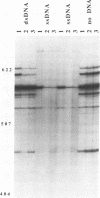
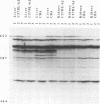
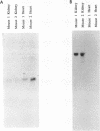
Abstract
Fingerprinting of RNA populations was achieved using an arbitrarily selected primer at low stringency for first and second strand cDNA synthesis. PCR amplification was then used to amplify the products. The method required only a few nanograms of total RNA and was unaffected by low levels of genomic double stranded DNA contamination. A reproducible pattern of ten to twenty clearly visible PCR products was obtained from any one tissue. Differences in PCR fingerprints were detected for RNAs from the same tissue isolated from different mouse strains and for RNAs from different tissues from the same mouse. The strain-specific differences revealed are probably due to sequence polymorphisms and should be useful for genetic mapping of genes. The tissue-specific differences revealed may be useful for studying differential gene expression. Examples of tissue-specific differences were cloned. Differential expression was confirmed for these products by Northern analysis and DNA sequencing uncovered two new tissue-specific messages. The method should be applicable to the detection of differences between RNA populations in a wide variety of situations.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. Industry surprised by firm US stance on biodiversity treaty. Nature. 1992 Jun 11;357(6378):428–428. doi: 10.1038/357428b0. [DOI] [PubMed] [Google Scholar]
- Barbu V., Dautry F. Northern blot normalization with a 28S rRNA oligonucleotide probe. Nucleic Acids Res. 1989 Sep 12;17(17):7115–7115. doi: 10.1093/nar/17.17.7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikaraishi D. M. Complexity of cytoplasmic polyadenylated and nonpolyadenylated rat brain ribonucleic acids. Biochemistry. 1979 Jul 24;18(15):3249–3256. doi: 10.1021/bi00582a009. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hassouna N., Michot B., Bachellerie J. P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984 Apr 25;12(8):3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M. S. An 'equalized cDNA library' by the reassociation of short double-stranded cDNAs. Nucleic Acids Res. 1990 Oct 11;18(19):5705–5711. doi: 10.1093/nar/18.19.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Pardee A. B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992 Aug 14;257(5072):967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. H., Bedigian H. G., Bouchard G., Denial T., Kosowsky M., Norberg R., Pugh S., Sargeant E., Turner R., Paigen B. Multilocus markers for mouse genome analysis: PCR amplification based on single primers of arbitrary nucleotide sequence. Mamm Genome. 1992;3(2):55–64. doi: 10.1007/BF00431247. [DOI] [PubMed] [Google Scholar]
- Van Ness J., Maxwell I. H., Hahn W. E. Complex population of nonpolyadenylated messenger RNA in mouse brain. Cell. 1979 Dec;18(4):1341–1349. doi: 10.1016/0092-8674(79)90244-7. [DOI] [PubMed] [Google Scholar]
- Welsh J., Liu J. P., Efstratiadis A. Cloning of PCR-amplified total cDNA: construction of a mouse oocyte cDNA library. Genet Anal Tech Appl. 1990 Feb;7(1):5–17. doi: 10.1016/0735-0651(90)90038-h. [DOI] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Genomic fingerprinting using arbitrarily primed PCR and a matrix of pairwise combinations of primers. Nucleic Acids Res. 1991 Oct 11;19(19):5275–5279. doi: 10.1093/nar/19.19.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., Petersen C., McClelland M. Polymorphisms generated by arbitrarily primed PCR in the mouse: application to strain identification and genetic mapping. Nucleic Acids Res. 1991 Jan 25;19(2):303–306. doi: 10.1093/nar/19.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., Pretzman C., Postic D., Saint Girons I., Baranton G., McClelland M. Genomic fingerprinting by arbitrarily primed polymerase chain reaction resolves Borrelia burgdorferi into three distinct phyletic groups. Int J Syst Bacteriol. 1992 Jul;42(3):370–377. doi: 10.1099/00207713-42-3-370. [DOI] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]