 bp/μs.
Incorporation of a transverse electric field is expected to cause a substantial change in the translocation kinetics by adding a lateral electrostatic force on the negatively charged polynucleotides in the nanochannel23,24,25. The Iion curve acquired under Vtrans = 0.5 V revealed DNA-associated channel blockade events at a relatively low frequency of 1.4 Hz (Fig. 2a). A slight increase in Iion base level from that at Vtrans = 0 V suggests a crosstalk between the trans-channel and the transverse ionic conductance26 (see also Supplementary Fig. S5).
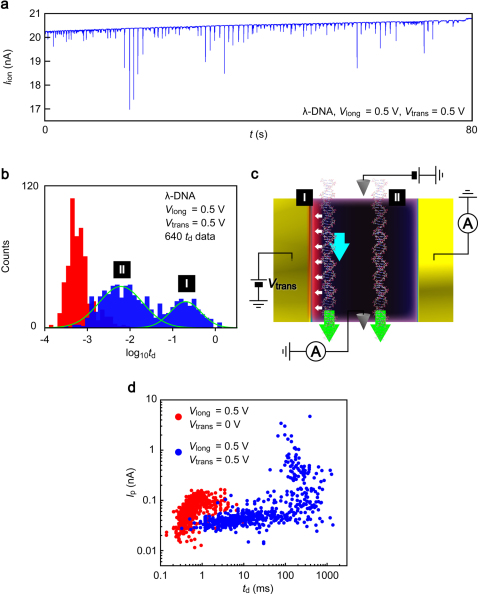
Figure 2. Transverse electric field induced dragging of DNA in a nanochannel.
(a) A Iion – t curve obtained in a 10 nM λ-DNA solution under Vlong = 0.5 V with the transverse voltage Vtrans = 0.5 V. Current blockades of various Ip and td were observed. The event frequency, or the DNA capture rate, was 1.4 Hz. (b) The trapping duration histogram constructed with 640 td data extracted from current blockade events detected at Vtrans = 0.5 V (blue). Data for Vtrans = 0 V are also displayed for comparison (red). td is shown in a logarithmic scale. Solid curves are the Gaussian fit to the histogram. (c) DNA translocation kinetics in an electrode-embedded nanochannel with a transverse field. Green arrows denote electrostatic forces on DNA with Vlong = 0.5 V applied along the channel. (I) Polynucleotides tends to be attracted to the positively charged electrode wall when enter the positive side of the channel (white arrows). In this case, strong affinity between nucleotides and gold induces large friction against DNA permeation that more than cancels the forward electroosmotic drag force (sky blue arrow). (II) On the other hand, many polynucleotide chains translocate at around center of the nanochannel as the transverse trapping field is screened in the electrolyte solution. (d) The blockade current Ip versus the translocation duration td scatter plots for Vtrans = 0 V (red) and Vtrans = 0.5 V (blue). The constant Ip band at around 30 pA that extends for td > 100 ms is characteristic of molecular translocation events involving transverse-field-induced DNA-electrode interaction26. In addition, the Ip ∝ td relation at Vtrans = 0 V cannot be explained by the constant event charge deficit model19. This indicates that DNA-electrode interaction already plays a role on the translocation at Vtrans = 0 V25.
To shed light on the transverse field effects on DNA translocation, a td histogram is constructed for the Iion – t curve obtained at Vtrans = 0.5 V (Fig. 2b). We find two peaks at 6 ms and 200 ms, which correspond to the translocation speed of 8.1 bp/μs and 0.24 bp/μs, respectively; the latter indicates more than two orders of magnitude reduction from the DNA velocity at Vtrans = 0 V. One possible explanation for the bimodal distribution is the existence of two distinct molecular pathways in the nanochannel with anti-parallel electroosmotic flow (Fig. 2c and Fig. S7). Negatively charged polynucleotides are anticipated to be attracted to the positive side of the embedded electrodes once a part of the strand approaches the electrode surface via Brownian motion, and are forced to flow within a confined nanospace defined by the screening length in 0.1 M KCl solution along the channel wall where there is forward electroosmotic dragging that facilitates the translocation15,24,25 (Fig. 2c, I). The polymer is also given an increased chance to experience large friction against the electrophoretic driving force via the electrically-induced Au electrode-DNA interactions20,27,28,29. This transverse field induced resistance overwhelms the anionic electroosmotic dragging considering the strong nonspecific affinity of DNA to Au11,27,28,29,30 as well as the electrostatic interaction between the DNA and the positively charged electrode, and causes the orders of magnitude decrease in the translocation velocity. On the other hand, the spatially confined trapping field allows large number of polynucleotide chains to electrophoretically flow at center of the nanochannel (Fig. 2c, II). These two hydrodynamic mechanisms account for the two distinct td states observed in Fig. 2b. Further theoretical and experimental insights are required to identify the actual DNA translocation dynamics in the nanochannel.
The role of the transverse field for reducing a DNA translocation speed is clearly shown by the scattering plot in Fig. 2d. It is noticeable that there is a region where Ip is almost constant while td varies extensively for the blockade events at Vtrans = 0.5 V. The low-Ip band was also observed in the recent nanopore experiments, which was explained by a cause of polynucleotide-pore interactions8,16. This further implies a contribution of field-induced Au-DNA interaction on the long translocation times. It is also inferred from the figure that the transverse field has another function to unfold DNA as large portion of the plots at Vtrans = 0.5 V are clustered at around the non-folded Ip level of 20 pA.
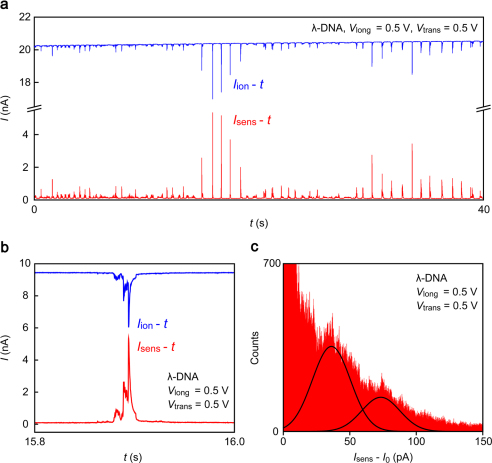
The DNA translocation events were simultaneously detected by Isens measurements (Fig. 3a). Iion and Isens display a synchronous variation where the former is blocking and the latter is enhancing13 (Fig. 3b, see also Supplementary Fig. S8). Moreover, the histogram of Isens demonstrates two broad peaks at integer multiples of 36 pA (Fig. 3c) that may correspond to transverse ionic conductance states in the channel occupied with zero- and one-fold DNA (Supplementary Fig. S9).
Figure 3. Crosstalk between trans-channel and transverse ionic pathways.
(a) The transverse current Isens concurrently measured with Iion under Vlong = 0.5 V and Vtrans = 0.5 V conditions. The background current is about 100 pA. Positive pulse-like signals were found instead of current blockades. (b) Direct comparison of the Iion blockade and simultaneously detected Isens spike. The line shapes of these curves are almost identical to each other though the sign is opposite. c, A current histogram built with the Isens - t curve. The base level I0 is subtracted from Isens. Solid curves are Gaussian distributions centered at 36 pA and 73 pA.
Discussion
The positive Isens transients are obviously too large for tunnelling current flowing through the 50 nm gap. Instead, it is attributable to the n-folded ion density in the charge-induced electric double layer of DNA with n folds31 that provides increased number of current-carrying ions across the transverse electrodes. Nevertheless, it is also probable that interaction between Iion and Isens demonstrated by the non-negligible influence of the trans-channel electric field on the steady-state transverse current (Supplementary Fig. S6) may also be a cause of the concomitant rise of the transverse current considering the very similar line shapes of Iion and Isens signals27. While the ability of the transverse field to slow down DNA translocation may greatly facilitates achieving sequencing by tunnelling current, the crosstalk between the trans-channel and transverse ion passages needs to be avoided, as the weak tunnelling current signatures, typically lower than 100 pA, would be buried in the transmitted ionic current blockade signals. However, the smaller the nanochannel or nanopore becomes, the lower the ionic conductance. The ion conductivity is also adjustable by the salt concentrations. Therefore, the coupling issue is expected to be resolved by employing appropriate experimental conditions to minimize the ionic current contributions to the transverse tunnelling components.
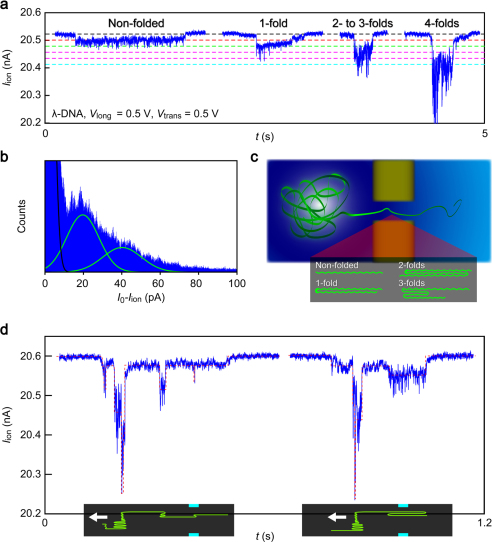
The field-retarded translocation improves the spatial resolution of nanopore detection. We observed Iion signals with long plateaus and steps at a multiple integer level of 20 pA (Fig. 4a). In fact, we find a pronounced peak and a shoulder in addition to the base level peak at zero in a histogram constructed with the Iion trace after subtracting the quasi-linear background current (Fig. 4b). The peak profile can be fitted with two Gaussian distributions at 20 pA and 41 pA (red curves in Fig. 4b). This peculiar feature indicates the existence of distinct DNA conformations. The magnitude of current blockage is roughly given by the channel versus molecule volume ratio3. For completely stretched double-stranded DNA of diameter 2 nm, this ratio is 0.1%. As the base level current is in a range of 20 to 21 nA, the blockade should be as large as 20 pA. When DNA is folded one time, the effective volume inside the channel doubles and so does the blockade current3,32. By analogy, the characteristic current states at 20 × n pA (n = 1, 2, 3 ···) can thus be assigned to DNA molecules with (n – 1) folds (Fig. 4a,c)32. This enables the identification of the local folding states within an individual polynucleotide chain by referring to the plateau current levels (Fig. 4d). Fluorescence imaging of stained DNA in a nanochannel has been reported to give similar results33. Nonetheless, the real-time single-molecule profiling capability demonstrated here is not only an electric analogue of fluorescence microscopy but with better time resolution. With further efforts to clarify the influence of the transverse field on DNA conformations and current blockages, it may find applications as a useful tool for studying atomistic biomolecule dynamics in a confined nanospace.
Figure 4. Real time electrical identification of local folds in a single-molecule DNA.
(a) Magnified views of long events with plateaus at 20 pA × n (n = 1, 2, 3 ···) that correspond to translocation of DNA with n-folds through the channel. (b) A current histogram constructed with the Iion trace. The background current I0 is subtracted from Iion. Black line is a fit to the pronounced peak at 0 A that originates from the base level current with 10 pA noise. Green lines are the Gaussian fits to the two peaks observed at 20 pA and 41 pA. These current states are attributed to ion blockade by non- and single-folded DNA in the channel. (c) Schematic illustration depicting translocation of DNA having some folds. (d) Examples of identification of local folds in an individual DNA by the Iion landscape. The schematic models describe DNA topologies assessed from the Iion profiles. Red dotted lines are guide to the eyes for Iion steps at integer multiples of 20 pA. White arrows indicate direction of DNA flow through a nanochannel.
Methods
Fabrication of electrode-embedded nanochannels
A 0.5 mm thick 30 mm square piece of substrate was cut from a silicon wafer coated with a 700 nm thick thermally grown SiO2 layer. Microelectrodes were patterned on the SiO2 surface by using a photolithography method with a photo-resist AZ5206E. Subsequently, Pt (2 nm)/Au (30 nm)/Pt (2 nm) layer was deposited by a radio-frequency (RF) magnetron sputtering. The thin Pt films served as adhesion layers. The sample was then immersed in a N,N-dimethylformamide for 8 hours and sonicated for several minutes for lift-off. Following the preparation of microscale electrodes, we delineated a pair of nanoelectrodes using an electron beam (EB) lithography technique with a resist ZEP520A-7 utilizing L-shaped pattern as alignment markers. After that, Pt (2 nm)/Au (30 nm)/Pt (2 nm)/SiO2 (30 nm) multi-layered film was deposited by the RF sputtering. As a result, we obtained an electrode gap of size 200 nm × 64 nm × 50 nm (length × height × width) after the lift-off process. Furthermore, a two-leveled fluidic channel was fabricated by first depositing SiO2 (30 nm)/Cr (100 nm) layer on the substrate. Arrays of squares were then patterned at the both sides of the nanoelectrodes using the EB lithography processes. An etching mask was obtained by wet-etching the exposed Cr region in a in a diluted hydrochloric acid solution of ceric ammonium nitrate (15 wt%). The sample was exposed to deep isotropic reactive ion etching (CF4, 100 W) to etch out SiO2. By removing the residual Cr layer, we acquired micro-pillars of 500 nm height. These posts were used as a spacer for circumventing roof collapse during PDMS sealing. Next, we again deposited Cr (25 nm) and made a mask for a nanochannel using the RF sputtering deposition, EB lithography, and wet etching processes. Finally, the substrate was dry-etched to form a 400 nm channel with 60 nm height. The fabrication scheme is summarized in Supplementary Fig. S2.
Nanochannel sealing
SU-8 photo-resist was spin-coated on a Si wafer and baked at 95°C for 45 minutes. An SU-8 mold for making PDMS fluidic channels was then obtained by using a photolithography method. After that, PDMS (SYLGARD184) was cured on the mold at 70°C for 1 hour. As a result, a PDMS block with two grooves of 0.4 mm width formed at the bottom was acquired. Between the grooves had a gap of size 0.2 mm, which was used to seal the electrode channel structure formed on a Si substrate. The sealing was implemented by treating the surface of the sample substrate and the PDMS block with oxygen plasma for activation and subsequently attached to each other to form permanent bonds.
Ionic current measurements
A tris-HCl 10 mM, EDTA 1 mM buffer solution was prepared with Milli-Q water (Milli-pore). We used this to obtain a 0.1 M KCl solution of pH = 8.0. For single-molecule translocation detection, 48.5 kbp λ-DNA (Takara Bio) was added at a concentration of 10 nM. The salt solution was introduced into the channel via holes drilled in the PDMS block attached on the sample. Ag/AgCl electrodes (BAS) inserted into additional two holes at both sides of the channel were used to measure the ionic current flowing through the electrode channel. Two cables were connected to the electrode pads for monitoring transverse currents across the nanoelectrodes. Measurements of the trans-channel and the transverse ionic current were performed by using a home-built preamplifier and a digitizer PXI-5922 (National Instruments) at a sampling rate of 1 MHz.
Author Contributions
M. Tsutsui, M. Taniguchi, and T. K. planned and designed experiments. M. Tsutsui and S. R. fabricated electrode-embedded nanochannel samples. M. Tsutsui and M. Taniguchi exhibited ionic current measurements. M. Tsutsui, Y. H., and M. F. performed data analyses. M. Tsutsui, M. Taniguchi, and T. K. co-wrote paper.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science (JSPS) through its “Funding Program for World-Leading Innovative R&D on Science and Technology”.
References
- Dekker C. Solid-state nanopores. Nat. Nanotechnol. 2, 209–215 (2007). [DOI] [PubMed] [Google Scholar]
- Branton D. et al. The potential and challenges of nanopore sequencing. Nat. Biotech. 26, 1146–1153 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwolak M. & Di Ventra M. Colloquim: Physical approaches to DNA sequencing and detection. Rev. Mod. Phys. 80, 141–165 (2008). [Google Scholar]
- Kasianowicz J. J., Brandin E., Branton D. & Deamer D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Nat. Acad. Sci. 93, 13770–13773 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J., Wu H.-C., Jayasinghe L., Patel A., Reid S. & Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 4, 265–270 (2009). [DOI] [PubMed] [Google Scholar]
- Lieberman K. R., Cherf G. M., Doody M. J., Olasagasti F., Kolodji Y. & Akeson M. Processive replication of single DNA molecules in a nanopore catalysed by phi29 DNA polymerase. J. Am. Chem. Soc. 132, 17961–17972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G. F. et al. DNA translocation through graphene nanopores. Nano Lett. 10, 3163–3167 (2010). [DOI] [PubMed] [Google Scholar]
- Garaj S., Hubbard W., Reina A., Kong J., Branton D. & Golovchenko J. A. Graphene as a subnanometre trans-electrode membrane. Nature 467, 190–193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerqvist J., Zwolak M. & Di Ventra M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 6, 779–782 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. & Chou S. Y. Nanogap detector inside nanofluidic channel for real-time label-free DNA analysis. Nano Lett. 8, 1472–1476 (2008). [DOI] [PubMed] [Google Scholar]
- Tsutsui M., Taniguchi M., Yokota K. & Kawai T. Identifying single nucleotides by tunnelling current. Nat. Nanotechnol. 5, 286–290 (2010). [DOI] [PubMed] [Google Scholar]
- Huang S. et al. Identifying single bases in a DNA oligomer with electron tunnelling. Nat. Nanotechnol. 5, 868–873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A. P. et al. DNA tunneling detector embedded in a nanopore. Nano Lett. 11, 279–285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui M., Sakon R., Iizumi Y., Okazaki T., Taniguchi M. & Kawai T. Single-molecule sensing electrode embedded in-plane nanopore. Sci. Rep. 1, 46 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser U. F. et al. Direct force measurements on DNA in a solid-state nanopore. Nat. Phys. 2, 473–477 (2006). [Google Scholar]
- Fologea D., Uplinger J., Thomas B., McNabb D. S. & Li J. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 5, 1734–1737 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal S. M., Akin D. & Bashir R. Solid-state nanopore channels with DNA selectivity. Nat. Nanotechnol. 6, 253–260 (2011). [DOI] [PubMed] [Google Scholar]
- Joseph D. Bronzino J. D. The Biomedical Engineering Handbook: Second Edition (CRC press, Boca Raton, 2000). [Google Scholar]
- Smeets R. M. M., Keyser U. F., Krapf D., Wu M. –Y., Dekker, N. H. & Dekker, C. Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 6, 89–95 (2006). [DOI] [PubMed] [Google Scholar]
- Lu Bo., Albertorio F. ,. Hoogerheide D. P. & Golovchenko J. A. Origins and consequences of velocity fluctuations during DNA passage through a nanopore. Biophys. J. 101, 70–79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravishanker G., Auffinger P. ,. Langley D. R. ,. Jayaram B. ,. Young M. A. & Beveridge D. L. Treatment of Counterions in Computer Simulations of DNA. Rev. Comput. Chem. 11, 317–372 (1997). [Google Scholar]
- Fologea D., Gershow M., Ledden B., McNabb D. S., Golovchenko J. A. & Li J. Detecting single stranded DNA with a solid state nanopore. Nano Lett. 5, 1905–1909 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerqvist J., Zwolak M. & Di Ventra M. Influence of the environment and probes on rapid DNA sequencing via transverse electronic transport. Byophys. J. 93, 2384–2390 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y., Liu J., Zhang B. & Qian S. Field effect regulation of DNA translocation through a nanopore. Anal. Chem. 82, 8217–8225 (2010). [DOI] [PubMed] [Google Scholar]
- He Y., Tsutsui M., Fan C., Taniguchi M. & Kawai T. Controlling DNA translocation through gate modulation of nanopore wall surface charges. ACS Nano 5, 5509–5518 (2011). [DOI] [PubMed] [Google Scholar]
- Albrecht T. How to understand and interpret current flow in nanopore/electrode devices. ACS Nano 5, 6714–6725 (2011). [DOI] [PubMed] [Google Scholar]
- Takeishi S. et al. Observation of electrostatically released DNA from gold electrodes with controlled threshold voltages. J. Chem. Phys. 120, 5501 (2004). [DOI] [PubMed] [Google Scholar]
- Wei R., Pedone D., Zürner A., Döblinger M. & Rant U. Fabrication of metallized nanopores in silicon nitride membranes for single-molecule sensing. Small 6, 1406 (2010). [DOI] [PubMed] [Google Scholar]
- Erdmann M., David R., Fornof A. R. & Gaub H. E. Electrically induced bonding of DNA to gold. Nat. Chem. 2, 745–749 (2010). [DOI] [PubMed] [Google Scholar]
- Kimura-Suda H., Petrovykh D. Y., Tarlov M. J. & Whitman L. J. Base-dependent competitive adsorption of single-stranded DNA on gold. J. Am. Chem. Soc. 125, 9014–9015 (2003). [DOI] [PubMed] [Google Scholar]
- Wong C. T. A. & Muthukumara M. Polymer capture by electro-osmotic flow of oppositely charged nanopores. J. Chem. Phys. 126, 164903 (2007). [DOI] [PubMed] [Google Scholar]
- Steinbock L. J., Otto O., Chimerel C., Gornall J. & Keyser U. F. Detecting DNA folding with nanocapillaries. Nano Lett. 10, 2493–2497 (2010). [DOI] [PubMed] [Google Scholar]
- Reccius C. H., Stavis S. M., Mannion J. T., Walker L. P. & Craighead H. G. Conformation, length, and speed measurements of electrodynamically stretched DNA in nanochannels. Biophys. J. 95, 273–286 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information