Abstract
Leishmania parasites lack a purine biosynthetic pathway and depend on surface nucleoside and nucleobase transporters to provide them with host purines. Leishmania donovani possess two closely related genes that encode high affinity adenosine-pyrimidine nucleoside transporters LdNT1.1 and LdNT1.2 and that transport the toxic adenosine analog tubercidin in addition to the natural substrates. In this study, we have characterized a drug-resistant clonal mutant of L. donovani (TUBA5) that is deficient in LdNT1 transport and consequently resistant to tubercidin. In TUBA5 cells, the LdNT1.2 genes had the same sequence as wild-type cells. However, because LdNT1.2 mRNA is not detectable in either wild-type or TUBA5 promastigotes, LdNT1.2 does not contribute to nucleoside transport in this stage of the life cycle. In contrast, the TUBA5 cells were compound heterozygotes at the LdNT1.1 locus containing two mutant alleles that encompassed distinct point mutations, each of which impaired transport function. One of the mutant LdNT1.1 alleles encoded a G183D substitution in predicted TM 5, and the other allele contained a C337Y change in predicted TM 7. Whereas G183D and C337Y mutants had only slightly elevated adenosine Km values, the severe impairment in transport resulted from drastically (≈20-fold) reduced Vmax values. Because these transporters were correctly targeted to the plasma membrane, the reduction in Vmax apparently resulted from a defect in translocation. Strikingly, G183 was essential for pyrimidine nucleoside but not adenosine transport. A mutant transporter with a G183A substitution had an altered substrate specificity, exhibiting robust adenosine transport but undetectable uridine uptake. These results suggest that TM 5 is likely to form part of the nucleoside translocation pathway in LdNT1.1
Purine salvage pathways are critical to the survival of parasitic protozoa that lack the ability to synthesize the purine ring de novo (1). Parasite nucleoside transporters located on the plasma membrane perform the crucial function of transporting preformed purine nucleosides from the host into the parasite, the first step in the salvage process. These transporters are also relevant pharmacologically because they mediate the uptake of important anti-leishmanial agents such as allopurinol riboside that are analogs of the natural substrates (2).
Leishmania donovani promastigotes (the parasite form within the gut of the sandfly vector) possess two biochemically and genetically distinct nucleoside transport activities, one for the uptake of adenosine and pyrimidine nucleosides (LdNT1) (3) and the other for the transport of the purine nucleosides inosine and guanosine (LdNT2) (4). LdNT1 and LdNT2 also mediate the membrane permeation of the cytotoxic adenosine and inosine analogs tubercidin and formycin B, respectively (5). A mutant L. donovani clone, TUBA5, that is deficient in LdNT1 transport activity has been isolated by mutagenesis of WT parasites with N-methyl-N-nitroso-N′-nitroguanidine, followed by selection in tubercidin (5). The TUBA5 cell line is incapable of transporting tubercidin, adenosine, and pyrimidine nucleosides but transports inosine and guanosine normally (5).
The genes coding for the LdNT1 transporters were isolated by virtue of their ability to restore tubercidin sensitivity to the transport-deficient TUBA5 cell line (3). Two closely related and tandemly arranged genes, LdNT1.1 and LdNT1.2, were identified that encoded permeases that lacked obvious cleavable signal sequences and exhibited high apparent affinities for their substrates (3). Whereas LdNT1.1 was abundantly expressed in both wild-type (WT) and TUBA5 promastigotes, there was no detectable LdNT1.2 transcript in promastigotes of either cell type (3). Consequently, all of the nucleoside transport activity in promastigotes is contributed by the LdNT1.1 genes. Because L. donovani is a diploid organism, both copies of LdNT1.1 must either be inactive or encode nonfunctional proteins. Previously, it has been demonstrated that the TUBA5 line has not suffered any large deletion or rearrangement at the NT1 locus that would disrupt the LdNT1.1 and LdNT1.2 ORFs (3). Furthermore, the presence of a full-length LdNT1.1 transcript in the TUBA5 promastigotes suggested that the mutations that lead to drug resistance did not interfere with either transcription, RNA processing, or RNA stability (3).
The present study was undertaken to understand the genetic basis for the transport-deficient and drug-resistant phenotype of the TUBA5 cell line. Whereas both LdNT1.2 genes were WT, single but distinct point mutations were identified within the ORFs of the two LdNT1.1 genes that each produced functionally inactive transporters. One of these mutated LdNT1.1 alleles encoded a transporter with a Gly-183 → Asp (G183D) substitution in predicted transmembrane segment 5 (TM 5) and the other a transporter with a Cys-337 → Tyr (C337Y) substitution in predicted TM 7. Both mutations dramatically lowered the Vmax values of transport but had little effect on the apparent affinity of the transporter for adenosine. Interestingly, when Gly-183 was replaced by Ala, the resulting permease retained the ability to transport adenosine but was severely impaired in its ability to transport the pyrimidine nucleoside uridine. Thus, Gly-183 plays an important role in determining the substrate selectivity of LdNT1.1. These results suggest that a forward genetic approach can be a very powerful tool in identifying residues critical for permeation and for governing substrate specificity of nucleoside transporters.
Materials and Methods
Materials.
[2,8,5-3H]adenosine (54.4 Ci/mmol) and [5,6-3H]uridine (39.5 Ci/mmol) was purchased from NEN. Monoclonal anti-green fluorescent protein (GFP) antibodies were obtained from CLONTECH. All other chemicals and reagents were of the highest commercial quality available.
Parasite Cell Culture.
The TUBA5 (5) strain of L. donovani was cultured at 26°C in DME-L (6) containing 10% FCS. TUBA5 transfectants were propagated in DME-L supplemented with either 100 μg/ml hygromycin B or G418. Genomic DNA was isolated from TUBA5 and DI700 strains as previously described (7).
Cloning and Sequencing of LdNT1.1 and LdNT1.2 Genes from TUBA5 Parasites.
Previous results (G.V., unpublished work) indicated that both LdNT1.1 and LdNT1.2 genes were contained within a single ≈17-kb HindIII fragment. Therefore, a phage library of HindIII-digested L. donovani TUBA5 genomic DNA was constructed in the Lambda DASH II vector (Stratagene) according to protocols supplied by the manufacturer. The library was screened with the LdNT1.1 ORF as probe, and positive clones were isolated by using standard protocols (8). Phage DNA from two of these clones was isolated by using the Phage Midiprep kit (Qiagen, Chatsworth, CA) and digested with EcoRI and XbaI to release a 7.5-kb fragment containing the LdNT1.1 gene and a 3.5-kb fragment containing the LdNT1.2 gene (3). Each of these fragments from both phage clones was subcloned into pBluescript for sequencing. LdNT1.1 and LdNT1.2 were then sequenced from both strands by the Oregon Health Sciences University Microbiology Research Core Facility by using a model 377 Applied Biosystems automated fluorescence sequencer (Perkin–Elmer).
PCR amplifications of the LdNT1.1 and LdNT1.2 ORFs from TUBA5 genomic DNA were performed by using primers specific for each gene within the 5′ and 3′ untranslated regions and Pfu Turbo polymerase (Stratagene). All amplified fragments were subcloned by using the Zero Blunt TOPO PCR Cloning kit (Invitrogen) and sequenced as described above.
Site-Directed Mutagenesis and Construction of Plasmids.
Mutagenesis was performed by using the QuikChange Site-Directed Mutagenesis kit (Stratagene) and WT LdNT1.1 subcloned in PCRR-Blunt II-TOPO (Invitrogen) as template. Mutagenic primers were designed to incorporate both the desired mutation and a silent change to introduce a restriction site to facilitate screening. Mutagenized constructs were analyzed both by restriction digestion and sequencing. For expression in TUBA5 cells, WT and mutated LdNT1.1 genes were excised from PCRR-Blunt II-TOPO with BamHI and EcoRV and subcloned into the BglII/SmaI sites of the leishmanial expression vector pX63Hyg (9).
To generate GFP fusions at the amino terminus of LdNT1.1 (GFP-LdNT1.1), ORFs of the WT and mutated LdNT1.1 genes were amplified by PCR by using forward and reverse primers containing BamHI restriction sites. After restriction digestion, the ORFs were subcloned into the BamHI site of the pXGGFP+2′ vector (10). All constructs were transfected into TUBA5 parasites as described (11).
Uptake Assays.
Time courses of [3H]adenosine and [3H]uridine uptake were performed as reported (12). Briefly, parasites were incubated with radiolabeled substrate for various times and then centrifuged through a cushion of dibutylphthalate. For kinetic analysis, initial rates of uptake at each substrate concentration were determined by linear regression analysis over the linear portion of the time course. These data were fitted to the Michaelis-Menton equation by least-squares analysis by using the grafit program (Erithacus Software, Horley, Surrey, U.K.).
Preparation of Cell Lysates and Immunoblots.
Total cell lysates were prepared from TUBA5 parasites as described (13). Appropriate volumes of lysates containing comparable amounts of protein were mixed with equal volumes of Laemmli sample buffer (8), heated at 65°C for 5 min, separated on 8% SDS polyacrylamide gels, and electroblotted onto nitrocellulose by standard methods (8). The blots were blocked with 5% nonfat dried milk in TBST (20 mM Tris, pH 7.6/150 mM NaCl/0.05% Tween 20) for 1 h and then incubated with the monoclonal anti-GFP antibody for 2 h. Blots were developed with a horseradish peroxidase-conjugated secondary goat anti-mouse (1:10,000) IgG and the chemiluminescence kit (Pierce) according to the manufacturer's instructions and exposed to XAR-5 film (Eastman Kodak).
Fluorescence Localization.
For localization of the GFP-tagged LdNT1.1 proteins, parasites were pelleted, washed in ice-cold PBS, and resuspended at a density of ≈107 cells/ml. Cells were attached to poly-l-lysine-coated coverslips and fixed with 100% methanol for 5 min. Coverslips were rinsed with PBS and mounted on slides in fluoromount-G (Southern Biotechnology Associates). Images were acquired and deconvolved by using the Deltavision Image Restoration System from Applied Precision (Issaquah, WA).
Results
Cloning and Sequencing of LdNT1 Genes from TUBA5 Parasites.
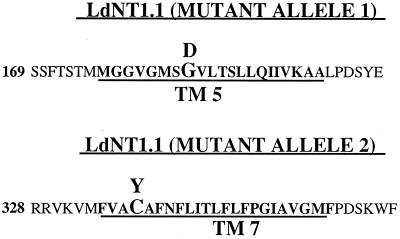
To determine whether mutations within the LdNT1 ORFs were responsible for the transport-deficient and tubercidin-resistant phenotype, LdNT1.1 and LdNT1.2 from the TUBA5 cell line were cloned and sequenced. The LdNT1.1 ORF from one of two independently isolated genomic clones contained a single G → A transition at nucleotide position 548 and thus encoded a transporter with a G183D substitution in predicted TM 5 (Fig. 1). Interestingly, the LdNT1.1 ORF from the second clone contained a G → A transition at a different position within the ORF, nucleotide 1010, and encoded a transporter with a C337Y missense substitution in predicted TM 7 (Fig. 1). These results indicated that both LdNT1.1 genes had acquired single but distinct point mutations, resulting in transporters with nonconservative amino acid substitutions. In contrast, the LdNT1.2 genes from both phage clones contained WT sequences. To confirm these results, LdNT1.1 and LdNT1.2 were amplified from TUBA5 genomic DNA by PCR and sequenced. Four of the five independently amplified LdNT1.1 genes encoded the C337Y mutation, whereas the fifth encoded the G183D substitution. All four independently amplified LdNT1.2 genes had the WT sequence.
Figure 1.
Location of missense mutations in the TUBA5 LdNT1.1 transporters. The positions of the G183D and C337Y mutations in the LdNT1.1 transporters encoded by the two mutant alleles of the TUBA5 cell line are indicated. The lines below the sequences indicate the positions of the predicted transmembrane segments (3).
Functional Characterization of the G183D and C337Y Transporters.
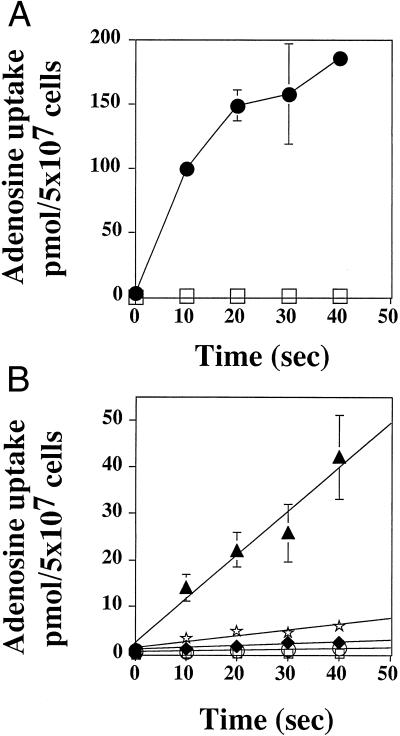
To determine whether each of the G183D and C337Y mutations was sufficient to produce a nonfunctional transporter, each mutation was individually introduced into WT LdNT1.1 by site-directed mutagenesis, and TUBA5 parasites transfected with these mutagenized constructs were assayed for [3H]adenosine uptake. Whereas TUBA5 parasites expressing WT LdNT1.1 exhibited robust uptake of radiolabeled adenosine, parasites expressing G183D and C337Y showed greatly reduced [3H]adenosine transport capabilities (Fig. 2 A and B). Moreover, whereas WT LdNT1.1 restored the sensitivity of TUBA5 parasites to tubercidin, TUBA5 parasites overexpressing G183D and C337Y transporters continued to display the drug resistance phenotype (data not shown). These results confirmed that the G183D and C337Y mutations were responsible for both the transport-deficient and drug-resistant phenotype of the TUBA5 cells.
Figure 2.
Functional characterization of G183 and C337 mutants. TUBA5 parasites expressing (A) WT LdNT1.1 (●) or vector pX63Hyg (□) and (B) G183A (▴), G183N (⋆), G183D (♦), C337Y (○), or pX63Hyg (□) were tested for uptake of 1 μM [3H]adenosine. Results are expressed as mean ± SD (n = 2).
Because both mutated transporters showed some residual transport activity when overexpressed in TUBA5 parasites, we were able to characterize them kinetically. G183D- and C337Y-mediated uptake of adenosine was saturable with apparent Km values of 0.77 ± 0.13 μM (n = 2) and 0.9 ± 0.17 μM (n = 2), respectively. These values are somewhat higher than the Km value of WT LdNT1.1 (0.17 ± 0.09 μM) (3), but this modest increase in Km cannot explain the transport-deficient phenotype. To determine whether the Vmax values of the mutant transporters were reduced, GFP fusion proteins were constructed in which ORFs of WT LdNT1.1, G183D, and C337Y were fused to the carboxyl terminus of GFP and expressed in the TUBA5 parasites. The results of substrate saturation experiments performed with the GFP fusion proteins are shown in Table 1. Consistent with earlier findings, GFP-G183D and GFP-C337Y displayed only slight increases in the apparent Km values for adenosine, compared with GFP-WT, but significantly lower Vmax values. Western blots probed with a monoclonal anti-GFP antibody revealed that GFP-C337Y was expressed at slightly higher levels (1.5-fold) than GFP-WT, whereas GFP-G183D levels were about 2-fold lower (data not shown). Vmax values normalized to GFP fusion protein levels are shown in Table 1. Thus GFP-G183D and GFP-C337Y have ≈18- and ≈23-fold lower Vmax values compared with GFP-WT, respectively, confirming that the mutations primarily affect Vmax.
Table 1.
Kinetic parameters for WT and mutant transporters
| Km, μM | Vmax, pmol/108 cells/s | Vnmax, pmol/108 cells/s | |
|---|---|---|---|
| WT | 0.38 ± 0.02 | 3.24 ± 0.22 | 3.24 ± 0.22 |
| G183D | 2.3 ± 0.45 | 0.09 ± 0.008 | 0.18 ± 0.006 |
| C337Y | 1.99 ± 0.3 | 0.22 ± 0.014 | 0.14 ± 0.008 |
Substrate saturation curves for adenosine were obtained for TUBA5 parasites expressing GFP-WT, GFP-G183D, and GFP-C337Y as described in Materials and Methods. Each value is the mean ± SD of two independent determinations. V
max values are Vmax values normalized to protein levels determined by Western blot analysis as described in the text.
Subcellular Localization of GFP-WT, GFP-G183D, and GFP-C337Y.
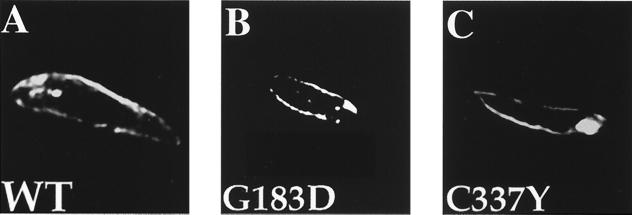
The reduction in Vmax values could reflect either the failure of G183D and C337Y to properly target to the cell surface or the innate impairment of transporter function. To determine whether the two mutants were correctly targeted to the cell surface, the subcellular locations of GFP-WT, GFP-G183D, and GFP-C337Y were monitored by using GFP fluorescence. As shown in Fig. 3A, GFP-WT was localized to the plasma membrane of the parasite cell body but excluded from the flagellum. A control TUBA5 cell line expressing GFP alone showed diffuse staining throughout the cytoplasm (data not shown). Like GFP-WT, both GFP-G183D and GFP-C337Y were targeted to the plasma membrane of the parasite (Fig. 3 B and C). Furthermore, there was little or no staining of the endoplasmic reticulum and Golgi apparatus, indicating that these mutations did not cause substantial misfolding and retention within internal membranes. These results suggest that the G183D and C337Y mutations produced the TUBA5 phenotype by affecting some aspect of the transporter cycle itself.
Figure 3.
Deconvolved fluorescence images of TUBA5 parasites overexpressing (A) GFP-WT, (B) GFP-G183D, and (C) GFP-C337Y.
Gly183 Is a Determinant of Substrate Selectivity.
To analyze the function of G183 further, site-directed mutagenesis was used to replace this residue with Ala (G183A) or Asn (G183N). TUBA5 parasites expressing G183A and G183N transporters were assayed for their ability to take up [3H]adenosine. Fig. 2B shows that G183A transported adenosine robustly whereas the G183N mutant was severely impaired in transport. Thus, although G183 was not essential for adenosine transport, amino acids with bulky (G183N) and/or charged (G183D) side chains were not tolerated at this position. Similarly, a C337S mutant was capable of transporting adenosine, albeit at lower rates than WT, suggesting that C337 per se was not essential for function, whereas the C337F mutation completely abolished transport, indicating that size restrictions operated at this amino acid position as well (data not shown).
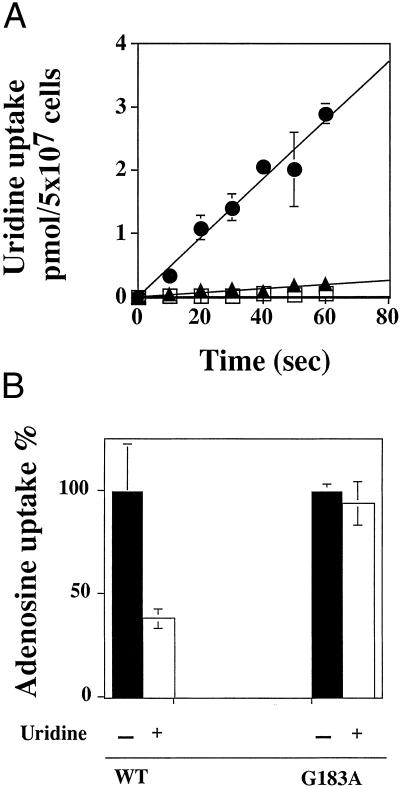
The G183A mutant was also assayed for [3H]uridine transport capability. Unexpectedly, G183A failed to transport [3H]uridine significantly above background levels (Fig. 4A). To determine whether this inability to transport uridine stemmed from diminished uridine binding, the ability of unlabeled uridine at a 160-fold excess to inhibit [3H]adenosine uptake mediated by WT LdNT1.1 and G183A was investigated. As shown in Fig. 4B, [3H]adenosine (0.25 μM) uptake by WT LdNT1.1 was substantially reduced by unlabeled uridine (40 μM). In marked contrast, unlabeled uridine at this concentration failed to significantly inhibit G183A-mediated [3H]adenosine uptake, confirming that the G183A mutation impaired the ability of the transporter to bind uridine. These results strongly indicated that G183 was essential for uridine but not adenosine transport.
Figure 4.
Characterization of G183A-mediated uridine uptake. (A) TUBA5 parasites expressing WT (●), G183A (▴), or vector pX63Hyg (□) were assayed for 1 μM [3H]uridine uptake. Results are expressed as mean ± SD (n = 2). (B) Uptake of [3H]adenosine (250 nM) by TUBA5 parasites expressing WT or G183A transporters was determined over a 10-s period in the absence or presence of uridine (40 μM). Results are expressed as the percentage of uptake in the absence of uridine, and each value is the mean ± SD (n = 3).
Discussion
The transport-deficient and tubercidin-resistant TUBA5 cell line was isolated from WT L. donovani by chemical mutagenesis followed by selection in drug (5). Biochemical characterization of this mutant cell line revealed normal levels and activities of intracellular purine salvage enzymes, suggesting that tubercidin resistance was due to impaired transport of the drug across the plasma membrane (5). This study has revealed that the molecular basis for this drug-resistant phenotype is point mutations within the two copies of the genes encoding the LdNT1.1 adenosine/pyrimidine nucleoside transporter that impair transport capability. Both G183D and C337Y permeases transported adenosine at very low levels when overexpressed in TUBA5 parasites. Kinetic analyses revealed that this impaired transport resulted from an ≈20-fold decrease in Vmax values for adenosine compared with WT LdNT1.1. Because both mutant transporters were correctly targeted to the plasma membrane, this reduction in Vmax likely resulted from a defect in the translocation cycle itself. Not surprisingly, there were no mutations within the ORFs encoding the LdNT1.2 transporter. Because these genes are not expressed at detectable levels in WT and TUBA5 promastigotes (3), there would be no strong selection pressure in the presence of tubercidin to acquire inactivating mutations.
To probe further the role of these residues in LdNT1.1 function, several C337 and G183 mutants were analyzed for transport proficiency. Whereas neither residue per se was essential for adenosine transport, replacement of either with residues that were larger than the WT residue (G183N and C337F) essentially abolished transport. These results also suggested that the loss of function of G183D in the TUBA5 cell line was not due solely to the introduction of a negative charge into TM 5 but could be attributed, at least partly, to the bulkier Asp side chain. The precise mechanism, however, whereby Asp or Asn residues at position 183 abrogate transport function cannot be definitively ascertained in the absence of a high resolution structure. The replacement of Gly with Ala (G183A) was well tolerated in terms of adenosine transport capability. Although G183A-mediated adenosine transport occurred at a lower rate than WT, it was still significantly higher (≈40-fold) than background rates (Fig. 2 A and B). In marked contrast, G183A failed to transport uridine significantly over background (Fig. 4A). Competition experiments revealed that a 160-fold excess of unlabeled uridine significantly inhibited WT but not G183A-mediated adenosine transport (Fig. 4B), suggesting that this mutant was impaired in its ability to bind uridine. Clearly, G183 is essential for pyrimidine nucleoside transport by LdNT1.1.
The absence of any structural information also makes it difficult to deduce exactly the mechanism by which G183 influences substrate selectivity. Glycine residues contribute extensively to helical packing interactions in membrane proteins (14, 15). Sequence motifs containing a pair of glycine residues located three residues apart (GXXXG) have been found to mediate high affinity associations, and these motifs occur frequently in transmembrane segments of membrane proteins (16, 17). Glycine residues in these motifs promote interhelical associations by providing a flat surface against which side chains of other residues can pack (17). Glycine also imparts flexibility to the polypeptide chain, a property that is often essential for function (16). For instance, conserved glycine residues in TM V of the Escherichia coli lactose permease are believed to contribute to the conformational flexibility of the substrate binding site (18).
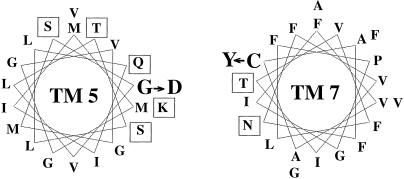
The helical wheel diagram of the LdNT1.1 TM 5 shows that it is distinctly amphipathic, with G183 located approximately in the center of the hydrophilic face (Fig. 5). This location of G183 makes its involvement in interhelical packing interactions less likely. On the other hand, amphipathic helices often line substrate permeation pathways in transporters, with the hydrophilic residues forming specific interactions with substrates (19–21). Therefore, it is plausible that G183 lies within the substrate permeation pathway in LdNT1.1, particularly given its role in determining substrate selectivity. There are four glycine residues in TM 5 of LdNT1.1 (Fig. 1). It is possible that these residues provide conformational flexibility to TM 5 that is necessary for nucleoside binding and translocation or that G183 is part of a substrate binding pocket that cannot accommodate bulkier side chains. There are several examples of glycine residues contributing to a substrate or ligand binding pocket. Gly-121 in rhodopsin, for example, is part of the retinal binding pocket and is believed to form a cavity for the packing of the C9 methyl group of retinal (22, 23). Replacement of this residue with bulkier side chains increases steric interactions with the chromophore and alters the spectral properties of the mutants (22, 23). Similarly, three transmembrane glycine residues in the Ca2+-ATPase appear to play important roles in Ca2+ binding and/or translocation (24). Alteration of C337 to a bulky tyrosine or phenylalanine virtually eliminates activity, whereas smaller side chains can be accommodated without eliminating transport. The fact that C337 is located in a hydrophilic patch of a largely hydrophobic helix (Fig. 4) suggests that this amino acid might also line the permeation pathway. Future studies by using chemical modification approaches such as the “substituted cysteine accessibility method” (19) should permit an evaluation of this potential explanation for these two loss-of-function mutations.
Figure 5.
Helical wheel representation of TMs 5 and 7 of LdNT1.1. Residues of TM 5 and TM 7 are shown on a helical wheel of 3.6 residues per turn. Amino acids with polar side chains are indicated within squares. The positions of the G183D (G → D) and C337Y (C → Y) mutations in TMs 5 and 7, respectively, are also indicated.
LdNT1.1 is a member of a growing family of known nucleoside transporters including the human and rat equilibrative nucleoside transporters hENT1 (25), hENT2 (26), rENT1, and rENT2 (27), and several nucleoside permeases from Leishmania (4), trypanosomes (28, 29), and Plasmodium (30, 31). All members of this family exhibit significant sequence identity to each other, and the predicted topology model encompassing 11 transmembrane helices (25) is either the optimal prediction or at least a plausible model for all family members. Hence, structurally significant features of LdNT1.1, such as the topological regions of the protein that constitute the substrate “permeation pathway,” are likely to be similar in the related permeases. A region encompassing TMs 3–6 of hENT1 has been implicated in substrate binding (32), and recent studies suggest that TM 4 of rENT2 forms part of the substrate translocation pathway through the rat transporter (33). Thus, the identification of TM 5 as a likely component of substrate permeation through LdNT1.1 is consistent with the prior functional studies on hENT1 and rENT2 and the notion that similar topological domains may be involved in ligand permeation by other members of the equilibrative nucleoside transporter family. We anticipate that structure-function studies of the type reported here will illuminate common structural features of all family members.
Multiple sequence alignments indicate that only 16 amino acid residues are conserved among all human and parasitic nucleoside transporters (34). These residues are likely to be important for transporter structure and/or function and are the obvious candidates for analysis by reverse genetic strategies. However, G183 and C337 are not among these highly conserved residues. That a forward genetic strategy led to the identification of these amino acids as important in LdNT1.1-mediated nucleoside permeation underscores the usefulness of this approach in structure/function analysis. Such forward genetic approaches offer a powerful tool to identify residues that are critical for substrate selectivity and/or permeation but that are not conserved among all of the members of a transporter family. The isolation of multiple transport-deficient mutants with distinct mutations within the nucleoside transporter genes should allow a detailed analysis of structure/function relationships in these parasite transporters.
Acknowledgments
We thank Aurelie Snyder for the fluorescent images presented in this paper. This work was supported by National Institutes of Health Grants AI44138 (to S.M.L.) and AI23682 (to B.U.). S.M.L. and B.U. are recipients of the Burroughs Wellcome Fund Scholar Awards in Molecular Parasitology. G.V. received an N. L. Tartar Research Fellowship from the Oregon Health Sciences University.
Abbreviations
- LdNT1.1
L. donovani nucleoside transporter 1.1
- LdNT1.1, the gene encoding LdNT1.1
TM, transmembrane segment
- GFP
green fluorescent protein
- WT
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Berens R L, Krug E C, Marr J J. In: Purine and Pyrimidine Metabolism. Marr J J, Müller M, editors. New York: Academic; 1995. pp. 89–117. [Google Scholar]
- 2.Marr J J, Berens R L. Mol Biochem Parasitol. 1983;7:339–356. doi: 10.1016/0166-6851(83)90016-6. [DOI] [PubMed] [Google Scholar]
- 3.Vasudevan G, Carter N S, Drew M E, Beverley S M, Sanchez M A, Seyfang A S, Ullman B, Landfear S M. Proc Natl Acad Sci USA. 1998;95:9873–9878. doi: 10.1073/pnas.95.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter N S, Drew M E, Sanchez M A, Vasudevan G, Landfear S M, Ullman B. J Biol Chem. 2000;275:20935–20941. doi: 10.1074/jbc.M002418200. [DOI] [PubMed] [Google Scholar]
- 5.Iovannisci D M, Kaur K, Young L, Ullman B. Mol Cell Biol. 1984;4:1013–1019. doi: 10.1128/mcb.4.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iovannisci D M, Ullman B. J Parasitol. 1983;69:633–636. [PubMed] [Google Scholar]
- 7.Landfear S M, Wirth D F. Mol Biochem Parasitol. 1985;15:61–82. doi: 10.1016/0166-6851(85)90029-5. [DOI] [PubMed] [Google Scholar]
- 8.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 9.Cruz A, Coburn C M, Beverley S M. Proc Natl Acad Sci USA. 1991;88:7170–7174. doi: 10.1073/pnas.88.16.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha D S, Schwarz J K, Turco S J, Beverley S M. Mol Biochem Parasitol. 1996;77:57–64. doi: 10.1016/0166-6851(96)02580-7. [DOI] [PubMed] [Google Scholar]
- 11.Kapler G M, Coburn C M, Beverley S M. Mol Cell Biol. 1990;10:1084–1094. doi: 10.1128/mcb.10.3.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aronow B, Kaur K, McCartan K, Ullman B. Mol Biochem Parasitol. 1987;22:29–37. doi: 10.1016/0166-6851(87)90066-1. [DOI] [PubMed] [Google Scholar]
- 13.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Warren S. Current Protocols in Immunology. New York: Wiley; 1999. [Google Scholar]
- 14.Eilers M, Shekar S C, Shieh T, Smith S O, Fleming P J. Proc Natl Acad Sci USA. 2000;97:5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javadpour M M, Eilers M, Groesbeek M, Smith S O. Biophys J. 1999;77:1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russ W P, Engleman D M. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 17.Senes A, Gerstein M, Engleman D M. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 18.Weinglass A B, Kaback R. Proc Natl Acad Sci USA. 1999;96:11178–11182. doi: 10.1073/pnas.96.20.11178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan R-T, Maloney P C. Cell. 1993;75:37–44. [PubMed] [Google Scholar]
- 20.Yan R-T, Maloney P C. Proc Natl Acad Sci USA. 1995;92:5973–5976. doi: 10.1073/pnas.92.13.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Giacomini K M. J Biol Chem. 1999;274:2298–2302. doi: 10.1074/jbc.274.4.2298. [DOI] [PubMed] [Google Scholar]
- 22.Han M, Lin S W, Smith S O, Sakmar T P. J Biol Chem. 1996;271:32330–32336. doi: 10.1074/jbc.271.50.32330. [DOI] [PubMed] [Google Scholar]
- 23.Han M, Groesbeek M, Sakmar T P, Smith S O. Proc Natl Acad Sci USA. 1997;94:13442–13447. doi: 10.1073/pnas.94.25.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen J P, Vilsen B. J Biol Chem. 1992;267:2767–2774. [PubMed] [Google Scholar]
- 25.Griffiths M, Beaumont N, Yao S Y M, Sundaram M, Boumah C E, Davies A, Kwong F Y P, Coe I, Cass C E, Young J D, Baldwin S A. Nat Med. 1997;3:89–93. doi: 10.1038/nm0197-89. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths M, Yao S Y M, Abidi F, Phillips S E V, Cass C E, Young J D, Baldwin S A. Biochem J. 1997;328:739–743. doi: 10.1042/bj3280739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao S Y M, Ng A M L, Muzyka W R, Griffiths M, Cass C E, Baldwin S A, Young J D. J Biol Chem. 1997;272:28423–28430. doi: 10.1074/jbc.272.45.28423. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez M A, Ullman B, Landfear S M, Carter N S. J Biol Chem. 1999;274:30244–30249. doi: 10.1074/jbc.274.42.30244. [DOI] [PubMed] [Google Scholar]
- 29.Mäser P, Sütterlin C, Kralli A, Kaminsky R. Science. 1999;285:242–244. doi: 10.1126/science.285.5425.242. [DOI] [PubMed] [Google Scholar]
- 30.Carter N S, Ben Mamoun C, Liu W, Silva E O, Landfear S M, Goldberg D E, Ullman B. J Biol Chem. 2000;275:10683–10691. doi: 10.1074/jbc.275.14.10683. [DOI] [PubMed] [Google Scholar]
- 31.Parker M D, Hyde R J, Yao S Y M, McRobert L, Cass C E, Young J D, Baldwin S A. Biochem J. 2000;349:67–75. doi: 10.1042/0264-6021:3490067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundaram M, Yao S Y M, Ng A M L, Griffiths M, Cass C E, Baldwin S A, Young J D. J Biol Chem. 1998;273:21519–21525. doi: 10.1074/jbc.273.34.21519. [DOI] [PubMed] [Google Scholar]
- 33.Yao S Y M, Sundaram M, Chomey E G, Cass C E, Baldwin S A, Young J D. Biochem J. 2001;353:387–393. doi: 10.1042/0264-6021:3530387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter N S, Landfear S M L, Ullman B. Trends Parasitol. 2001;17:142–145. doi: 10.1016/s1471-4922(00)01806-7. [DOI] [PubMed] [Google Scholar]