Abstract
Improving the ability of DNA-based vaccines to induce potent Type1/Th1 responses against intracellular pathogens in large outbred species is essential. Rhodoccocus equi and equine infectious anemia virus (EIAV) are two naturally occurring equine pathogens that also serve as important large animal models of neonatal immunity and lentiviral immune control. Neonates present a unique challenge for immunization due to their diminished immunologic capabilities and apparent Th2 bias. In an effort to augment R. equi- and EIAV-specific Th1 responses induced by DNA vaccination, we hypothesized that a dual promoter plasmid encoding recombinant equine IL-12 (rEqIL-12) would function as a molecular adjuvant. In adult horses, DNA vaccines induced R. equi- and EIAV-specific antibody and lymphoproliferative responses, and EIAV-specific CTL and tetramer-positive CD8+ T lymphocytes. These responses were not enhanced by the rEqIL-12 plasmid. In neonatal foals, DNA immunization induced EIAV-specific antibody and lymphoproliferative responses, but not CTL. The R. equi vapA vaccine was poorly immunogenic in foals even when co-administered with the IL-12 plasmid. It was concluded that DNA immunization was capable of inducing Th1 responses in horses; dose and route were significant variables, but rEqIL-12 was not an effective molecular adjuvant. Additional work is needed to optimize DNA vaccine-induced Th1 responses in horses, especially in neonates.
Keywords: VapA, Gag p15/p26, Equine tetramers
1. Introduction
Conceptually, DNA-based vaccines offer many of the benefits of live vaccines without the inherent risks, such as reversion to virulence [1,2]. Some of the more compelling features of DNA vaccination are safety of manufacturing and handling, stability, and de novo synthesis of antigens identical to those produced during an active infection. Unlike virus vectored vaccines, the plasmid backbone of a DNA vaccine does not stimulate immune responses that may limit the ability to boost with a homologous virus. Importantly, in vivo expression of foreign genes encoded by DNA vaccines has been found to be effective at stimulating cellular immunity, notably the Type 1 responses (secretion of IFNγ and induction of cytotoxic T lymphocytes) that are required for immune clearance of many intracellular pathogens.
For these reasons and others, DNA-based immunization is being investigated for the prevention of a number of infectious diseases that have resisted traditional approaches and for which antibody alone is not sufficient. Although most studies have been performed in laboratory mice, DNA vaccination can also be effective in larger outbred domestic species. In the horse, targeted pathogens include West Nile Virus (WNV) [3], equine influenza virus [4,5], equine arteritis virus (EAV) [6], equine herpes virus-1 (EHV-1) [7–10], vesicular stomatitis virus (VSV) [6,11], rabies [12] and equine infectious anemia virus (EIAV) [13] (Table 1). EIAV is also a model for human immunodeficiency virus-1 (HIV-1). Cytotoxic T lymphocytes (CTL) that recognize and lyse infected cells are critical for the control of both EIAV and HIV-1. Although DNA vaccines are being aggressively explored for HIV-1 because of their ability to induce CD8+ CTL, a distinct drawback (especially outside of small laboratory animals) has been their diminished potency compared to live attenuated vaccines and viral vectors [2]. A primary focus therefore, has been the development of methods to increase immune responses using alternative delivery methods, improved plasmid design (e.g. codon optimization), genetically encoded molecular adjuvants, and prime-boost strategies.
Table 1.
Experimental DNA vaccines in the horse
| Equine pathogen | Ref. | Gene(s)/plasmid | Dose | Route | Adjuvant or carrier or [prime-boost] | No. DNA immunizations | Measured response to DNAa | Comments |
|---|---|---|---|---|---|---|---|---|
| Rabies | [12] | GP-G | 200 μg | i.m. | No adjuvant vs. Al(PO4) vs. DMRIE-DOPE | 2× | NtAb(+) | Strong adjuvant effect (DMRIE-DOPE); not tested for protection |
| EAV | [6] | Orf2, Orf5, Orf7 | 35 μg and 1.2 mg | Gene gun and i.m. | IL-2 plasmid (200 μg i.m.) Gene gun and i.m. were delivered simultaneously (10 skin sites and 4 i.m. sites, respectively); no adjuvant | 4× | NtAb(+) | Mixture of 7 plasmids (6× EAV + IL-2); not tested for protection |
| EIAV | [13] | SU | 250 μg | i.d. | – | 5–6× | Ab(+); LP(+); NtAb(−); CTL(−) | Codon optimized; not considered protective (no CTL/NtAb induced) |
| VSV | [11] | GP-G | 250 μg | i.d. | ±Oligonucleotides | 2× | NtAb(+) | No oligonucleotide effect |
| WNV | [3] | prM/E | 1.0 mg | i.m. | – | 1× | NtAb(+); Vir(−) | Produces virus like particles (VLP) |
| Influenza | [59] | NP, HA | 50 μg | Gene gun | Skin and mucosa (40 sites) [boost 2× w/rMVA] | 1× | Ab(−), LP(−) | After rMVA boost: Ab(+), LP(+) and IFNγ (+) |
| [58] | NP, HA | 50 μg | Gene gun | Skin and mucosa (40 sites) [boost 2× w/rMVA] | 1× | Ab(−); LP(−); IFN3 (−) | After rMVA-HA: Ab(+), LP(+); IFNγ (+); Pro(+), DNA priming not required | |
| [5] | HA | 100 μg + 75 μg | i.n. + gene gun | ±Cholera toxin (mucosal adjuvant) + skin and mucosa (60 sites) | 2× + 2× | Ab(+); Vir(NS); Pro(+) | Cholera toxin induced mucosal IgA, which is thought to play a role in protection | |
| [60] | HA | 37.5 μg | Gene gun | ±IL-6 plasmid skin and mucosa (60 sites) | 3× | Ab(+); LP(+); IFN3(+); VS(±); Pro(+) | Site of DNA administration affected regional distribution of response; IL-6 may have shifted response to Th2 | |
| [4] | HA | 12 or 19 μg | Gene gun | Skin (24 sites); or skin and mucosa (38 sites) | 3× | Ab(+); Pro(+) | Complete protection with skin + mucosal route | |
| EHV-1 | [10] | gB, gC, gD | 25 μg total | Gene gun | Skin and mucosa (60 sites) | 4× | NtAb(−); Ab(±); LP(−); CTL(−); Pro(−) | Limited immune response |
| [8] | gB, gC, gD | 500 μg of each | i.m. | No adjuvant vs. Al(PO4) vs. carbopol vs. DMRIE-DOPE and GM-CSF plasmid | 2× | Ab(−); VS(±); Vir(+) | ↑ NtAb in group DMRIE-DOPE and GM-CSF; in separate trial also tested recombinant canary pox (ALVAC) | |
| [7] | gD | 500 μg | i.m. | [1× = prime-boost with gD recombinant protein boost] | 2× or 1× | Ab(−), NtAb(−) | Minimal response to DNA, prime-boost response equivalent to protein alone | |
| [9] | gD | 500, 200, or 50 μg | i.m. | – | 3× | Ab(+); NtAb(+) | All horses had pre-existing Ab titers; after gD DNA, 8/15 had increased anti-gD Ab and 9/15 had increased NtAb | |
| R. equi | [20] | VapA | 1.0 mg | i.d. and i.t. | [rVapA recombinant protein + Ribi™ adjuvant] | 2× + protein boost | Ab(±); LP(±); IFNγ(−) | Poorly immunogenic in neonates |
Test (+, −, ±): NtAb, neutralizing antibody; Ab, antibody titer; CTL, cytotoxic T lymphocytes; IFNγ, interferon-gamma; LP, antigen-specific lymphoproliferation; V, viremia (−, prevented viremia); VS, virus shedding (−, inhibited virus shedding); Pro (+, protection against clinical disease); ±, in some animals; NS, not significant; rMVA, modified vaccinia Ankara virus.
DNA vaccination holds additional promise as an effective immunization method in neonates [14]. Beyond their potential to avoid interference by maternal antibody, DNA vaccines can overcome the T helper 2 (Th2) bias inherent to early life and have been shown to induce adult-like Type 1 responses. Rhodococcal pneumonia is an important cause of morbidity and mortality in young horses and a disease for which an effective vaccine is sorely needed [15]. The agent Rhodococcus equi is a facultative intracellular bacterium closely related to Mycobacterium species. Although equine rhodococcal pneumonia occurs almost exclusively between 2 and 5 months of age, most foals are exposed to R. equi during the first few weeks of life. Therefore, the first dose of an effective vaccine would likely need to be administered shortly after birth—a time when the animal’s immune system is immature relative to adults. In particular, neonatal foals are reported to have diminished abilities to produce IFNγ compared to adult horses and, like neonates of other species, may have diminished abilities to produce CTL [16,17]. Importantly, evidence from our laboratory and others suggest that a Type 1 immune response, which includes both T helper 1 (Th1)-like CD4+ T lymphocytes and CTL, is required for immunity against R. equi [18,19]. In other words, a vaccine that protects foals against rhodococcal pneumonia must induce the types of immune responses that neonates seem least capable of mounting. This represents a formidable challenge.
We previously tested a plasmid encoding R. equi virulence-associated protein A (VapA) as a candidate DNA immunogen in horses [20]. VapA is an immunodominant surface-exposed molecule that is encoded by the R. equi virulence-associated plasmid, required for virulence, and postulated to be a target of protective Type 1 immune responses. In adult horses, vapA DNA vaccine administration by a combination of intradermal and intrabroncheal routes induced a strong humoral and cellular recall response in both peripheral blood and pulmonary lymphocytes. This strategy however, failed to consistently induce antibody responses or detectable T lymphocyte responses in immunized foals. In experiments described here, we tested the hypothesis that co-immunization of neonatal foals with plasmids encoding equine IL-12 (rEqIL-12) and R. equi vapA would induce strong Type 1 immune responses, as characterized by VapA-specific lymphoproliferation and IFNγ production. Interleukin-12 (IL-12) is a heterodimeric cytokine produced mainly by macrophages, dendritic cells and B lymphocytes. It has potent effects on the induction and maturation of both CD4+ and CD8+ T lymphocytes. Importantly, the role of IL-12 in the development of protective Type 1 immune responses against related bacteria (e.g. Mycobacterium species) has been well established [21]. A relative IL-12 deficiency (i.e. an impaired capacity for production of IL-12 compared to adults) is one of a number of immunologic “defects” described in early life. Although it is by no means the only defect, the use of IL-12 as an adjuvant has been shown to have immunomodulatory effects in neonates [22].
We also investigated the effects of IL-12, dose, and route on equine immune responses to DNA vaccination using a plasmid encoding EIAV Gag p15 and p26. Previous experience with this codon-optimized plasmid had shown it to be poorly immunogenic in horses (unpublished data). We hypothesized that IL-12 and the routes previously used to administer the experimental VapA DNA vaccine would significantly increase the potency of the p15/p26 plasmid. Importantly, the EIAV system allowed us to examine the induction of epitope-specific CTL and tetramer-positive CD8+ T lymphocytes in both adult horses and neonatal foals.
2. Materials and methods
2.1. Cloning and expression of biologically active equine IL-12 (rEqIL-12)
Peripheral blood mononuclear cells (PBMC) used for p40 subunit cloning were washed with HBSS, suspended in PBMC growth medium (RPMI 1640 + 10% fetal bovine serum + 50 μM β-mercaptoethanol) at 1 × 106 ml−1; then stimulated with 0.0075% (wt/vol) of Staphylococus Aureus Cowan strain (Pansorbin, Calbiochem, La Jolla, CA) for 18–24 h at 37 °C + 5% CO2 before harvest and mRNA isolation. Unstimulated PBMC for the constitutively expressed p35 subunit cloning were used directly for mRNA isolation. For the p40 subunit, a library was made using Lambda Zap II (Stratagene, La Jolla, CA) which was then screened by hybridization with a p40-specific 32P labeled probe. The probe was produced using degenerate primers (based on the bovine IL-12 sequence NM174356) and nested PCR. The product of the nested reaction was cloned and sequenced for p40 specificity, and then labeled with 32P. In vivo excision into pBluescript SK (−) phagemid vector (Stratagene) was performed for positive clones. 5-Prime and 3-prime RACE using primers based on the published EqIL-12 sequence, accession number Y11130, was used to construct a clone for the p35 subunit [23]. Sequences of isolated clones were confirmed by dye-terminator automated sequencing at the Laboratory for Biotechnology and Bioanalysis at Washington State University. Subcloning of the two subunits into the pBudCE4 dual expression vector (Invitrogen, Carlsbad CA) to make pIL-12 was accomplished by a PCR reaction using EqIL-12-specific primers with 5′ and 3′ extensions containing restriction enzyme sites, additional nucleotides for reading frame adjustments, and a Kozak consensus sequence [24] (Table 2). Following the PCR reaction, the products were digested with the appropriate enzymes and ligated into pBudCE4. Again, sequences of the inserted genes and proper phasing were confirmed by dye terminator automated sequencing.
Table 2.
Equine specific IL-12 primers 1
 |
Restriction endonuclease sequences are in bold, Kozak sequences are italicized and shaded, and GC-clamp sequences are underlined.
NIH 3T3 cells were transfected using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). Twenty-four-well plates were seeded with 5 × 104 3T3 cells in DMEM + 10% FBS and incubated overnight at 37 °C + CO2. The next day 0.5, 0.6, or 0.7 μg of pIL-12 or pBudCE4 in 50 μl Opti-MEM serum-free medium (Invitrogen) was added to 50 μl of Opti-MEM containing 2.5 μl of Lipofectamine 2000 and incubated at room temperature for 20 min. Medium was removed from the cells, the DNA/Lipofectamine 2000 mixture added and plates were then incubated at 37 °C + CO2 overnight. Cells were trypsinized and passaged the next day into 25 cm2 flasks with selection medium of DMEM + 10% FBS + 300 μg/ml Zeocin. Cells were monitored daily, fed as needed with fresh medium, and amplified for cell stocks. Non-transfected cell controls died within 7–10 days.
rEqIL-12 purified under native conditions from medium of pIL12-transfected cells using 6Xhis binding ProBond columns (Invitrogen) and imidazole elution was electrophoresed in 4–20% Tris–HCl gels for Western Blot analysis. Proteins were transferred to nitrocellulose filters and probed with anti-V5 or anti-myc (Invitrogen) antibody followed by incubation with HRPO conjugated goat anti-mouse IgG secondary antibody (KPL, Gaithersburg, MD). Proteins were visualized by addition of chemiluminescence reagents and exposure to X-ray film.
A functional biologic assay similar to the method used for human and mouse IL-12 was performed to determine rEqIL-12 induction of cell proliferation and the effective concentration of IL-12 to give a 50% increase in thymidine uptake (EC50) [25,26]. PBMC were seeded into 175 cm2 flasks at 5 × 106 ml−1 in PBMC growth medium + 10 μg/ml PHA-P (Sigma–Aldrich, St. Louis, MO) and stimulated for 3 days at 37 °C + 5% CO2. After the 3-day stimulation, cells were harvested, washed and suspended in the same medium but without PHA-P and with 20 μg/ml rHuIL-2 (Sigma–Aldrich); then incubated another 4 days. Cells were then washed 3× with HBSS, suspended in PBMC growth medium + 5% horse serum at 2 × 106 ml−1, and seeded into 96-well plates at 100 μl per well. Fifty microlitre dilutions of conditioned medium collected from pIL-12-transduced cells were added to respective wells in triplicate for each dilution. Negative control wells with pBudCE4-transduced cell conditioned medium only and positive control wells with 10 μg/ml Con A were also performed. Plates were incubated at 37 °C + 5% CO2 for 3 days. Fifty microlitre of media containing 0.25 μCi 3H thymidine was added to each well for the last 16 h of incubation. Cells were then harvested using a Tomtec cell harvester and thymidine uptake counts measured using a Wallac Betaplate scintillation counter. EC50 results were expressed as units per ml using the formula:
2.2. Construction of the R. equi pVR1055vapA
Cloning and confirmed expression of the recombinant plasmid DNA containing the vapA gene in the sense orientation, pVR1055vapA was previously described [20]. The endotoxin level in the purified plasmid was <0.1 EU/μg DNA as determined using the Limulus amoebocyte lysate assay [KQCL kit, BioWhittaker, Walkersville, MD].
2.3. Construction of a codon-optimized plasmid expressing EIAV Gag p15 and p26
The EIAV gag p15 and p26 sequences (EIAVWSU5 nt 475–1551; GenBank accession no. AF247394) were codon-optimized for Equidae genes. Equidae and EIAV codon usage was obtained from http://www.kazusa.or.jp/codon/ and the gag p15 and p26 genes were optimized and synthesized commercially by GenScript Corp. (Piscataway, NJ). The optimized Gag genes plus a Kozak consensus sequence for optimum eukaryotic expression were inserted into the multiple cloning site of the VR-1055 eukaryotic expression plasmid vector (Vical, San Diego, CA). The resulting plasmid vector was designated VR-p15/p26. Equine kidney cells were transfected with VR-p15/p26 using Lipofectamine 2000 (Invitrogen). After 48 h, total RNA was extracted and RT-PCR performed to confirm transcription of the construct. Gag p15 and p26 protein expression in the transfected cells was confirmed by immunoblot [27] (data not shown). The Gag-GW12 CTL epitope (GSQKLTTGNCNW) [28], is contained within Gag p15 (and thus encoded by VR-p15/p26). This epitope is presented by the MHC class I molecule 7–6 associated with ELA-A1 haplotype [29], and Gag-GW12 CTL are identified by the 7–6/Gag-GW12 tetramer [30]. A large preparation of endotoxin-free VR-p15/p26 was obtained using a Plasmid Giga Kit (Qiagen Inc., Valenica, CA).
2.4. Preparation of R. equi antigens
R. equi 33701 soluble antigen (SRA) and recombinant VapA antigen (rVapA) were prepared as previously described [31], as was native vapA enriched antigen (nVapA) [32]. Soluble antigen derived from Corynebacterium pseudotuberculosis (SCA) was also prepared as previously described [31] and used as a negative control. All proteins were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining and immunoblotting, and quantified by the bicinchoninic acid (BCA) method.
2.5. Immunizations with R. equi vapA and rEqIL-12
Three adult ponies H636, H642 and H615 were immunized with 1 mg of pVR1055vapA and 1 mg of IL-12 plasmid (day 1) and DNA boosted using the same regimen 14 days later (day 14). Three additional control ponies, H607, H624 and H563 received 1 mg of pVR1055vapA and 1 mg of the control plasmid (pBudCE4 without the IL-12 inserts) at the same time points. Animals were inoculated with 0.5 mg of each plasmid by the intradermal (i.d.) route and 0.5 mg of each plasmid by the intratracheal (i.t.) route. For the i.d. injections, 0.5 mg of each plasmid combination diluted in 1 ml of Hanks solution, was injected at 5 sites on each side of the neck (100 μl/site). For the i.t. inoculations, 0.5 mg of each plasmid combination diluted in 5 ml of saline was delivered via injection into the trachea between tracheal rings in the mid neck region using a 12 cm3 syringe and 18.5 gauge needle.
In order to characterize the primary immune response elicited by vapA DNA with IL-12 DNA as an adjuvant and compare these results with foals previous vaccinated with vapA DNA alone [20], four foals ≤7 days of age were vaccinated (day 1) with pVR1055vapA and the IL-12 plasmid. Both vaccines were administered i.d. (following the same protocol used in the adult ponies) and intranasally (i.n.), as previously described [20]. Briefly, a flexible tube was used to deliver 0.5 mg of plasmid DNA in 1 ml of Hanks solution. All foals received a second DNA vaccination 2 weeks later (day 15) consisting of both constructs and using the same dose and route as before. On day 30 all foals received a protein boost with purified rVapA. The protein boost was delivered via i.n. and i.d. routes. For the i.n. inoculation, 0.1 mg of protein was administered in 1 ml of Hanks solution. For the i.d. inoculation, 0.5 ml of Hanks solution containing 0.1 mg of protein was emulsified in 0.5 ml of RIBI adjuvant (Ribi MPL/TDM adjuvant, Corixa Corp., Seattle) and injected in 5 sites on each side of the neck just below the mane (100 μl/site).
2.6. R. equi pulmonary challenge of the vapA immunized animals
Preparation and pulmonary challenge using virulent R. equi was as previously described [31,33]. Briefly, the R. equi ATCC 33701 virulent strain was kept as a frozen stabilate. After reconstitution and selection of a single colony, bacteria was grown in Brain Heart Infusion medium (BHI, DIFCO Laboratories, Detroit, Michigan) for 16 h at 37 °C with shaking. Following centrifugation the pellet was washed twice with phosphate buffered saline (PBS). Bronchoalveolar lavage fluid (BALF) was obtained for analysis just prior to challenge. For adult ponies the right lung was inoculated with 2 × 107 R. equi 33701 in 1 ml of PBS followed by flushing of the endoscope with 15 ml of air before removal. Adult ponies were challenged 4 weeks following the second vaccination. The challenge dose for foals was 104 organisms per animal and was administered into the right lung via an endoscope as a 20 ml suspension in PBS. Pony foals were challenged 2 weeks following the rVapA protein boost. This low challenge dose was based on a previous report of experimental infection in foals with R. equi ATCC 33701 strain [34] and was used because our objective was to evaluate immune responses with and without IL-12 DNA as an adjuvant and not to induce disease to evaluate protection.
2.7. Immunizations with EIAV gag p15/p26 and EqIL-12 plasmids
Four adult horses with the ELA-A1 haplotype were used in these experiments. Two horses received the EIAV VR-p15/p26 plasmid plus the pIL-12 plasmid and two horses received the EIAV VR-p15/p26 plasmid plus the empty vector control, pBudCE4. The routes of immunization were identical to the R. equi vapA DNA-immunized adult ponies but the dose of each immunogen was increased. Horses were inoculated with 2.5 mg of each plasmid by the i.d. route and 2.5 mg of each plasmid by the i.t. route on day 1 and boosted on day 15, and days 43 and 57 or 57 and 71. All horses were sampled prior to the first vaccination, 14 days after the second and third vaccinations, and 7 days following the fourth.
In order to examine the ability of DNA vaccination to stimulate immune response in neonates, three foals with the ELA-A1 haplotype were immunized within the first week of life (designated day 1) with the EIAV VR-p15/p26 plasmid using the same routes and doses as for the adult horses on days 1, 15, 36 and 50. Foals were sampled prior to the first immunization, 14 days following the second and third immunizations, and 7 days following the fourth. The immunization protocols are summarized in Table 3.
Table 3.
Summary of DNA vaccination protocols
| Animal ID | Age | Immunogen | Dose and route | Comments | Boost |
|---|---|---|---|---|---|
| A. H636, H642, H615 | Adult | vapA + IL-12 (1 mg each, mixed) | 0.5 mg i.d. 0.5 mg i.t. |
i.d. divided between 10 sites i.t. single injection |
Day 14—same protocol as primary |
| B. H607, H624, H563 | Adult | vapA + control plasmid | Same as (A) | Same as (A) | Same as (A) |
| C. H670, H669, H672, H673 | Foals | vapA + IL-12 (1 mg each, mixed) | 0.5 mg i.d. 0.5 mg i.n. |
i.d. divided between 10 sites i.n. one dose Prime-Boost* |
Day 14—same (DNA) Day 30—rVapA protein*: i.d. 10 μg × 10 sites (100 μg total) + Ribi™ adjuvant; +100 μg protein i.n. |
| D. H629, H633, A2166, JJ, MM | Foals | vapA plasmid [20] | Same as (C) | Same as (C) | Same as (C) |
| E. H630, H632, A2167, JH, Elly | Foals | vapA control plasmid [20] | Same as (C) | Same as (C) | Same as (C) |
| F. A2201, A2185 | Adult | p15/p26 + IL-12 (5 mg each, mixed) | 2.5 mg i.d. 2.5 mg i.t. |
i.d. divided between 10 sites i.t. single injection |
Day 15, 43–57, 57–71 |
| G. A2190, A2192 | Adult | p15/p26 + control | Same as (F) | Same as (F) | Same as (F) |
| H. A2222, A2223, A2224 | Foals | p15/p26 alone (no IL-12) | Same as (F) | Same as (F) | Day 15, 36, 50 |
DNA prime + boost with recombinant protein.
2.8. Isolation of PBMC and bronchoalveolar lavage fluid (BALF) cells
BAL and cell isolation were performed as previously described [33]. After each BAL, horses were placed in a stall and monitored daily for changes in rectal temperature, respiration, and pulse as determined by physical exam and auscultation of the lungs. After isolation, equine PBMC and BALF cells were resuspended in growth medium consisting of RPMI 1640 medium [Hyclone, Logan, UT] containing 25 mM HEPES [Sigma–Aldrich] with 10% heat-inactivated and filtered normal horse serum [Invitrogen], 0.05 mM 2-mercaptoethanol [Sigma–Aldrich], 2 mM L-glutamine [Invitrogen], and 50 μg/ml gentamicin [Invitrogen] [33]. For adult horses, commercial serum was not used. Instead, serum was collected in bulk from each horse prior to immunization, filtered through a 0.2 μm pore-size filter, heat inactivated for 30 min at 56 °C, and frozen in 50 ml aliquots at −20 °C. Thawed autologous serum was added to complete medium prior to each assay.
2.9. Lymphocyte proliferation assays
The proliferative responses of cells obtained from peripheral blood and BAL fluid were evaluated by measurement of the uptake of [methyl-3H]-thymidine [31]. For R. equi antigen stimulation, cells were assayed in quadruplicate as previously reported by co-culture with soluble R. equi antigen (SRA), nVapA (native VapA), rVapA (recombinant VapA), and SCA (soluble C. pseudotuberculosis antigen, as a negative control) [31]. For EIAV, cells were stimulated with EIAV Gag peptides containing broadly recognized T helper epitopes as follows: a pool of three p26 peptides (Gag p26 221–245, Gag p26 241–261, Gag p26 250–269) and a p15 (Gag p15 13–32) peptide [35,36]. Procedures for EIAV antigen stimulation were identical to those for R. equi antigen stimulation [31]. For all lymphocyte proliferation assays, cells were cultured without stimulation (i.e. in medium alone) as a negative control. The positive control consisted of cells stimulated with pokeweed mitogen (PWM), a known T-lymphocyte mitogen in horses [37]. Data were expressed as the mean counts per minute (cpm) of replicate wells. The stimulation index (SI) for each antigen was derived by dividing the mean cpm for that antigen by the mean cpm of medium alone.
2.10. Quantification of cytokine mRNA
For analysis of antigen-specific cytokine expression, PBMC and BAL cells were incubated with antigen for 24 h and RNA lysate extracted and stored, as previously described [31]. Complementary DNA (cDNA) was generated using reverse transcriptase and the products obtained were used in real-time PCR to measure expression of equine interferon-gamma (IFNγ), equine interleukin-4 (IL-4) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [31]. All samples were analyzed in triplicate [38]. After PCR amplification, data acquisition was performed using the iCycler iQ™ Multicolor Real-Time PCR Detection System, version 3.1 [Bio-Rad, Hercules, CA]. Transcript levels were determined for each sample by comparing the threshold cycle values for each cytokine to the corresponding standard curves. The transcript levels for IFNγ and IL-4 transcripts were normalized to the GAPDH transcript level for the same sample. In each real-time PCR reaction, mixtures containing no DNA were included to control for extraneous DNA in the reagents.
2.11. Determination of anti-R. equi and anti-VapA antibodies
R. equi- and VapA-specific antibodies in serum were analyzed using previously described ELISA’s [20,31]. To detect total IgG antibodies bound, plates were incubated with peroxidase-conjugated caprine antibodies (1:10,000; KPL) directed against horse IgG. To detect the specific antibody isotypes bound, the plates were incubated with anti-equine IgGa (CVS48), IgGb (CVS39), IgG(T) (CVS40), IgA (7/C8) [39], or IgM 1.9/3.2 (VMRD) murine monoclonal antibodies. These plates were washed with PBS-Tween and incubated with a peroxidase-conjugated anti-mouse IgG (KPL). Serum from a known high responder and normal horse serum were used as standard positive and negative controls on each plate, respectively, and to generate a standard curve for correction of interplate variability [38].
2.12. CTL assay
CTL assays were performed as described [28,30,40] with modifications. Briefly, PBMC were isolated and stimulated with peptide-pulsed monocytes. For peptide stimulations, seven peptide pools were used, each containing 10–14 synthetic peptides covering the entire EIAVWSU5 Gag p15 and p26 proteins [40]. Peptide pool 1 contained the Gag-GW12 peptide (Gag p15 21–32), against which CD8+ T cells identified by the 7–6/Gag-GW12 tetramer (above) are directed. Peptide pool 1 also contained the p15 13–32 peptide used in proliferation assays (above). Peptide pools 5 and 6 contained the three Gag p26 peptides used in proliferation assays (above). Peptide pools (final concentration of each peptide was 103 nM) and PBMC were incubated for 2 h at 37 °C with occasional mixing before centrifugation at 250 × g for 10 min. PBMC were resuspended to 2 × 106 ml−1 in RPMI 1640 medium with 10% FBS, 20 mM HEPES, 10 μg/ml gentamicin, and 10 μM 2-ME. One ml of resuspended cells was added to each well of a 24-well plate and incubated for 1 wk at 37 °C before use in CTL assays. CTL activity was measured in freshly isolated and stimulated PBMC with a 17-h 51Cr release assay using equine kidney (EK) target cells obtained by biopsy [41]. Target cells were pulsed with the same peptide pools used for stimulation, at a final concentration of 104 nM for each peptide. The formula, %specific lysis = [(E − S)/(M − S)] × 100, was used, where E is the mean of three test wells, S the mean spontaneous release from three target cell wells without effector cells, and M is the mean maximal release from three target cell wells with 2% Triton X-100 in distilled water. The E:T cell ratio was 50:1, and each well contained ~30,000 target cells. Only assays with a spontaneous target cell lysis of <30% were used. The standard error (S.E.) of percent specific lysis was calculated using a formula that accounts for the variability of E, S, and M [42]. Significant lysis was defined as the percent specific lysis of peptide-pulsed target cells that was >10% and also >3S.E. above the nonpulsed target cells.
2.13. Tetramer analysis—Gag-specific CD8+ T lymphocytes
The 7–6/Gag-GW12 tetramer was used to identify Gag-GW12-specific CD8+ T lymphocytes in freshly isolated and stimulated PBMC as described [30] with modifications. Briefly, the murine anti-equine CD3 monoclonal antibody F6G [43,44] and the murine anti-equine CD8 monoclonal antibody HT14A [45] were directly labeled with Alexa Fluor 647 and 488 dyes, respectively, using the Alexa Fluor Monoclonal Antibody Labeling Kit (Invitrogen). Gag-GW12 peptide stimulations were done as described above using 103 nM Gag-GW12. Freshly isolated or peptide-stimulated PBMC were stained first with PE-conjugated 7–6/Gag-GW12 tetramer for 30 min at 37 °C, washed, then directly labeled F6G and HT14A were added for 15 min at 4 °C. For analysis of stimulated PBMC, propidium iodide staining was performed to assess cell viability. Three-color (or four-color for stimulated PBMC) analysis was performed on live CD3-gated lymphocytes using a FACSort flow cytometer (Becton Dickinson, Franklin Lakes, NJ), with Cell Quest and Paint-A-Gate Pro software.
2.14. Detection of EIAV-specific antibody responses
Serum antibodies against EIAV Gag p26 were detected using the Equine Infectious Anemia Virus Antibody ELISA Test Kit (VMRD, Inc., Pullman, WA).
2.15. Animals
All horses and ponies were from a closed herd maintained at Washington State University. This herd is free of EIAV infection based on annual serotesting for antibodies against EIAV Gag p26. All experiments involving these animals were approved by the Washington State University Institutional Animal Care and Use Committee. Throughout the study, horses and ponies were housed in individual stalls or small outside paddocks. Foals remained with their dams for the duration of the foal studies. No cases of rhodococcal pneumonia had been previously diagnosed in foals housed at any of these sites for at least 3 years preceding this study. At the completion of the studies all horses, ponies and foals were returned to the breeding herd. Adult ponies H636, H642, H615, H607, H624 and H563 (aged 5, 4, 10, 12, 12 and 9 years, respectively) and pony foals H670, H669, H673 and H672 were used in the R. equi IL-12 vaccination experiments. Two-year-old Arabian horses A2185, A2190, and A2192, yearling Arabian horse A2201, and Arabian horse foals A2222, A2223, and A2224 were used in the EIAV vaccination experiments. Serotesting at the conclusion of the study confirmed that no animals were exposed to EIAV during vaccination experiments. All horses and foals immunized with EIAV gag p15/p26 had the ELA-A1 MHC class I haplotype as determined serologically by lymphocyte microcytotoxicity [46–48], using reagents kindly provided by Dr. Ernest Bailey (University of Kentucky, Lexington, KY).
Prior to all immunization and BAL procedures blood was obtained via jugular venipuncture and samples were submitted to the Washington State University Clinical Pathology Laboratory for determination of complete blood counts and fibrinogen concentrations. Following immunization and BAL procedures all animals were monitored daily for changes in rectal temperature, respiration, and pulse as determined by physical examination and auscultation of the lungs.
3. Results
3.1. Expression and biological activity of rEqIL-12
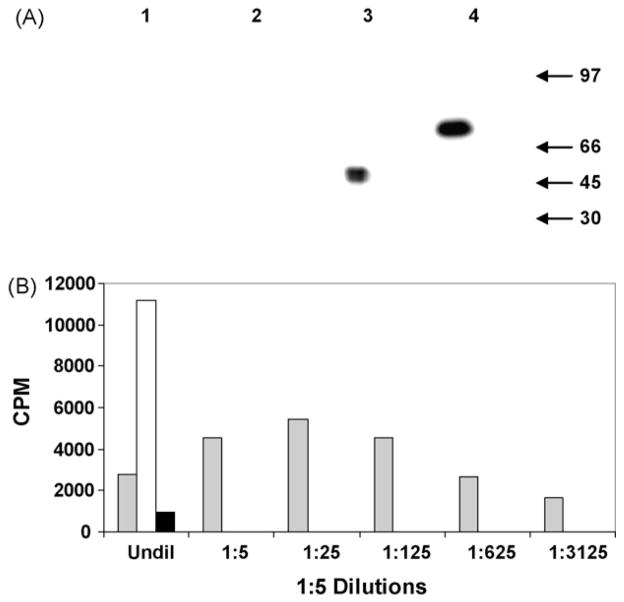
NIH 3T3 cells transfected with pIL-12 and the empty vector pBudCE4, were selected by G418 antibiotic resistance and analyzed for expression of rEqIL-12 by Western blot analysis (Fig. 1A). Using reducing condition and an anti-V5 antibody specific for the p35 subunit tag, the IL-12 p35 subunit could be seen migrating as a single band at about 35 kD. Under non-reducing conditions the IL-12 heterodimer migrated as expected at about 70 kD (Fig. 1A: Lanes 3 and 4, respectively). No bands were detected in the empty vector negative control samples (Fig. 1A: Lanes 1 and 2). An anti-myc antibody specific for the p40 subunit tag detected a similar 70 kD in the culture medium of pIL-12 transfected cells, but only under native conditions (data not shown). Repeated blots with anti-myc failed to detect a product when gels were run under denaturing conditions.
Fig. 1.
(A) Immunoblot of purified rEqIL12 reacted with Anti-V5 MAb. pBudCE4 control reduced (Lane 1), pBudCE4 non-reduced (Lane 2), pIL12 reduced (Lane 3), pIL12 non-reduced (Lane 4). Migration of molecular weight markers in kilodaltons is indicated by arrows on right side of panel. (B) Functional activity of rEqIL12 was measured using a thymidine uptake proliferation assay. Serial dilutions of media from pIL12 or pBudCE4-transduced 3T3 cells were added to PHA-P stimulated PBMC then harvested 3 days later and counted. rEqIL12, pBudCE4, Con A.
Recombinant EqIL-12 induction of cell proliferation and titration of this activity was evaluated using a tritiated thymidine uptake assay (Fig. 1B). Although proliferative responses to the pBudCE4 control remained at a low background level, conditioned medium from pIL-12 transfected cells had high activity for induction of cell proliferation, resulting in titers of 50–80 units per ml. A possible inhibitory effect was seen at the highest concentrations tested. This diminished proliferation could be due to an excess of p40 homodimer in the conditioned medium [49–51]. The effect was quickly extinguished upon dilution and a high level of proliferation observed.
3.2. Antigen-specific responses in vapA-immunized adult horses
To determine whether vapA DNA co-administration with rEqIL-12 could enhance immune responses in adult horses, 3 adult ponies were immunized with the vapA and rEqIL-12 plasmids (Group 1) and 3 adult ponies were immunized with the vapA construct plus the empty pBudCE4 vector control (Group 2). nVapA-specific antibody responses in serum were determined just prior to vaccination (day 0), at 2 and 4 weeks following a DNA boost, and at 1 and 2 weeks post-pulmonary challenge with virulent R. equi. nVapA-specific total IgG and isotype responses demonstrated significant titers post-vaccination and pre-challenge, indicating that the vapA DNA construct was expressed in vivo and induced an antibody response. However, there was no significant difference in total IgG or isotype-specific nVapA titers between the 2 groups (data not shown).
VapA-specific cytokine and lymphoproliferative responses were examined in cells recovered from peripheral blood and BALF at the same time points evaluated for specific antibody. Cytokine and lymphoproliferative responses to nVapA, SRA, rVapA were measured, using PWM as a positive control. Media alone and the SCA antigen served as negatives controls. Antigen-induced IL-4 and IFNγ expression in both peripheral blood and BALF cells were inconsistent, with large variations in the responses among animals and across time points. No significant differences in antigen-specific cytokine responses were detected between the 2 groups (data not shown). Post-vaccination antigen-specific lymphoproliferation measurements failed to demonstrate a significant response in any of the vapA-immunized ponies regardless of whether they were co-immunized with rEqIL-12 or the empty control vector. Both peripheral blood and BALF cells from all ponies showed a significant stimulation index following pulmonary challenge with R. equi compared to the antigen and media controls—but there was no evidence of a significant IL-12 effect (data not shown).
3.3. Antigen-specific responses in vapA-immunized neonatal foals
The apparent lack of an IL-12 effect in vapA-immunized adult ponies could reflect previous antigen exposure. R. equi is considered ubiquitous in the equine environment, and IL-12 may not significantly enhance an established memory response. In order to characterize primary immune responses generated by pVR1055vapA and co-administration with rEqIL-12, four neonatal pony foals were immunized with both constructs followed by a protein boost as described previously and detailed in Section 2 [20]. VapA-specific cytokine, lymphoproliferative and antibody responses were evaluated just prior to immunization, 2 weeks post-protein boost (day of challenge with virulent R. equi) and 2 weeks post-challenge. The results were compared to results in a previous study in our laboratory of 5 foals immunized with the vapA construct alone using the same dose, route and timing of immunization [20]. Foals in the previous study were not challenged with R. equi after immunization. In the previous study we observed significant VapA-specific IgG titers in 2 of 5 foals but failed to detect any antigen-specific cytokine or lymphoproliferative responses compared to the 5 foals that received the control plasmid [20]. Our hypothesis was that the rEqIL-12 molecular adjuvant would increase the number of foals responding and enhance the Type 1 responses that are relevant to immune clearance of R. equi.
In the experiments reported here, vapA and rEqIL-12 DNA co-administration failed to increase the number of foals that developed a primary immune response to VapA, as measured by antigen-specific cytokine production, lymphoproliferation and antibody titers (data not shown). There were no significant differences for the measured parameters between foals in this study immunized with both vapA and rEqIL-12 DNA and foals in the previous study immunized with the vapA construct alone (data not shown).
3.4. EIAV immunization of adult horses
Two possible conclusions from the VapA data were (a) that the rEqIL-12 molecular adjuvant had no significant biological effect in horses or (b) that the rEqIL-12 was unable to overcome the relative immaturity of the neonatal immune response. In order to better determine whether rEqIL-12 DNA co-administration could enhance primary immune responses in adult horses, we tested its ability to augment immune responses in naive adult horses immunized with an EIAV p15/p26 DNA vaccine. Previous experience with the EIAV VR-p15/p26 vaccine failed to demonstrate any CTL responses to 4 intramuscular (i.m.) injections at 5.0 mg per dose (unpublished data, RH Mealey). We hypothesized that the rEqIL-12 molecular adjuvant would promote a primary immune response to the otherwise weakly to non-immunogenic EIAV VR-p15/p26 construct. For these experiments we elected to use the higher 5.0 mg dose of EIAV vaccine used in previous VR-p15/p26 vaccination experiments and the combined i.d/i.t. routes to match the route used in the vapA DNA immunizations of ponies.
Arabian horses were matched at a single A1 allele in order to measure peptide-specific CTL responses and quantitate tetramer-positive CD8+ T lymphocytes. Two horses were immunized with EIAV VR-p15/p26 DNA plus the rEqIL-12 plasmid and 2 horses were immunized with EIAV VR-p15/p26 DNA and the pBudCE4 empty vector control. A total of 5.0 mg of EIAV DNA were administered using the i.t. (2.5 mg) and i.d. (2.5 mg) routes. Horses were immunized 4 times on days 1, 15, 43 or 57 and 57 or 71, as in previous VR-p15/p26 experiments. Peripheral blood lymphocytes were evaluated for tetramer staining, antigen-specific CTL, and lymphoproliferative responses just prior to vaccination and at 14 days post-second vaccination, 14 days post-third vaccination and 7 days post-fourth vaccination. Serum was evaluated for EIAV p26-specific antibody at the same time points.
3.5. Induction of EIAV Gag-specific CD8+ T lymphocytes and CTL
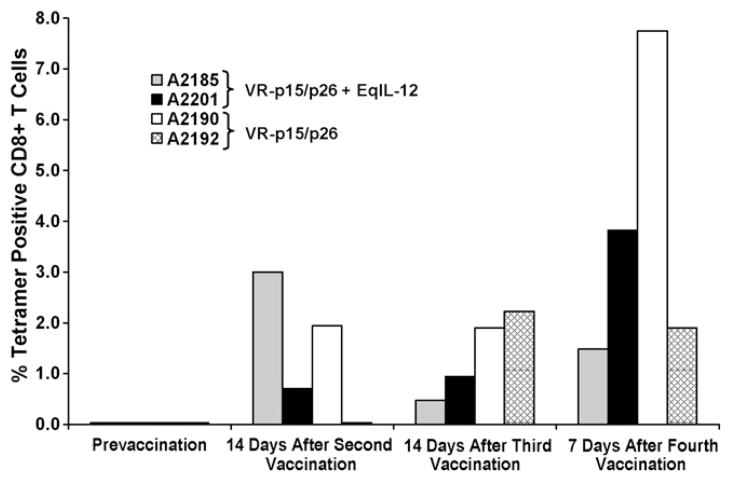
For horses A2185 and A2201, which received VR-p15/p26 plus rEqIL-12, 7–6/Gag-Gw12 tetramer-positive CD8+ T lymphocytes were detected in Gag-GW12-stimulated PBMC beginning 2 weeks after the second vaccination (Fig. 2). For A2185, frequencies of Gag-GW12-specific CD8+ T lymphocytes in stimulated PBMC ranged from 0.5 to 3.0% during the study period, while those for A2201 ranged from 0.7 to 3.8%. Tetramer-positive CD8+ T lymphocytes were also detected in stimulated PBMC from horses A2190 and A2192 (which received VR-p15/p26 without rEqIL-12), beginning 14 days after the second vaccination for A2190, and 14 days after the third vaccination for A2192 (Fig. 2). During the study period, frequencies of Gag-GW12-specific CD8+ T lymphocytes in stimulated PBMC ranged from 1.9 to 7.7% in A2190, and 0–2.2% in A2192.
Fig. 2.
Tetramer analysis: Immunization with VR-p15/p26 with or without EqIL-12 induced Gag-GW12-specific CD8+ T cells. PBMC obtained from vaccinated horses A2185, A2201, A2190, and A2192 at the indicated time-points were stimulated for 1 week with Gag peptide pool 1, then labeled with the 7–6/Gag-GW12 tetramer, and percent tetramer positive CD8+ cells were determined on CD3+ gated lymphocytes using flow cytometry. A2185 and A2201 were immunized with VR-p15/p26 + EqIL-12; A2190 and A2192 were immunized with VR-p15/p26 without Eq-IL12.
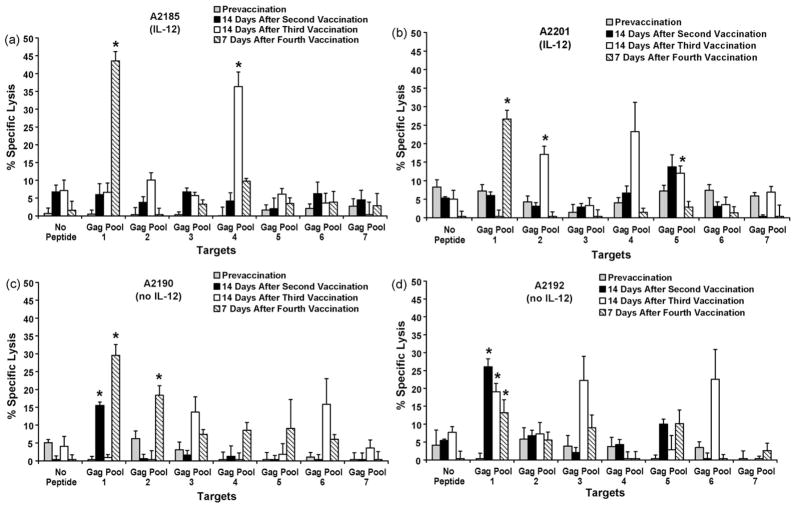
All four horses developed Gag-specific CTL activity at various time-points following immunization with VR-p15/p26 (Fig. 3). For A2185, CTL in stimulated PBMC recognized epitopes in Gag pools 1 and 4 (Fig. 3a), while CTL from A2201 recognized epitopes in Gag pools 1, 2, and 5 (Fig. 3b). For A2190, CTL recognized epitopes in Gag pools 1 and 2 (Fig. 3c), while CTL from A2192 recognized epitopes in Gag pools 1 and 3 (Fig. 3d).
Fig. 3.
Cytotoxic T lymphocytes: Immunization with VR-p15/p26 with or without EqIL-12 induced Gag-specific CTL in adult horses. PBMC obtained at the indicated time-points were stimulated for 1 week with Gag peptide pools 1–7 and CTL activity determined on Gag peptide pool-pulsed EK targets for vaccinated horses (a) A2185, (b) A2201, (c) A2190, and (d) A2192. Error bars are S.E. E:T ratio, 50:1. Significant specific lysis is indicated with an asterisk. A2185 and A2201 were immunized with VR-p15/p26 + EqIL-12; A2190 and A2192 were immunized with VR-p15/p26 without Eq-IL12.
Taken together, these results indicated that co-administration of rEqIL-12 did not enhance the ability of VR-p15/p26 DNA to induce CTL. In fact, the highest frequency of 7–6/Gag-GW12 tetramer positive CD8+ T lymphocytes was observed in A2190, which did not receive rEqIL12 (Fig. 2). Moreover, CTL activity appeared earlier in A2190 and A2192, which received VR-p15/p26 DNA alone (Fig. 3).
3.6. EIAV lymphoproliferation and antibody results
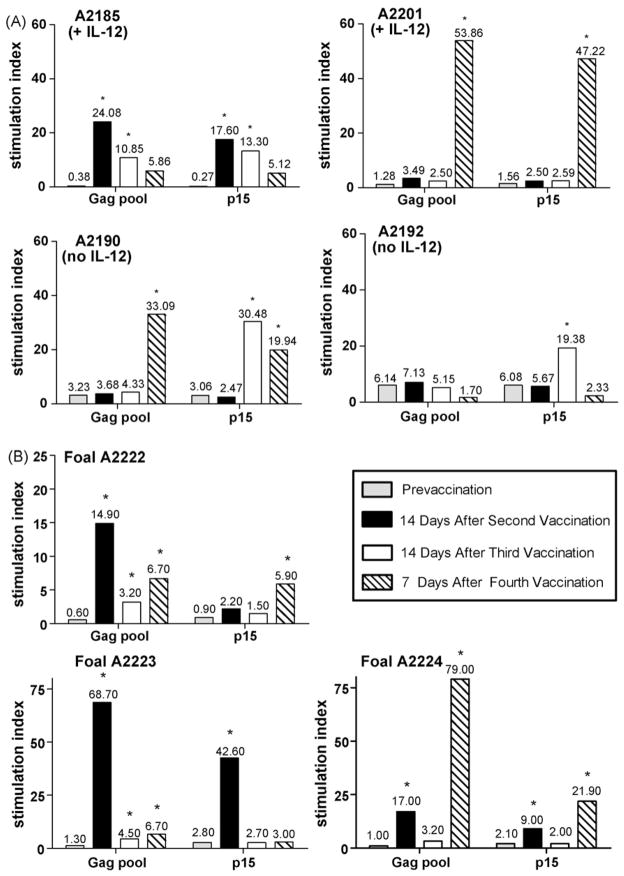
Peripheral blood cells were stimulated with a pool of 3 overlapping Gag p26 peptides (Gag p26 221–245, Gag p26 241–261, Gag p26 250–269) and with the p15 13–32 peptide. The three Gag p26 peptides were also present in Gag peptide pools 5 and 6 (used in CTL assays), while the p15 peptide was also present in Gag peptide pool 1 (used in CTL assays). This p15 peptide also contained the CTL epitope peptide Gag-GW12 (p15 21–32). These four peptides are known to contain broadly recognized T helper epitopes [35,36]. A post-immunization SI was considered significant if it was at least 3× the pre-immunization SI for that antigen and if the corresponding PWM SI was 10 or greater [35]. Cells from 3 of the 4 horses showed significant post-vaccination proliferation to both Gag pool and p15 peptides compared to pre-immunization levels, although significant and peak stimulation occurred at different post-vaccination time points for particular horses and antigens (Fig. 4A). Cells from the remaining horse (A2192) also showed significant post-vaccination stimulation to p15 peptide, but did not show a post-vaccination response to Gag pool peptides. Both control horses (no IL-12) and one of the rEqIL-12 co-immunized horses seroconverted to EIAV p26 at the 2 weeks post-second vaccination time point. The remaining rEqIL-12 co-immunized horse seroconverted at the 2 weeks post-third vaccination time point.
Fig. 4.
Lymphoproliferation: Immunization of adult horses with VR-p15/p26 with or without rEqIL-12 and immunization of foals with VR-p15/p26 alone induced EIAV antigen-specific lymphoproliferative responses. Peripheral blood mononuclear cells obtained at the indicated time points were stimulated for 5 days with Gag peptide pool and p15 peptide. (A) Horses A2185 and A2201 were co-immunized with rEqIL-12 DNA and horses A2190 and A2192 were co-immunized with the pBudCE4 vector control. (B) Foals were immunized with VR-p15/p26 alone. An asterisk (*) indicates an SI of at least 3× the pre-immunization SI for that antigen and a corresponding PWM SI of 10 or greater.
In summary, adult horses immunized 4 times with 5.0 mg per dose of EIAV VR-p15/p26 using the i.t. and i.d. routes developed antigen-specific immune responses as measured by tetramer-positive staining of CD8 lymphocytes, peptide-specific CTL and lymphoproliferative responses, and EIAV p26 antibody titers. However, co-administration with rEqIL-12 did not correlate with earlier or enhanced responses for any of the measured parameters. In previous experiments, 4 injections of the 5.0 mg dose of EIAV VR-p15/p26 DNA using the i.m. route failed to induce any detectible immune responses. Thus, although rEqIL-12 failed to have an effect on EIAV p15/p26 immune responses measured in experiments reported here, the i.t. and i.d. routes of EIAV DNA vaccine delivery consistently induced antigen-specific responses in adult horses whereas i.m. delivery of the same DNA vaccine using an identical dose, frequency and timing was not immunogenic.
3.7. Antigen-specific responses in EIAV-immunized foals
To determine whether the same routes, dose and immunogen that stimulated a primary EIAV response in adult horses could also stimulate a primary response in neonatal foals, 3 foals were immunized with the EIAV VR-p15/p26 DNA vaccine. Foals did not receive rEqIL-12 DNA, as co-administration of this plasmid had not affected any of the measured parameters in the adult EIAV VR-p15/p26-vaccinated horses or in any of the R. equi vapA DNA-vaccinated animals.
Unlike the adult horses, PBMC from EIAV-vaccinated foals failed to show a significant CTL response to any of the Gag peptide pool-pulsed target cells (data not shown). Furthermore, no tetramer-positive CD8+ T lymphocytes were detected in the peripheral blood of immunized foals. However, as with cells from EIAV-vaccinated adult horses, cells from all 3 foals did show a significant post-vaccination lymphoproliferative response to both Gag pool and p15 peptides compared to preimmunization levels (Fig. 4B). Peak peptide stimulation occurred at different post-vaccination time points for particular foals and antigens, as was also observed using PBMC from the adult horses. All 3 EIAV-vaccinated foals seroconverted to EIAV p26 11–17 days after the third vaccination time point, confirming a humoral response to vaccination. This seroconversion occurred later than seroconversion of 3 of the 4 EIAV-vaccinated adult horses. When retested 12 months later, none of the DNA immunized foals continued to be seropositive for EIAV p26, whereas 2 of the 4 immunized adult horses (A2190 and A2192) remained positive. Both of these horses were seronegative when tested again 18 months post-vaccination. Interestingly, the two persistently seropositive adults were the two that did not receive the rEqIL-12 plasmid.
4. Discussion
Despite their theoretical advantages, apparent capabilities in mice, and movement into human clinical trials, the promise of DNA vaccines has not yet been realized. Notably, the ability of DNA vaccination to induce robust immune responses outside of small laboratory animal models has been disappointing [2]. For example, DNA immunization experiments in humans, non-human primates, and many domestic animals commonly require milligram amounts of DNA to induce detectable responses.
HIV provides a compelling example. Since a live attenuated vaccine is probably unacceptable, considerable effort has been invested in enhancing the efficacy of DNA vaccines in humans and non-human primate models [2]. In one example, a DNA vaccine successfully primed for potent CTL responses in rhesus monkeys challenged with SHIV (a hybrid virus composed of an SIV core and an HIV envelope). Although the vaccine did not prevent SHIV infection, it controlled viral loads, preserved CD4+ T cell counts, and prevented disease and mortality due to AIDS. However, the immunization regimen involved at least three 5 mg i.m. doses of a DNA vaccine and required augmentation with either 5 mg of another plasmid encoding an IL-2/Fc hybrid (interleukin-2 fused with the Fc portion of IgG) or co-administration of the purified IL-2/Fc fusion protein [52,53]. This study and others illustrate the methods that have been explored to improve the potency of DNA vaccines (reviewed in Hokey & Weiner) [2]. Those methods can be subdivided into delivery mechanisms (e.g. alternate routes, gene guns, electroporation, novel formulations or carriers), plasmid design (e.g. promoters, plasmid backbone, leader sequences), adjuvants (e.g. cytokines, costimulatory molecules, TLR ligands, etc.) and others. One important approach has been the use of the cytokine IL-12 as an adjuvant. For example, co-administration of a dual promoter plasmid encoding IL-12 (similar to the rEqIL-12 plasmid employed here) was shown to increase the priming efficiency of a SHIV DNA vaccine in primates and to diminish the dose of an SIV DNA vaccine required to induce cellular immune responses [54,55].
In the present study, we utilized two equine model systems (R. equi VapA and EIAV) to further explore DNA vaccination in large mammals. Our initial hypothesis was that co-administration of a dual promoter plasmid encoding equine IL-12 would enhance the potency of experimental DNA vaccines in the horse. Specifically, the use of IL-12 as molecular adjuvant would increase the ability of the VapA DNA vaccine to induce Type 1 cellular immune responses in neonatal foals and it would significantly improve the potency of a p15/p26 Gag vaccine. The latter EIAV DNA vaccine had previously been ineffective at inducing CTL and tetramer-positive CD8+ T lymphocytes, which are critical for protective immunity. Although IL-12 was shown to be expressed and biologically active in vitro, we were unable to detect a significant adjuvant effect in foals or adult horses. There are several possible explanations. One important consideration is in vivo expression from the IL-12 plasmid, as expression in vitro does not guarantee expression in the horse. The recombinant product may not have been expressed at sufficient levels to act as an effective adjuvant, or may have considerably less biological activity in vivo—at least in conjunction with these two DNA vaccines. One improvement might be expression of IL-12 as a bicistronic message from a single promoter [23,56]. The encoded recombinant fusion protein would include a linker that increases the likelihood of forming an appropriate, biologically active heterodimer and diminishes the possibility of forming inactive or inhibitory homodimers. Nevertheless, dual promoter IL-12 plasmids similar to this one have worked as significant molecular adjuvants in other systems [54,55,57]. In neonates, which have a number of immunologic “defects” when compared to adults, IL-12 alone may also be insufficient to improve the number of foals that respond to DNA vaccination. Although IL-12 production is impaired, the low frequency of APC, diminished APC function, and other limitations in neonates may mean that additional immunostimulation will be required. Perhaps IL-12 will be better used in conjunction with other adjuvants to direct a developing neonatal immune response toward a Th1 cytokine profile and induction of CTL.
In contrast to our previous studies using intramuscular injection, the EIAV p15/p26 Gag vaccine was shown to induce peptide specific CTL activity and tetramer-positive CD8+ T lymphocytes when administered via the same intradermal/intratracheal route used to deliver the VapA DNA vaccine. Again, the IL-12 plasmid had no enhancing effect. The dose was high (5 mg total divided between the two routes) and we have not yet tested whether this route improves potency, as manifested by the ability of a lower dose to produce the same effect. Intradermal delivery of DNA vaccines is postulated to improve uptake of plasmid or plasmid encoded antigen-by-antigen presenting cells (APC), especially compared to injection into skeletal muscle [2]. However, a subsequent study in which the EIAV p15/p26 plasmid was delivered by the intradermal route alone was unable to induce similar immune responses (unpublished data). These data suggest that the intratracheal component, which was originally intended to provide for uptake of R. equi VapA-expressing plasmid by APC in the respiratory tract, is more important than expected. This route may provide for uptake at inductive sites in the upper or lower respiratory tree, or in the pharynx. The intratracheal route proved to be easy and straightforward and, in contrast to a number of other DNA vaccines in the horse (notably influenza and EHV-1), involved a single injection site [4,5,10,58–60] (see Table 1). However, it would be an unusual method of immunization in any species and was intended here as an experimental “proof of concept”. It may be that stimulation of immune cells in more accessible inductive sites such as the pharynx (e.g. intranasal and/or aerosol) will produce similar effects and provide a more practical approach. The development of DNA vaccination strategies in non-murine species seems increasingly empirical; further experiments will be required to better sort the many potential variables.
We have previously shown that tetramer analysis is more sensitive than CTL assays for detection of antigen-specific CD8+ T cells during acute EIAV infection, and that tetramer and CTL assay results do not always correlate [30]. Interestingly, CTL directed against an epitope(s) in Gag peptide pool 1 were detected early in adult horse A2192 with a CTL assay, at a time point (14 days after the second vaccination) when tetramer positive CD8+ T cells were not observed. However, it was likely that that these early CTL recognized an epitope(s) within Gag peptide pool 1 other than Gag-GW12, and that Gag-GW12-specific CTL arose later (as evidenced by the positive tetramer results 14 days after the third vaccination). Also of interest, detectable lymphoproliferative responses in the four adult horses lagged behind the appearance of tetramer positive CD8+ T cells and CTL. Peptides known to contain broadly recognized T helper epitopes were used to stimulate PBMC for these proliferation assays [35,36]. However, because only four such peptides were used, it is possible that early proliferative responses against helper epitopes in other EIAV proteins were missed.
The same dose and route of EIAV Gag p15/p26 DNA vaccine that induced CTL and tetramer-positive CD8+ T lymphocytes in adult horses was unable to do the same in neonates. However, there was an immune response to DNA vaccination in all 3 foals as evidenced by seroconversion to EIAV p26 and peptide-specific lymphoproliferation. This latter finding differs from VapA immunized foals which received a virtually identical plasmid. The only notable changes were dose (5 mg pVR-p15/p26 versus 1 mg pVR1055vapA), route (intratracheal versus intranasal) and codon optimization for p15/p26. In VapA immunized foals, only a subset of foals seroconverted and antigen-specific lymphoproliferation was typically not detected. Co-administration of IL-12 plasmid made no difference. We were also unable to reliably detect IFNγ production by PBMC from immunized foals after stimulation with VapA antigen (measured by real time RT-PCR). The markedly diminished response of foals to the EIAV DNA vaccine and the poor responses of foals to VapA DNA immunization in general likely reflect the inherently decreased immunologic capabilities of neonates compared to adults in all species. This “relative immunodeficiency” of early life and the well described neonatal Th2 bias are among the immunization problems for which DNA vaccines have been advocated as a solution [14,61]. It may be that a better optimized/more potent DNA vaccine will be able to induce strong Type 1 immune responses in young foals. However, it may also be that the “immunologic immaturity” of foals will not be easily overcome using a DNA immunization strategy.
In general, the experience with DNA vaccines in horses has been similar to other large, outbred species (reviewed in Table 1). Although the doses have typically not been as large as utilized in primates, each dose is commonly divided between multiple sites and horses have often been boosted multiple times. A variety of delivery routes and several adjuvants and carriers have been tested. In most cases, DNA immunization has not been very effective—especially when the goal has been to induce cellular immune responses such as CTL and IFNγ production, and when the DNA vaccine was given alone. Responses have been improved with prime-boost strategies, especially when the boost involves a live replicating vector like modified vaccinia Ankara (rMVA) or canary pox virus (ALVAC) [8,58,59]. In the few studies demonstrating immune protection, ponies vaccinated with equine influenza haemagglutinin (HA) were protected against an experimental challenge [4,5,58]. However, the live rMVA vector expressing HA was protective with or without DNA priming [58]. It seems clear that DNA vaccines in the horse need significantly more work and that optimization may be very different for each disease or antigen. Identification of immunoprotective antigens is also an obstacle that is likely to be particularly problematic for pathogens that have large genomes and for which an immune response to just one or two antigens may not be sufficient. A prime-boost strategy employing a live recombinant virus appears most promising, although it remains to be seen if such a strategy will be effective in all situations—such as neonatal foals.
The most notable exception to the experience with DNA vaccination in larger mammals has been flavivirus vaccines [3,62,63]. An equine West Nile Virus vaccine is one of only 2 approved DNA vaccines in the United States and it does not require a prime—heterologous live virus boost strategy. A single i.m. injection of a plasmid expressing the WNV pre-membrane (prM) and envelope (E) proteins protected ponies against a mosquito vectored challenge [3]. DNA vaccination induced neutralizing antibody and prevented viremia, fever, and clinical disease. Similar results have been demonstrated in mice [3,64]. There are probably several reasons for the startling effectiveness of DNA vaccines against flaviruses like WNV and Japanese encephalitis virus. The most important may be that cells transformed with a plasmid expressing the flavivirus coat proteins secrete the antigens in the form of virus-like particles (VLP), which are essentially “empty” virions lacking viral nucleic acid [64]. The assembled VLP are highly immunogenic, replicate the native proteins authentically, and likely foster spread of antigen from injected myocytes to professional APC. Also important, neutralizing antibody alone is protective and is seemingly easy to elicit. Passive transfer of E protein specific neutralizing antibody can protect recipients from flavivirus induced encephalitis [3,65]. Most of the E protein epitopes that elicit virus-neutralizing antibody are conformational and these epitopes appear to be faithfully reproduced in cells transformed with prM/E plasmids. So the apparent disparity by which DNA vaccines are highly effective against flavivirus infection may reflect unique features of the system. In horses, DNA vaccination to prevent WNV infection has seemed to offer few advantages over more traditional immunization approaches. The company that designed the vaccine and acquired approval has not marketed it, as their killed virus product is effective, safe, and has been easier and less expensive to produce given existing infrastructure. DNA vaccination may not offer significant advantages for other pathogens when neutralizing antibody is sufficient and relatively easily induced by more tried and true methods.
In conclusion, it seems clear that the strong T cell responses to DNA vaccines demonstrated in mice will be more difficult to achieve in larger, outbred mammals. At a minimum, considerable experimentation will be required to improve potency and optimize immune responses in these species. Moreover, optimization may differ between host species and for different pathogens. The experiments reported here, especially when considered in the context of other equine DNA vaccine trials, demonstrate the potential effects of dose, route, and plasmid design (codon optimization) in the horse. Although an equine IL-12 plasmid had no detectable adjuvant effect, improved plasmid design (e.g. a single promoter and bicistronic mRNA) could produce better results. Importantly, foals consistently responded poorly to DNA vaccination raising the possibility that a DNA approach would require further modification to effectively overcome the limitations of the immune system inherent to the neonatal and perinatal periods.
Acknowledgments
Funding was provided by the Grayson-Jockey Club Research Foundation, the Cadeau Foundation, and NIH Grants AI058787 and AI067125. The authors gratefully acknowledge the outstanding animal care provided by Emma Karel and Lori Fuller.
References
- 1.Gurunathan S, Klinman DM, Seder RA. DNA vaccines: immunology, application, and optimization. Annu Rev Immunol. 2000;18:927–74. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 2.Hokey DA, Weiner DB. DNA vaccines for HIV: challenges and opportunities. Springer Semin Immunopathol. 2006;28(3):267–79. doi: 10.1007/s00281-006-0046-z. [DOI] [PubMed] [Google Scholar]
- 3.Davis BS, Chang GJ, Cropp B, Roehrig JT, Martin DA, Mitchell CJ, et al. West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J Virol. 2001;75(9):4040–7. doi: 10.1128/JVI.75.9.4040-4047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lunn DP, Soboll G, Schram BR, Quass J, McGregor MW, Drape RJ, et al. Antibody responses to DNA vaccination of horses using the influenza virus hemagglutinin gene. Vaccine. 1999;17(18):2245–58. doi: 10.1016/s0264-410x(98)00496-4. [DOI] [PubMed] [Google Scholar]
- 5.Soboll G, Nelson KM, Leuthner ES, Clark RJ, Drape R, Macklin MD, et al. Mucosal co-administration of cholera toxin and influenza virus hemagglutinin-DNA in ponies generates a local IgA response. Vaccine. 2003;21(21–22):3081–92. doi: 10.1016/s0264-410x(03)00161-0. [DOI] [PubMed] [Google Scholar]
- 6.Giese M, Bahr U, Jakob NJ, Kehm R, Handermann M, Muller H, et al. Stable and long-lasting immune response in horses after DNA vaccination against equine arteritis virus. Virus Genes. 2002;25(2):159–67. doi: 10.1023/a:1020109801925. [DOI] [PubMed] [Google Scholar]
- 7.Foote CE, Love DN, Gilkerson JR, Rota J, Trevor-Jones P, Ruitenberg KM, et al. Serum antibody responses to equine herpesvirus 1 glycoprotein D in horses, pregnant mares and young foals. Vet Immunol Immunopathol. 2005;105(1–2):47–57. doi: 10.1016/j.vetimm.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Minke JM, Fischer L, Baudu P, Guigal PM, Sindle T, Mumford JA, et al. Use of DNA and recombinant canarypox viral (ALVAC) vectors for equine herpes virus vaccination. Vet Immunol Immunopathol. 2006;111(1–2):47–57. doi: 10.1016/j.vetimm.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Ruitenberg KM, Love DN, Gilkerson JR, Wellington JE, Whalley JM. Equine herpesvirus 1 (EHV-1) glycoprotein D DNA inoculation in horses with pre-existing EHV-1/EHV-4 antibody. Vet Microbiol. 2000;76(2):117–27. doi: 10.1016/s0378-1135(00)00237-6. [DOI] [PubMed] [Google Scholar]
- 10.Soboll G, Hussey SB, Whalley JM, Allen GP, Koen MT, Santucci N, et al. Antibody and cellular immune responses following DNA vaccination and EHV-1 infection of ponies. Vet Immunol Immunopathol. 2006;111(1–2):81–95. doi: 10.1016/j.vetimm.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Cantlon JD, Gordy PW, Bowen RA. Immune responses in mice, cattle and horses to a DNA vaccine for vesicular stomatitis. Vaccine. 2000;18(22):2368–74. doi: 10.1016/s0264-410x(00)00007-4. [DOI] [PubMed] [Google Scholar]
- 12.Fischer L, Minke J, Dufay N, Baudu P, Audonnet JC. Rabies DNA vaccine in the horse: strategies to improve serological responses. Vaccine. 2003;21(31):4593–6. doi: 10.1016/s0264-410x(03)00504-8. [DOI] [PubMed] [Google Scholar]
- 13.Cook RF, Cook SJ, Bolin PS, Howe LJ, Zhou W, Montelaro RC, et al. Genetic immunization with codon-optimized equine infectious anemia virus (EIAV) surface unit (SU) envelope protein gene sequences stimulates immune responses in ponies. Vet Microbiol. 2005;108(1–2):23–37. doi: 10.1016/j.vetmic.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Bot A, Bona C. Genetic immunization of neonates. Microbes Infect. 2002;4(4):511–20. doi: 10.1016/s1286-4579(02)01566-6. [DOI] [PubMed] [Google Scholar]
- 15.Meijer WG, Prescott JF. Rhodococcus equi. Vet Res. 2004;35(4):383–96. doi: 10.1051/vetres:2004024. [DOI] [PubMed] [Google Scholar]
- 16.Breathnach CC, Sturgill-Wright T, Stiltner JL, Adams AA, Lunn DP, Horohov DW. Foals are interferon gamma-deficient at birth. Vet Immunol Immunopathol. 2006;112(3–4):199–209. doi: 10.1016/j.vetimm.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Patton KM, McGuire TC, Hines MT, Mealey RH, Hines SA. Rhodococcus equi-specific cytotoxic T lymphocytes in immune horses and development in asymptomatic foals. Infect Immun. 2005;73(4):2083–93. doi: 10.1128/IAI.73.4.2083-2093.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hines SA, Stone DM, Hines MT, Alperin DC, Knowles DP, Norton LK, et al. Clearance of virulent but not avirulent Rhodococcus equi from the lungs of adult horses is associated with intracytoplasmic gamma interferon production by CD4+ and CD8+ T lymphocytes. Clin Diagn Lab Immunol. 2003;10(2):208–15. doi: 10.1128/CDLI.10.2.208-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patton KM, McGuire TC, Fraser DG, Hines SA. Rhodococcus equi-infected macrophages are recognized and killed by CD8+ T lymphocytes in a major histocompatibility complex class I-unrestricted fashion. Infect Immun. 2004;72(12):7073–83. doi: 10.1128/IAI.72.12.7073-7083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez AM, Hines MT, Palmer GH, Knowles DP, Alperin DC, Hines SA. Analysis of anamnestic immune responses in adult horses and priming in neonates induced by a DNA vaccine expressing the vapA gene of Rhodococcus equi. Vaccine. 2003;21(25–26):3815–25. doi: 10.1016/s0264-410x(03)00329-3. [DOI] [PubMed] [Google Scholar]
- 21.Ottenhoff TH, Verreck FA, Hoeve MA, van de Vosse E. Control of human host immunity to mycobacteria. Tuberculosis (Edinb) 2005;85(1–2):53–64. doi: 10.1016/j.tube.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Pertmer TM, Oran AE, Madorin CA, Robinson HL. Th1 genetic adjuvants modulate immune responses in neonates. Vaccine. 2001;19(13–14):1764–71. doi: 10.1016/s0264-410x(00)00388-1. [DOI] [PubMed] [Google Scholar]
- 23.McMonagle EL, Taylor S, van Zuilekom H, Sanders L, Scholtes N, Keanie LJ, et al. Production of biologically active equine interleukin 12 through expression of p35, p40 and single chain IL-12 in mammalian and baculovirus expression systems. Equine Vet J. 2001;33(7):693–8. doi: 10.2746/042516401776249426. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44(2):283–92. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 25.Chizzonite R, Truitt T, Podlaski FJ, Wolitzky AG, Quinn PM, Nunes P, et al. IL-12: monoclonal antibodies specific for the 40-kDa subunit block receptor binding and biologic activity on activated human lymphoblasts. J Immunol. 1991;147(5):1548–56. [PubMed] [Google Scholar]
- 26.Gately MK, Desai BB, Wolitzky AG, Quinn PM, Dwyer CM, Podlaski FJ, et al. Regulation of human lymphocyte proliferation by a heterodimeric cytokine, IL-12 (cytotoxic lymphocyte maturation factor) J Immunol. 1991;147(3):874–82. [PubMed] [Google Scholar]
- 27.McGuire TC, O’Rourke KI, Baszler TV, Leib SR, Brassfield AL, Davis WC. Expression of functional protease and subviral particles by vaccinia virus containing equine infectious anaemia virus gag and 5’ pol genes. J Gen Virol. 1994;75(Pt 4):895–900. doi: 10.1099/0022-1317-75-4-895. [DOI] [PubMed] [Google Scholar]
- 28.Mealey RH, Zhang B, Leib SR, Littke MH, McGuire TC. Epitope specificity is critical for high and moderate avidity cytotoxic T lymphocytes associated with control of viral load and clinical disease in horses with equine infectious anemia virus. Virology. 2003;313(2):537–52. doi: 10.1016/s0042-6822(03)00344-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGuire TC, Leib SR, Mealey RH, Fraser DG, Prieur DJ. Presentation binding affinity of equine infectious anemia virus CTL envelope and matrix protein epitopes by an expressed equine classical MHC class I molecule. J Immunol. 2003;171(4):1984–93. doi: 10.4049/jimmunol.171.4.1984. [DOI] [PubMed] [Google Scholar]
- 30.Mealey RH, Sharif A, Ellis SA, Littke MH, Leib SR, McGuire TC. Early detection of dominant Env-specific and subdominant Gag-specific CD8+ lymphocytes in equine infectious anemia virus-infected horses using major histocompatibility complex class I/peptide tetrameric complexes. Virology. 2005;339(1):110–26. doi: 10.1016/j.virol.2005.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez AM, Hines MT, Palmer GH, Alperin DC, Hines SA. Identification of pulmonary T-lymphocyte and serum antibody isotype responses associated with protection against Rhodococcus equi. Clin Diagn Lab Immunol. 2002;9(6):1270–6. doi: 10.1128/CDLI.9.6.1270-1276.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan C, Prescott JF, Patterson MC, Nicholson VM. Molecular characterization of a lipid-modified virulence-associated protein of Rhodococcus equi and its potential in protective immunity. Can J Vet Res. 1995;59(1):51–9. [PMC free article] [PubMed] [Google Scholar]
- 33.Hines MT, Paasch KM, Alperin DC, Palmer GH, Westhoff NC, Hines SA. Immunity to Rhodococcus equi: antigen-specific recall responses in the lungs of adult horses. Vet Immunol Immunopathol. 2001;79(1–2):101–14. doi: 10.1016/s0165-2427(01)00258-6. [DOI] [PubMed] [Google Scholar]
- 34.Wada R, Kamada M, Anzai T, Nakanishi A, Kanemaru T, Takai S, et al. Pathogenicity and virulence of Rhodococcus equi in foals following intratracheal challenge. Vet Microbiol. 1997;56(3–4):301–12. doi: 10.1016/s0378-1135(97)00098-9. [DOI] [PubMed] [Google Scholar]
- 35.Fraser DG, Leib SR, Zhang BS, Mealey RH, Brown WC, McGuire TC. Lymphocyte proliferation responses induced to broadly reactive Th peptides did not protect against equine infectious anemia virus challenge. Clin Diagn Lab Immunol. 2005;12(8):983–93. doi: 10.1128/CDLI.12.8.983-993.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraser DG, Oaks JL, Brown WC, McGuire TC. Identification of broadly recognized. T helper 1 lymphocyte epitopes in an equine lentivirus. Immunology. 2002;105(3):295–305. doi: 10.1046/j.0019-2805.2001.01370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen G, Yeargan M, Costa LR, Cross R. Major histocompatibility complex class I-restricted cytotoxic T-lymphocyte responses in horses infected with equine herpesvirus 1. J Virol. 1995;69(1):606–12. doi: 10.1128/jvi.69.1.606-612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohler AK, Stone DM, Hines MT, Byrne BA, Alperin DC, Norton LK, et al. Rhodococcus equi secreted antigens are immunogenic and stimulate a type 1 recall response in the lungs of horses immune to R. equi infection. Infect Immun. 2003;71(11):6329–37. doi: 10.1128/IAI.71.11.6329-6337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheoran AS, Lunn DP, Holmes MA. Monoclonal antibodies to subclass-specific antigenic determinants on equine immunoglobulin gamma chains and their characterization. Vet Immunol Immunopathol. 1998;62(2):153–65. doi: 10.1016/s0165-2427(97)00162-1. [DOI] [PubMed] [Google Scholar]
- 40.Chung C, Mealey RH, McGuire TC. CTL from EIAV carrier horses with diverse MHC class I alleles recognize epitope clusters in Gag matrix and capsid proteins. Virology. 2004;327(1):144–54. doi: 10.1016/j.virol.2004.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGuire TC, Tumas DB, Byrne KM, Hines MT, Leib SR, Brassfield AL, et al. Major histocompatibility complex-restricted CD8+ cytotoxic T lymphocytes from horses with equine infectious anemia virus recognize Env and Gag/PR proteins. J Virol. 1994;68(3):1459–67. doi: 10.1128/jvi.68.3.1459-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siliciano RF, Keegan AD, Dintzis RZ, Dintzis HM, Shin HS. The interaction of nominal antigen with T cell antigen receptors. I. Specific binding of multivalent nominal antigen to cytolytic T cell clones. J Immunol. 1985;135(2):906–14. [PubMed] [Google Scholar]
- 43.Blanchard-Channell M, Moore PF, Stott JL. Characterization of monoclonal antibodies specific for equine homologues of CD3 and CD5. Immunology. 1994;82(4):548–54. [PMC free article] [PubMed] [Google Scholar]
- 44.Lunn DP, Holmes MA, Antczak DF, Agerwal N, Baker J, Bendali-Ahcene S, et al. Report of the Second Equine Leucocyte Antigen Workshop, Squaw valley, California, July 1995. Vet Immunol Immunopathol. 1998;62(2):101–43. doi: 10.1016/s0165-2427(97)00160-8. [DOI] [PubMed] [Google Scholar]
- 45.Kydd J, Antczak DF, Allen WR, Barbis D, Butcher G, Davis W, et al. Report of the First International Workshop on Equine Leucocyte Antigens, Cambridge, UK, July 1991. Vet Immunol Immunopathol. 1994;42(1):3–60. doi: 10.1016/0165-2427(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 46.Bailey E. Identification and genetics of horse lymphocyte alloantigens. Immunogenetics. 1980;11(5):499–506. doi: 10.1007/BF01567818. [DOI] [PubMed] [Google Scholar]
- 47.Bailey E. Population studies on the ELA system in American standard-bred and thoroughbred mares. Anim Blood Groups Biochem Genet. 1983;14(3):201–11. doi: 10.1111/j.1365-2052.1983.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 48.Terasaki PI, Bernoco D, Park MS, Ozturk G, Iwaki Y. Microdroplet testing for HLA-A, -B, -C, and -D antigens, The Phillip Levine Award Lecture. Am J Clin Pathol. 1978;69(2):103–20. doi: 10.1093/ajcp/69.2.103. [DOI] [PubMed] [Google Scholar]
- 49.Gillessen S, Carvajal D, Ling P, Podlaski FJ, Stremlo DL, Familletti PC, et al. Mouse interleukin-12 (IL-12) p40 homodimer: a potent IL-12 antagonist. Eur J Immunol. 1995;25(1):200–6. doi: 10.1002/eji.1830250133. [DOI] [PubMed] [Google Scholar]
- 50.Heinzel FP, Hujer AM, Ahmed FN, Rerko RM. In vivo production and function of IL-12 p40 homodimers. J Immunol. 1997;158(9):4381–8. [PubMed] [Google Scholar]
- 51.Kato K, Shimozato O, Hoshi K, Wakimoto H, Hamada H, Yagita H, et al. Local production of the p40 subunit of interleukin 12 suppresses T-helper 1-mediated immune responses and prevents allogeneic myoblast rejection. Proc Natl Acad Sci USA. 1996;93(17):9085–9. doi: 10.1073/pnas.93.17.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barouch DH, Fu TM, Montefiori DC, Lewis MG, Shiver JW, Letvin NL. Vaccine-elicited immune responses prevent clinical AIDS in SHIV(89. 6P)-infected rhesus monkeys. Immunol Lett. 2001;79(1–2):57–61. doi: 10.1016/s0165-2478(01)00266-8. [DOI] [PubMed] [Google Scholar]
- 53.Barouch DH, Santra S, Schmitz JE, Kuroda MJ, Fu TM, Wagner W, et al. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290(5491):486–92. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 54.Egan MA, Chong SY, Megati S, Montefiori DC, Rose NF, Boyer JD, et al. Priming with plasmid DNAs expressing interleukin-12 and simian immunodeficiency virus gag enhances the immunogenicity and efficacy of an experimental AIDS vaccine based on recombinant vesicular stomatitis virus. AIDS Res Hum Retroviruses. 2005;21(7):629–43. doi: 10.1089/aid.2005.21.629. [DOI] [PubMed] [Google Scholar]
- 55.Schadeck EB, Sidhu M, Egan MA, Chong SY, Piacente P, Masood A, et al. A dose sparing effect by plasmid encoded IL-12 adjuvant on a SIVgag-plasmid DNA vaccine in rhesus macaques. Vaccine. 2006;24(21):4677–87. doi: 10.1016/j.vaccine.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 56.Lee YL, Ye YL, Yu CI, Wu YL, Lai YL, Ku PH, et al. Construction of single-chain interleukin-12 DNA plasmid to treat airway hyperresponsiveness in an animal model of asthma. Hum Gene Ther. 2001;12(17):2065–79. doi: 10.1089/10430340152677412. [DOI] [PubMed] [Google Scholar]
- 57.Chong SY, Egan MA, Kutzler M, Megati S, Masood A, Roopchard V, et al. Comparative ability of plasmid IL-12 and IL-15 to enhance cellular and humoral immune responses elicited by a SIVgag plasmid DNA vaccine and alter disease progression following SHIV(89.6P) challenge in rhesus macaques. Vaccine. 2007 doi: 10.1016/j.vaccine.2006.11.070. [DOI] [PubMed] [Google Scholar]
- 58.Breathnach CC, Clark HJ, Clark RC, Olsen CW, Townsend HG, Lunn DP. Immunization with recombinant modified vaccinia Ankara (rMVA) constructs encoding the HA or NP gene protects ponies from equine influenza virus challenge. Vaccine. 2006;24(8):1180–90. doi: 10.1016/j.vaccine.2005.08.091. [DOI] [PubMed] [Google Scholar]
- 59.Breathnach CC, Rudersdorf R, Lunn DP. Use of recombinant modified vaccinia Ankara viral vectors for equine influenza vaccination. Vet Immunol Immunopathol. 2004;98(3–4):127–36. doi: 10.1016/j.vetimm.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Soboll G, Horohov DW, Aldridge BM, Olsen CW, McGregor MW, Drape RJ, et al. Regional antibody and cellular immune responses to equine influenza virus infection, and particle mediated DNA vaccination. Vet Immunol Immunopathol. 2003;94(1–2):47–62. doi: 10.1016/s0165-2427(03)00060-6. [DOI] [PubMed] [Google Scholar]
- 61.Siegrist CA. Neonatal and early life vaccinology. Vaccine. 2001;19(25–26):3331–46. doi: 10.1016/s0264-410x(01)00028-7. [DOI] [PubMed] [Google Scholar]
- 62.Chang GJ, Davis BS, Hunt AR, Holmes DA, Kuno G. Flavivirus DNA vaccines: current status and potential. Ann N Y Acad Sci. 2001;951:272–85. [PubMed] [Google Scholar]
- 63.Powell K. DNA vaccines—back in the saddle again? Nat Biotechnol. 2004;22(7):799–801. doi: 10.1038/nbt0704-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang GJ, Hunt AR, Davis B. A single intramuscular injection of recombinant plasmid DNA induces protective immunity and prevents Japanese encephalitis in mice. J Virol. 2000;74(9):4244–52. doi: 10.1128/jvi.74.9.4244-4252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang MJ, Wang MJ, Jiang SZ, Ma WY. Passive protection of mice, goats, and monkeys against Japanese encephalitis with monoclonal antibodies. J Med Virol. 1989;29(2):133–8. doi: 10.1002/jmv.1890290211. [DOI] [PubMed] [Google Scholar]