Abstract
We collected urban soil samples impacted by polycyclic aromatic hydrocarbons (PAHs) from a sorbent-based remediation field trial to address concerns about unwanted side-effects of 2% powdered (PAC) or granular (GAC) activated carbon amendment on soil microbiology and pollutant biodegradation. After three years, total microbial cell counts and respiration rates were highest in the GAC amended soil. The predominant bacterial community structure derived from denaturing gradient gel electrophoresis (DGGE) shifted more strongly with time than in response to AC amendment. DGGE band sequencing revealed the presence of taxa with closest affiliations either to known PAH degraders, e.g. Rhodococcus jostii RHA-1, or taxa known to harbor PAH degraders, e.g. Rhodococcus erythropolis, in all soils. Quantification by real-time polymerase chain reaction yielded similar dioxygenases gene copy numbers in unamended, PAC-, or GAC-amended soil. PAH availability assessments in batch tests showed the greatest difference of 75% with and without biocide addition for unamended soil, while the lowest PAH availability overall was measured in PAC-amended, live soil. We conclude that AC had no detrimental effects on soil microbiology, AC-amended soils retained the potential to biodegrade PAHs, but the removal of available pollutants by biodegradation was most notable in unamended soil.
Introduction
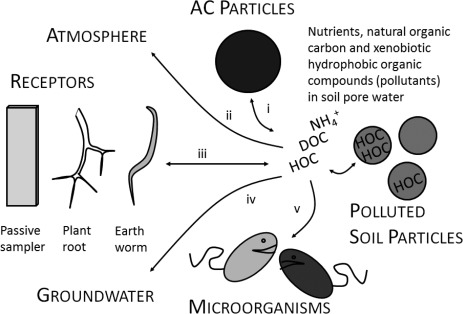
Activated carbon (AC) addition is currently being investigated as in situ technology to remediate polluted sediments and soil.1 A critical consideration for the application in aerobic soil is the long-term impact of AC on intrinsic microbial community structure and functioning, including the potential to biodegrade pollutants. Adding AC to soil will reduce the soil porewater concentration of hydrophobic organic compounds (HOCs)2 which will be adsorbed and strongly bound by the AC (Figure 1, process i). Pollutants bound in the micropores of AC become less accessible for biouptake by soil-dwelling organisms3−5 and plants4,6 in comparison with weakly bound or dissolved pollutants. AC amendment may thereby reduce the transfer of HOCs from the soil matrix into the terrestrial food-chain and also reduce phytotoxicity7 (Figure 1, process iii). Addition of AC to soil will also reduce HOCs leaching8 (Figure 1, process iv) and volatilization risks5 (Figure 1, process ii). Strong binding of HOCs by AC will, however, reduce the availability of HOCs to microorganisms with the ability to metabolize these compounds. A potential consequence could be the disappearance of HOC degraders from the predominant soil microbial community and/or the down-regulation of HOCs metabolism in favor of other carbon substrates. In soils with abundant HOCs degrading microorganisms, this may impair the pollution attenuation via biodegradation (Figure 1, process v).
Figure 1.
Illustration of the interlinkage of the various processes affecting the hydrophobic organic compounds (HOCs) fate in activated carbon (AC) amended soil. (i) AC amendment increases the sorption of HOCs and reduces the concentration of HOCs dissolved in porewater. Reduced HOC porewater concentrations typically imply (ii) less HOCs volatilization to the atmosphere,5 (iii) less biouptake of HOCs by earth worms and plants,7,21 and (iv) less HOCs leaching to groundwater.8 Reduced HOC porewater concentrations may also change soil microbial community structure and functioning, including (v) the HOCs biodegradation. Secondary effects of AC on other substrates such as dissolved organic carbon and nutrients concentrations,8 soil wettability, and physical soil structure may also change soil microbial community structure and functioning.
There are reports of differing effects of strong sorbent amendments on pollutant biodegradation processes. On the one hand, addition of AC to contaminated soils was shown to reduce spiked 14C phenanthrene metabolism to 14CO2 in laboratory batch experiments,9 and Karapanagioti et al.10 showed that slow sorption kinetics limited spiked phenanthrene biodegradation in sediment slurries containing coal particles. On the other hand, Vasilyeva et al.7 suggested that AC helped overcome toxicity of polychlorinated biphenyls to microbes, and Payne et al.11 also reported that addition of AC had a slight stimulatory effect on PCB dechlorination in sediment. Bushnaf et al.12 reported that reduced availability of monoaromatic hydrocarbons in biochar amended soil led to greater biodegradation of linear, branched and cyclic alkanes, and the total petroleum hydrocarbon vapor degradation was comparable in sandy soil with and without 2% biochar. In a direct comparison of AC amendment with biostimulation and bioaugmentation, Hale et al.13 found that 2% AC addition was more effective than biostimulation or bioaugmentation in further reducing the availability of an already strongly sequestered polycyclic aromatic hydrocarbon (PAH) pollution in River Tyne sediment.
The aim of this paper was to study effects of AC amendment on the predominant bacterial community structure, and its functioning with emphasis on the biodegradation of PAHs. To our knowledge, this is the first-ever investigation of the long-term effects of AC amendment on soil microbial communities under realistic field conditions. We report changes in the predominant bacterial community structure over a three year period following amendment of a PAH-polluted urban soil with granular or powdered activated carbon (GAC and PAC). For the samples collected in year 3, we compared the bacterial communities and their functioning in greater detail using molecular microbiological and chemical methods. We assess the combined effect of sorption and biodegradation on PAH availability in soil with and without GAC or PAC to better understand impacts on environmental risks at contaminated sites.
Materials and Methods
Soil Characterization and Sampling
Details of the field trials of AC-based soil remediation are reported by Hale et al.8 The soil used in the lysimeter experiment was excavated at a building site in Drammen, Norway, intermixed using an excavator bucket with 2% wet soil weight of PAC (SilCarbon TH90, average 20 μm grain size, with 80% < 45 μm) or GAC (SilCarbon 0.3–0.8, 300–800 μm grain size), and embedded in outdoor lysimeters with 25 m2 surface area and between 2.5 and 3 m depth. The soil initially had a total organic carbon content of 2.50 ± 0.04% dry weight, and amended soils contained the intended AC dose, although with considerable variability, 2.0 ± 1.0% for GAC or 2.4 ± 1.9% for PAC.8 Two series of soil samples for microbial analysis were taken immediately after the AC amendment of the soil, after 6 months, and after 3 years, and frozen at −20 °C with and without the addition of absolute ethanol 1:1 (v/v). For the year 3 samples, additional soil samples were stored in the cold room at 4 °C without ethanol addition for the batch experiments.
Total Microbial Cell Number
Ten μL of the year 3 sample stored in ethanol was added to 990 μL of filter-sterilized phosphate buffer saline (PBS, Oxoid) and the cells were stained by adding 50 μL of SYBR Gold nucleic acid stain in 100× concentrate in dimethyl sulfoxide (DMSO) (Invitrogen Ltd., Paisley, UK), wrapped in foil and incubated at room temperature for 30 min, after which they were filtered using a sterile Millipore filter holder and a 0.2-mm-pore-size black polycarbonate filter (diameter 25 mm; Millipore). The filters were transferred to glass microscope slides containing a drop of Citifluor (Citifluor Ltd., Canterbury, United Kingdom) antifadent to help adhesion to the slide. A further drop of Citifluor was placed on top of the filter, and a coverslip was carefully placed on top of the preparation. Total bacteria were determined by direct count under an oil-immersion objective (100× magnification) using an Olympus BX40 Epifluorescence microscope; 20 randomly chosen fields of view were counted using a dilution that yielded between 30 and 300 fluorescent cells having a clear outline and finite cell shape. The error between samples was estimated using the standard deviation of the mean from three replicates measured per treatment. Cell number per g of soil was determined by considering the numbers and the areas of fields of view on the filter membrane, the area of the membrane and the original sample dilution factor.14
Soil Respiration
Soil respiration was measured for the year 3 samples by monitoring CO2 concentrations in 50-mL crimp-top vials containing 15 g wet weight of either unamended, PAC-amended, or GAC-amended soil. For each soil type duplicate batches were monitored for 4 days at room temperature (20 °C). GC-MS analysis of CO2 was performed on a Fisons 8060 GC linked to a Fisons MD800 MS with a HP-PLOT-Q capillary column.
Bacterial Community Analysis
A fingerprinting method, denaturing gradient gel electrophoresis (DGGE), was carried out to determine similarities and differences between the predominant bacterial communities in the experimental treatments, and to identify selected members of these communities. Total bacterial DNA was extracted from 0.5-g (wet weight) aliquots of the stored (without ethanol) soil samples, taken at times 0 and 6 months and 3 years after the start of the lysimeter experiments. The DNA extraction was carried out using Fast DNA Soil Kit (Qbiogene) with a purification step, modified from the method of Griffiths et al.,15 added prior to the nucleic acid extraction in order to prevent the coextraction of compounds such as humic acids and clay minerals, which are known to inhibit PCR amplification. Briefly, the pretreatment consisted in the extraction of nucleic acids from the soil matrix by using hexadecyltrimethylammonium bromide (CTAB, Sigma-Aldrich) extraction buffer and phenol/chloroform/isoamyl alcohol (25:24:1, Sigma-Aldrich), followed by phenol removal using chloroform/isoamyl alcohol (24:1, Sigma-Aldrich). Primers 2 and 3, targeting the bacteria, were used to PCR amplify the V3 region of bacterial 16S rRNA gene fragments, as previously described by Muyzer et al.,16 and the PCR products were analyzed by DGGE as described previously.13 The DGGE images were normalized and interpreted using the image analysis software BioNumerics (Applied Maths NV, St. Martens-Latem, Belgium). Primer 6 (Primer-E Ltd., Plymouth, UK) was used to perform cluster analysis using the Pearson product-moment correlation coefficient and Analysis of Similarity (ANOSIM).
Band Sequencing
Dominant DGGE bands were excised from the gel, PCR-amplified using primers 2/3, purified and sequenced with primer 2 or 3 (3.2 pmol/μes/μL) using the ABI prism Big Dye Terminator Cycle Sequencing Ready reaction Kit and an ABI Prism 377 DNA sequencer (Applied Biosystems, USA) as previously described.13 Sequences were compared against the Ribosomal Database Project (RDP10) and GenBank databases using the BLAST algorithm to determine the closest matching sequence identity.
Quantitative Real-Time PCR
The abundances of specific ring-hydroxylating dioxygenase α-subunit (RHDα) genes were measured in each sample by real-time PCR using the following primer sets: (i) P6B (forward (f): 5′-TGGCGAACTCGTGTCGGCAC-3′; reverse (r): 5′- CGTCCAGRCAACCGAADAYC −3′), targeting a pdoA2/phdA clade that includes mostly Mycobacterium species and have M. vanbaalenii as a reference strain; (ii) P4 (f: 5′-CCGGAGACTTCCTGACGAC-3′; r: 5′-GCASACGAAYCGACGGGT-3′) targeting a ebdA1/etbA1/akbA1b clade that only includes Rhodococcus species and have R. jostii RHA1 as a reference strain; and (iii) P7B (f: 5′- CACBTGCAGCTAYCACG −3′; r: 5′- CATGTGGTCCATGTAGAAC −3′) targeting a ipbA1/bphA1 clade that includes mostly Rhodococcus and some Pseudomonas species and, in particular, includes several R. erythropolis strains. Real-time PCR experiments were conducted on a BioRad CFX96 (Hercules, CA) with a C1000 thermal cycler iCycler and software version 1.6 (BioRad CFX Manager). Ten μL of reaction mixture contained 3 μL of template DNA (or filtered sterile molecular biology water, Sigma-Aldrich, as negative control), and 15 pmol of each primer, with the SsoFast EvaGreen Supermix for the CFX96 (Biorad Laboratories Ltd.). The following temperature profile was used in the amplifications: step one heated to 98 °C (2 min), followed by 40 cycles of 2 s of denaturation at 98 °C, 5 s at the primer specific annealing temperature (58 °C for P6B, 60 °C for P4, and 59 °C for P7B). At the end of the real-time PCR, a melting curve was performed as a final step that consisted of the measurement of the SYBR Green signal intensities during a 0.2 °C temperature increment every 10 s from 65 to 95 °C. The corresponding plot of change in fluorescence versus temperature shows a single peak for every amplicon at its specific melting temperature. For every set of primers, calibration curves were obtained from standard DNA prepared from plasmids containing the cloned target sequences. The plasmid DNA concentration was quantified using Nanodrop 1000 spectrophotometer (Thermo Scientific). The copy number of standard plasmids was calculated according to plasmid size, insert lengths, and assuming a molecular mass of 660 Da per base pair. Stock solutions of standard DNA were prepared at a concentration of 109 copies of plasmid μL–1. For the calibration curves, DNA standards ranging from 109 to 101 target gene copies μL–1 were prepared by diluting the stock solutions. All the standard curves were linear (R2 > 0.99) over the 9 orders of magnitude (109 to 101 μL–1) of gene copy number. Every sample was run in triplicate and each experiment was repeated at least twice. In every run, the accuracy in the detection of the target gene in the soil samples was confirmed by comparing the melt curves and the agarose analysis of the qPCR products to the ones obtained with DNA extracted from model organisms (positive control) in order to avoid false detection in the environmental samples.
PAH Availability Assessed by Uptake by Polyethylene Samplers
A series of batch experiments was set up to study the uptake of available PAHs in passive samplers.13 Passive samplers are able to passively accumulate HOCs from contaminated water until partitioning equilibrium is established. Pollutants taken up by passive samplers are potentially also available for uptake by critical receptors such as plants and earth worms (Figure 1, process iii). To study the effect of sorption alone (Figure 1, process i) compared to the combined effect of sorption and biodegradation (Figure 1, processes i and v) on pollutant availability, batches were set up with and without sodium azide addition which inhibits PAH biodegradation.17,18 The microcosms comprised the following: 5 g (wet weight) of unamended soil, PAC-amended, or GAC-amended soil and 40 mL of water. The water content of the samples was between 16 and 21% wet weight. For each soil type triplicate batches were set up with and without 1 g/L sodium azide (Sigma Aldrich). A 0.15 ± 0.01 g clean polyethylene (PE) passive sampler was added to each batch. Blank controls were run in parallel and consisted of triplicate batches containing PE samplers and water with and without sodium azide. Batches were plugged with cotton balls and mixed at 100 rpm on an orbital shaker (IKA labortechnik, Germany) in a secondary dark containment consisting of a cardboard box with some holes punched into the lid for aeration. PE samplers were removed after 3 weeks and extracted twice for 24 h in 10 mL of 50:50 v:v hexane/acetone. A surrogate standard was added to the first extraction to monitor and correct for recoveries, which were 78 ± 11% for d10-phenanthrene, 83 ± 6% for d10-pyrene, 92 ± 4% for d12-benzanthracene, and 94 ± 8% for d12-benzperyl.
Sample Cleanup and PAH Analysis
Sample cleanup was performed with a silica gel column topped by sodium sulfate before GC-MS analysis with an Agilent 6850 Gas Chromatograph (DB-XLB column length 30 m, i.d. 25 mm, and 1 μm film thickness) coupled to an Agilent 5973 mass spectrometer as described by Hale et al.8
Results and Discussion
Total Microbial Cell Count and Soil Respiration
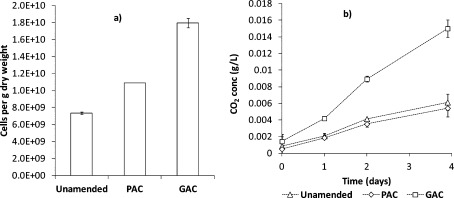
A statistically significant difference was observed for the total cell counts 3 years after the amendment, with GAC amended soil having the highest cell count of 1.8·1010 cells per gram of dry soil weight, 2.5 times higher than the unamended soil, which had the lowest cell count (Figure 2a). These values are in the higher range for most typical soil types (e.g., (19, 20)). The soil respiration rate was also a statistically significant factor of 2.6 and 2.7 higher in the GAC-amended soil compared to the unamended and PAC-amended soils (Figure 2b). Soil respiration resulted in a linear increase in the CO2 batch headspace concentration over the entire 4-day monitoring period for all soils (R2 values >0.98), and soil respiration could be quantified as 0.19 ± 0.02, 0.18 ± 0.01, and 0.49 ± 0.03 μg CO2 per g of dry soil per hour for unamended, PAC-, and GAC-amended soil, respectively. These observations coincide with field observations of better plant growth in GAC amended soil, see Jakob et al.21 The soil respiration rates were similar to the values found in another study on a hydrocarbon contaminated site.22 These results show that AC amendment overall was not detrimental to aerobic microbial activity despite previously reported enhanced DOC binding in AC amended soils.8
Figure 2.
(a) Total microbial cell count for the soil samples taken after 3 years and (b) soil respiration for the same samples. Error bars indicate the standard deviation for replicates defined in the method section. Invisible error bars are smaller than the lines or symbols in the figures.
Changes in the Predominant Bacterial Community Structure
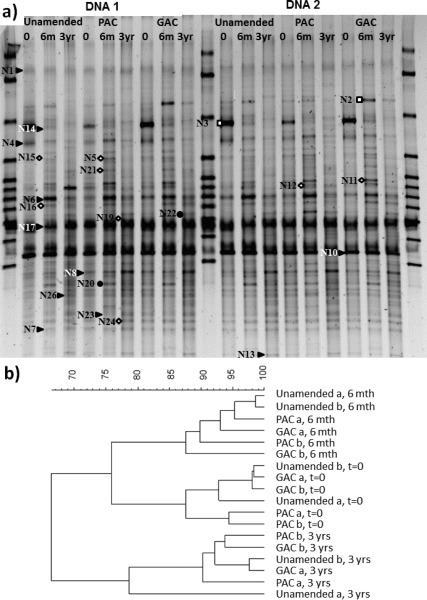
The predominant soil bacterial community structure, as assessed by DGGE and cluster analysis of amended and unamended soil after 0, 6, and 36 months (Figure 3), showed that all the samples from each particular time period clustered closely with other samples from the same time period, regardless of treatment. This indicates that the predominant bacterial community structure shifted more strongly with time than in response to PAC or GAC amendment (ANOSIM test for differences between time groups, Global R = 0.852, p < 0.01; for differences between treatments, Global R = −0.317, p = 0.97). Overall, similarity between predominant bacterial communities, determined by the Pearson product-moment correlation coefficient, was greater than 65% (Figure 3b). Duplicate DNA samples from the same time point and treatment showed higher similarity (>77% overall and average similarity coefficient >87%, Figure 3b), indicating good method reproducibility. The most abundant microbial species are important drivers of carbon and nutrient cycling in soil. The fairly high similarities of predominant bacterial community structures in AC amended and unamended soil observed in this study alleviates concerns about strong side-effects of AC amendment. The findings agree with field community surveys of macroinvertebrates in sediments where AC amendment generally had only weak effects on community structure and diversity,23,24 although clear effects on species abundance were observed in one field study.24
Figure 3.
(a) DGGE gel showing the location of excised bands with the correspondent affiliation to three classes of environmental bacteria (right solid triangle = actinobacteria; ◊ = proteobacteria; □ = bacteroidetes; ● = unknown). Closest matching sequences are reported in Table S1 in Supporting Information. DNA was extracted twice (DNA1 and DNA2) from the unamended and AC amended soils (PAC, GAC) at time zero, 6 months, and 3 years. (b) Dendrograms showing cluster analysis of the similarities (Pearson product moment correlation coefficient) between the different community compositions of the treatments over time, examined by DGGE.
Band Sequencing Results
Identification of the nearest matching neighbor to sequenced DGGE bands was carried out using the Basic Local Alignment Search Tool (BLAST) and classification was confirmed using the Classifier tool in the Ribosomal Database Project (RDP 10). Twenty-three of the twenty-six most predominant taxa were sequenced and classified into three main phyla, Bacteroidetes, Actinobacteria, and Proteobacteria (identified by different symbols, Figure 3 and Table S1 in Supporting Information) that harbor common environmental bacteria known to play an important role in the decomposition of organic materials.25−28 The three most abundant and prevalent taxa (bands N6, N10, and N17 respectively) belonged to the order Micrococcinaea within the Actinobacteria phylum, whose nearest neighbors were identified as an Curtobacterium, Arthrobacter, and Microbacterium species although with varied similarities (89–99%; Table S1 in Supporting Information). These bacteria are common soil inhabitants. The nearest-matching neighbor of several predominantly abundant (intense bands) taxa are of interest because of the reported widespread ability to degrade PAHs in members of these taxa (e.g., (29)): band N7 was 96% identical to a Mycobacterium sp. and 92% similar to the versatile PAH-degrading M. vanbaalenii PYR-1,30,31 band N8 was 95% similar to the versatile PAH-degrading Rhodococcus josttii strain RHA1,32 band N16 95% was identical to an uncultured Sphingomonas sp., and band N23 was 99% similar to a strain of the species Rhodococcus erythropolis, members of which are known to degrade biphenyls and isopropylbenzene (e.g., refs (33, 34)). These groups are often present in all soils (e.g., (35)), but appear to dominate more frequently in contaminated soils (e.g., (36, 37)), although to our knowledge it is unusual to obtain such a predominance of sequences matching mycolic acid-containing actinomycetes (e.g., rhodococci and mycobacteria). These bands were present at all times in bacterial communities from unamended, PAC-, and GAC-amended soil, but their relative intensity varied over time (see Table 1). The relative intensity of bands can provide a rough proxy of the proportional abundance of those taxa in the community. The relative intensities of these bands by year 3 were on average stronger than for the beginning of the study, but comparable between treatments, which would indicate a similar relative abundance of these putative PAH-degrading taxa in the soils. The R. jostii RHA-1-like sequence (band N8) showed a particularly marked increase in intensity between the start and end of the study, where it increased from about 3% to nearly 10% of the predominant bacterial community as expressed by relative intensity in AC amended and unamended soil.
Table 1. Summary of the Relative Intensities of Selected DGGE Excised Bands for Every Duplicate DNA Extraction and Their Average for Different Treatments and Time Point .
| N7 DGGE Mycobacterium-like sequence |
N8 DGGE Rhodococcus RHA1-like
sequence |
N23 DGGE Rhodococcus erythropolis-like
sequence |
||||||
|---|---|---|---|---|---|---|---|---|
| relative
band intensity % |
relative band intensity % |
relative band
intensity % |
||||||
| soil | time | sample | average | average | average | |||
| unamended | 0 | DNA1 | 1.87 | 1.52 ± 0.50 | 2.93 | 2.72 ± 0.30 | 1.59 | 1.43 ± 0.22 |
| DNA2 | 1.16 | 2.51 | 1.28 | |||||
| 6 months | DNA1 | 4.50 | 4.46 ± 0.05 | 3.26 | 3.30 ± 0.06 | 1.84 | 1.87 ± 0.04 | |
| DNA2 | 4.42 | 3.34 | 1.90 | |||||
| 3 years | DNA1 | 3.00 | 2.96 ± 0.06 | 8.36 | 8.92 ± 0.80 | 2.21 | 2.35 ± 0.20 | |
| DNA2 | 2.92 | 9.48 | 2.50 | |||||
| PAC | 0 | DNA1 | 3.48 | 3.15 ± 0.46 | 4.65 | 4.26 ± 0.55 | 2.22 | 2.16 ± 0.08 |
| DNA2 | 2.83 | 3.87 | 2.11 | |||||
| 6 months | DNA1 | 3.42 | 3.04 ± 0.54 | 3.34 | 3.33 ± 0.004 | 1.94 | 2.00 ± 0.08 | |
| DNA2 | 2.65 | 3.33 | 2.06 | |||||
| 3 years | DNA1 | 3.25 | 3.49 ± 0.34 | 9.37 | 9.26 ± 0.16 | 3.32 | 3.03 ± 0.40 | |
| DNA2 | 3.73 | 9.14 | 2.74 | |||||
| GAC | 0 | DNA1 | 1.96 | 1.84 ± 0.17 | 3.53 | 3.19 ± 0.50 | 1.66 | 1.57 ± 0.12 |
| DNA2 | 1.71 | 2.85 | 1.48 | |||||
| 6 months | DNA1 | 4.11 | 2.64 ± 2.08 | 3.33 | 2.71 ± 0.89 | 2.11 | 1.82 ± 0.40 | |
| DNA2 | 1.17 | 2.08 | 1.54 | |||||
| 3 years | DNA1 | 3.73 | 3.34 ± 0.55 | 8.55 | 7.90 ± 0.92 | 2.77 | 2.67 ± 0.15 | |
| DNA2 | 2.96 | 7.25 | 2.57 | |||||
Real-Time PCR Results
Real-time PCR is a valuable tool for the quantification of the functional PAH ring-hydroxylating dioxygenases.38,39 Dioxygenase systems add both atoms of molecular oxygen to the aromatic ring as a first step in the aerobic degradation of lower molecular weight PAHs, and dioxygenase gene quantification is therefore of interest in the assessment of PAH bioremediation potential.40,41 The RHDα gene copy numbers reported in Table 2 indicate that those represented by the reference Rhodococcus RHA1 species (from here-on called etbARHA1-RHDα) showed the highest concentrations (∼107–108 g–1), while those represented by a Rhodococcus erythropolis reference species (from here-on called ipdAReryth-RHDα) showed the lowest concentrations (∼105–106 g–1) for every treatment. Those represented by the reference Mycobacterium vanbaalenii PYR-1 species (from here-on called pdoA2PYR1-RHDα) had intermediate concentrations (∼106–107 g–1). Those putative aromatic ring-degrading bacteria (i.e., RHDα containing bacteria) analyzed accounted for between 0.003 and 1% of the total bacterial population if each bacterial cell is assumed to contain one gene copy. These values are similar to the absolute and relative abundance values reported by Cebron et al.39 who used quantitative PCR to target general dioxygenase gene populations in contaminated soils, but higher than those in studies that only targeted specific dioxygenase gene populations.42−44 The concentrations of each targeted gene fell within an order of magnitude between unamended soil, soil amended with PAC, and soil amended with GAC. Indeed, the predominant etbARHA1-RHDα gene population increased over the time of the trial, and to a greater extent in AC-amended compared to unamended lysimeters. This confirms that bacteria with the ability to degrade PAHs remain in PAC- and GAC-amended soil at levels comparable to the populations in unamended soil. The real-time PCR method measures gene numbers, or the potential to degrade PAHs, not gene expression, which may be down-regulated by some pollutant degraders.45 In other words, while the results indicate that the ability to synthesize dioxygenases was present in all bacterial communities isolated from the soil samples, regardless of AC amendment, this does not necessarily imply active synthesis of the enzymes. It is interesting to note that there was a significant correlation between targeted RHDα gene abundances and the concentration of those taxa found by DGGE analysis (relative intensity × total cell count) that had close affiliations to the reference species of those RHDα genes (Pearson’s correlation, r2 = 0.83, P < 0.01, n = 9). However, despite the close affiliation of these 16S rRNA gene fragments with the reference species for each targeted clade of RHDα, the similarities were often lower (e.g., 92–96%) than those cut-offs usually used for species level (97%) and it cannot be confidently concluded that these taxa would necessarily contain RHDα genes even if they were deemed to be similar taxa.
Table 2. Dioxygenase Gene Copy Number per gram of Wet Soil for Every Duplicate DNA Extraction Sample, for Unamended and AC-Amended Soils at Different Time Points.
| model organism |
Mycobacterium vanbaalenii |
Rhodococcus jostii RHA1 |
Rhodococcus erythropolis |
|||||
|---|---|---|---|---|---|---|---|---|
| targeted genes |
phdA, pdoA2 |
ebdA1, etbA1, akbA1b |
ipbA1, bphA1 |
|||||
| soil | time | sample | gene copy n/g wet soil | average | gene copy n/g wet soil | average | gene copy n/g wet soil | average |
| Unamended | 0 | DNA1 | 1.18 ± 0.13 × 107 | 13.9 ± 1.5 × 106 | 3.76 ± 0.35 × 107 | 37.2 ± 5.1 × 106 | 1.95 ± 0.70 × 106 | 2.39 ± 0.48 × 106 |
| DNA2 | 1.60 ± 0.17 × 107 | 3.69 ± 0.68 × 107 | 2.83 ± 0.23 × 106 | |||||
| 6 mths | DNA1 | 3.11 ± 0.42 × 106 | 3.1 ± 0.3 × 106 | 8.45 ± 0.04 × 107 | 77.8 ± 4.0 × 106 | 2.85 ± 0.33 × 106 | 2.77 ± 0.55 × 106 | |
| DNA2 | 3.11 ± 0.15 × 106 | 7.12 ± 0.74 × 107 | 2.69 ± 0.78 × 106 | |||||
| 3 yrs | DNA1 | 3.12 ± 0.44 × 106 | 3.3 ± 0.3 × 106 | 1.20 ± 0.09 × 108 | 119.0 ± 10.0 × 106 | 7.85 ± 0.85 × 105 | 0.71 ± 0.06 × 106 | |
| DNA2 | 3.45 ± 0.21 × 106 | 1.17 ± 0.10 × 108 | 6.43 ± 0.40 × 105 | |||||
| PAC | 0 | DNA1 | 1.98 ± 0.31 × 107 | 18.3 ± 1.8 × 106 | 4.43 ± 0.54 × 107 | 44.0 ± 4.5 × 106 | 2.72 ± 0.26 × 106 | 2.49 ± 0.39 × 106 |
| DNA2 | 1.69 ± 0.04 × 107 | 4.38 ± 0.36 × 107 | 2.26 ± 0.52 × 106 | |||||
| 6 mths | DNA1 | 3.60 ± 0.20 × 106 | 3.6 ± 0.5 × 106 | 7.75 ± 0.99 × 107 | 77.8 ± 8.8 × 106 | 6.34 ± 0.80 × 106 | 6.79 ± 0.61 × 106 | |
| DNA2 | 3.51 ± 0.70 × 106 | 7.80 ± 0.77 × 107 | 7.24 ± 0.42 × 106 | |||||
| 3 yrs | DNA1 | 4.47 ± 0.90 × 106 | 4.9 ± 0.6 × 106 | 1.79 ± 0.17 × 108 | 175.0 ± 10.0 × 106 | 1.55 ± 0.08 × 106 | 1.31 ± 0.05 × 106 | |
| DNA2 | 5.22 ± 0.26 × 106 | 1.71 ± 0.04 × 108 | 1.07 ± 0.03 × 106 | |||||
| GAC | 0 | DNA1 | 9.48 ± 0.40 × 106 | 10.4 ± 0.6 × 106 | 3.37 ± 0.12 × 107 | 34.7 ± 4.4 × 106 | 1.37 ± 0.30 × 106 | 1.57 ± 0.25 × 106 |
| DNA2 | 1.13 ± 0.90 × 107 | 3.57 ± 0.76 × 107 | 1.78 ± 0.22 × 106 | |||||
| 6 mths | DNA1 | 2.56 ± 0.81 × 106 | 2.5 ± 0.6 × 106 | 8.43 ± 0.13 × 107 | 79.6 ± 8.5 × 106 | 2.50 ± 0.16 × 106 | 2.37 ± 0.22 × 106 | |
| DNA2 | 2.45 ± 0.38 × 106 | 7.49 ± 0.43 × 107 | 2.23 ± 0.28 × 106 | |||||
| 3 yrs | DNA1 | 5.29 ± 0.90 × 106 | 5.7 ± 0.8 × 106 | 1.72 ± 0.23 × 108 | 177.0 ± 17.0 × 106 | 4.79 ± 0.32 × 105 | 0.67 ± 0.20 × 106 | |
| DNA2 | 6.12 ± 0.63 × 106 | 1.82 ± 0.13 × 108 | 8.76 ± 0.07 × 105 | |||||
Each quantification was performed in triplicate real-time PCR runs. phdA: iron–sulfur protein large subunit; pdoA2: putative PAH ring-hydroxylating dioxygenase large subunit; etbA1/ebdA1: ethylbenzene dioxygenase alpha subunit; akbA1b: alkylbenzene dioxygenase; ipbA1: isopropylbenzene-2,3-dioxygenase iron-sulphur protein subunit; bphA1: large subunit of biphenyl dioxygenase
Assuming a carbon content of 310 fg per cell46 and one etbARHA1-RHDα gene copy per cell, the biomass carbon corresponding to the gene copy numbers in Table 2 would be between 40 and 70 μg per g of dry soil, which is greater than the solid-phase PAH concentration and much greater than the freely dissolved PAH concentration of below 1 ng per g of soil.8 The comparison suggests that microorganisms with dioxygenase genes utilized other main carbon substrates and could therefore grow and persist long-term in the soil with and without AC amendment. The findings alleviate concerns that enhanced PAH binding by AC may cause a significant decrease in the abundance of putative PAH degraders in AC amended soils.
PAH-Uptake by Passive Samplers
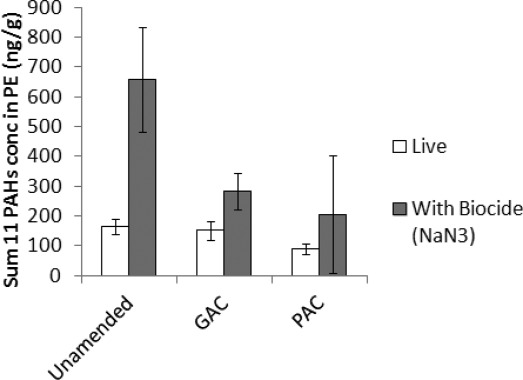
PE passive samplers were employed in batch systems with and without the biocide, sodium azide, and show that both sorption to the AC, and biodegradation, affected the availability of PAHs in the soil samples taken 3 years after the start of the lysimeter experiment (Figure 4). PAHs taken up by passive samplers are considered available to be taken up by organisms and plants, and tend to provide better correlations with ecotoxity assessments than total soil concentrations.47 The high standard deviation of the PAC-amended sterile soil measurements is likely due to small-scale heterogeneity in the distribution of the PAC8,48 or PAH pollution in soil,49 as one PE sampler had much higher uptake of PAHs than the other two replicates. Nevertheless, for the soil slurries with sodium azide, which is normally added to batches to illustrate the effect of sorption only,2 there was a statistically significant reduction in the PAH uptake by PE samplers for the AC amended soils, a 57% reduction for the GAC amendment, and a 69% reduction for PAC amendment compared to the unamended soil (t test, one-tailed, p < 0.01 for GAC and p < 0.05 for PAC). The effect of biodegradation is illustrated by statistically significant 75% and 46% reductions of the PAH uptake by PE passive samplers for unamended and GAC-amended soil, respectively, when comparing soil slurries with and without sodium azide (t test, one-tailed, p < 0.01 for unamended and p < 0.05 for GAC). The PAH compound pattern was also indicative of biodegradation; the uptake of the smaller two- and three-ring PAH compounds, which tend to be more readily biodegradable,40 was on average 99% lower in the unamended soil without, as compared to with, sodium azide (Table S2 in Supporting Information). Overall, PAH uptake was the lowest for PAC-amended soil without sodium azide, and our observations suggest active PAH biodegradation despite low PAH availability in both AC amended and unamended soil, alleviating concerns that AC amendment may result in down-regulation of PAH metabolism. However, the benefit of AC amendment was less clearly pronounced without biocide addition. For soils slurries without sodium azide, only the 44% reduction for PAC amended soil compared to the unamended soil was statistically significant (t test, one-tailed, p < 0.05). Therefore, for a realistic assessment of the long-term benefits of AC amendment under field conditions, laboratory pilot-trials should be conducted with both sterile and live soils to account for the pollution attenuation which may occur in control soil due to biodegradation.
Figure 4.
PAH uptake by passive samplers embedded for 3 weeks in slowly mixed slurries of unamended, PAC-. and GAC-amended soil sampled after 3 years. The sum of 11 of the 16 USEPA PAH compounds is shown (those with concentrations in PE above the analytical detection limit: acenaphtylene, acenaphthene, fluorene, phenanthrene, anthracene, fluoranthene, pyrene, benz(a)anthracene, chrysene, indeno(123 cd)pyrene, and benzo(ghi)perylene). Biocide inhibited batches contained 1 g/L sodium azide.
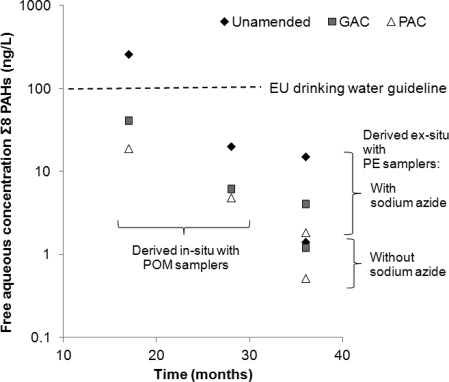
To put these results into context, we compare in Figure 5 free aqueous PAH concentrations Cw estimated from the PAH concentrations in polyethylene, Cpe, using PE-water partitioning coefficients Kpe and the relationship Cw = Cpe/Kpe,50 with free aqueous PAH concentrations reported by Hale et al.8 which were derived from passive polyoxymethylene (POM) samplers embedded field lysimeters. In-situ and ex-situ assessments are not exactly comparable because of differences in temperature and mixing regimes, which are more optimal under laboratory conditions, and therefore tend to facilitate pollutant mass transfer and biodegradation.48 Nevertheless, the combined data seem to show a continuing trend of decreasing free aqueous PAH concentrations in both AC amended and unamended soil over the 3-year remediation trial period, eventually falling to very low levels below EU guidance values for drinking water set out in EU directive 200/60/EC in all systems. This overall trend is the combined effect of all attenuation processes which comprise PAH sorption, biodegradation, chemical reactions, leaching, and volatilization.51 The difference between the accelerated pollution attenuation in the AC-amended systems and the natural pollution attenuation become smaller with time, and may eventually converge, when natural attenuation has depleted the readily available PAH pool to the same extend as AC amendment. According to our results, the biodegradation of available PAHs is a significantly contributing factor to this reduction in free aqueous PAH concentrations, even though total PAH concentration of 23 ± 15 μg per g of untreated dry soil21 were not measurably changed during the field experiment.8 AC amendment benefits, such as reduced PAH uptake by earthworms and plants21 and reduced PAH leaching,8 and AC amendment costs, need to be assessed in comparison with the working of intrinsic attenuation mechanisms, which may also reduce risks in the long term, in particular in soils with a high abundance of pollutant degrading microorganisms.
Figure 5.
Comparison of the free aqueous PAH concentrations derived from PE sampler concentrations measured in year 3 in ex-situ batch tests with the field data reported by Hale et al.,8 which were derived from POM samplers embedded in the lysimeters 17 and 28 months after the start of the experiments. Shown is the sum of those compounds of the 16 USEPA PAHs which could be quantified in both studies: phenanthrene, anthracene, fluoranthene, pyrene, benz(a)anthracene, chrysene, indeno(123 cd)pyrene, and benzo(ghi)perylene.
Acknowledgments
Funding for this research was provided by the Engineering and Physical Sciences Research Council (EPSRC) grants EP/F008473/1, EP/F012934/1, and EP/D079055/1. R.J.D. acknowledges funding for an RCUK Academic Fellowship (RES88007300) and an EPSRC Challenging Engineering award (EP/I025782/1). The Research Council of Norway funded the lysimeter trials through the “Active” project grant in their KMB program (project 192936). The recycling company Lindum Ressurs and Gjenvinning A/S provided field assistance. We thank Khaled M. Bushnaf for conducting the soil respiration experiments.
Supporting Information Available
Compilation of closest relatives and their similarities with the sequence of dominant bands excised from the DGGE gels and the PAH compound specific uptake by PE in soil slurries without compared to with sodium azide addition. This information is available free of charge via the Internet at http://pubs.acs.org/.
The authors declare no competing financial interest.
Supplementary Material
References
- Ghosh U.; Luthy R. G.; Cornelissen G.; Werner D.; Menzie C. A. In-situ Sorbent Amendments: A New Direction in Contaminated Sediment Management. Environ. Sci. Technol. 2011, 45, 1163–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandli R. C.; Hartnik T.; Henriksen T.; Cornelissen G. Sorption of native polyaromatic hydrocarbons (PAH) to black carbon and amended activated carbon in soil. Chemosphere 2008, 73, 1805–1810. [DOI] [PubMed] [Google Scholar]
- Fagervold S. K.; Chai Y. Z.; Davis J. W.; Wilken M.; Cornelissen G.; Ghosh U. Bioaccumulation of Polychlorinated Dibenzo-p-Dioxins/Dibenzofurans in E. fetida from Floodplain Soils and the Effect of Activated Carbon Amendment. Environ. Sci. Technol. 2010, 44, 5546–5552. [DOI] [PubMed] [Google Scholar]
- Langlois V.; Rutter A.; Zeeb B. Activated carbon immobilizes residual polychlorinated biphenyls in weathered contaminated soil. J. Environ. Qual. 2011, 40, 1130–1134. [DOI] [PubMed] [Google Scholar]
- Baker J.; Chang C. W.; Ghosh U.; Paul P.; Kjellerup B.. Application of Tools to Measure Pcb Microbial Dechlorination and Flux into Water during in-Situ Treatment of Sediments; SERDP, 2011; p 138. [Google Scholar]
- Hilber I.; Wyss G. S.; Mäder P.; Bucheli T. D.; Meier I.; Vogt L.; Schulin R. Influence of activated charcoal amendment to contaminated soil on dieldrin and nutrient uptake by cucumbers. Environ. Pollut. 2009, 157, 2224–2230. [DOI] [PubMed] [Google Scholar]
- Vasilyeva G. K.; Strijakova E. R.; Nikolaeva S. N.; Lebedev A. T.; Shea P. J. Dynamics of PCB removal and detoxification in historically contaminated soils amended with activated carbon. Environ. Pollut. 2010, 158, 770–777. [DOI] [PubMed] [Google Scholar]
- Hale S. E.; Elmquist M.; Brändli R.; Hartnik T.; Jakob L.; Henriksen T.; Werner D.; Cornelissen G. Activated carbon amendment to sequester PAHs in contaminated soil: A lysimeter field trial. Chemosphere 2012, 87, 177–184. [DOI] [PubMed] [Google Scholar]
- Rhodes A. H.; Carlin A.; Semple K. T. Impact of Black Carbon in the Extraction and Mineralization of Phenanthrene in Soil. Environ. Sci. Technol. 2008, 42, 740–745. [DOI] [PubMed] [Google Scholar]
- Karapanagioti H. K.; Gossard C. M.; Strevett K. A.; Kolar R. L.; Sabatini D. A. Model coupling intraparticle diffusion/sorption, nonlinear sorption, and biodegradation processes. J. Contam. Hydrol. 2001, 48, 1–21. [DOI] [PubMed] [Google Scholar]
- Payne R. B.; May H. D.; Sowers K. R. Enhanced Reductive Dechlorination of Polychlorinated Biphenyl Impacted Sediment by Bioaugmentation with a Dehalorespiring Bacterium. Environ. Sci. Technol. 2011, 45 (20), 8772–8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnaf K. M.; Puricelli S.; Saponaro S.; Werner D. Effect of biochar on the fate of volatile petroleum hydrocarbons in an aerobic sandy soil. J. Contam. Hydrol. 2011, 126, 208–215. [DOI] [PubMed] [Google Scholar]
- Hale S. E.; Meynet P.; Davenport R. J.; Jones M. D.; Werner D. Changes in polycyclic aromatic hydrocarbon availability in River Tyne sediment following bioremediation treatments or activated carbon amendment. Water Res. 2010, 44, 4529–4536. [DOI] [PubMed] [Google Scholar]
- Davenport R.; Curtis T.; Goodfellow M.; Stainsby F.; Bingley M. Quantitative use of fluorescent in situ hybridization to examine relationships between mycolic acid-containing actinomycetes and foaming in activated sludge plants. Microbiology 2000, 66, 1158–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. I.; Whiteley A. S.; O’Donnell A. G.; Bailey M. J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 2000, 66, 5488–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G.; de Waal E. C.; Uitterlinden A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen A.; Schmidt S.; T.K. H.; Henriksen S.; Jacobsen C. J.; Andersen O. Strong impact on the polycyclic aromatic hydrocarbon (PAH)-degrading community of a PAH-polluted soil but marginal effect on PAH degradation when priming with bioremediated soil dominated by Mycobacteria. Appl. Environ. Microbiol. 2007, 73, 1474–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. W.; Ong S. K.; Moorman T. B. Mineralization of PAHs in coal-tar impacted aquifer sediments and associated microbial community structure investigated with FISH. Chemosphere 2007, 69, 1563–1573. [DOI] [PubMed] [Google Scholar]
- Bloem J.; Veninga M.; Shepherd J. Fully automatic determination of soil bacterium numbers, cell volumes, and frequencies of dividing cells by confocal laser scanning microscopy and image analysis. Appl. Environ. Microbiol. 1995, 61, 1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W.; Coleman D.; Wiebe W. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci., U.S.A. 1998, 95, 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob L.; Hartnik T.; Henriksen T.; Elmquist M.; Brandli R.; Hale S. E.; Cornelissen G.. PAH-sequestration capacity of granular and powdered activated carbon amendments in soil, and their effects on earthworms and plants. Chemosphere 2011, not supplied. [DOI] [PubMed]
- Dawson J. J. C.; Godsiffe E. J.; Thompson I. P.; Ralebitso-Senior T. K.; Killhama K. S.; Patona G. I. Application of biological indicators to assess recovery of hydrocarbon impacted soils. Soil Biol. Biochem. 2007, 39, 164–177. [Google Scholar]
- Cho Y. M.; Ghosh U.; Kennedy A. J.; Grossman A.; Ray G.; Tomaszewski J. E.; Smithenry D. W.; Bridges T. S.; Luthy R. G. Field Application of Activated Carbon Amendment for In-Situ Stabilization of Polychlorinated Biphenyls in Marine Sediment. Environ. Sci. Technol. 2009, 43, 3815–3823. [DOI] [PubMed] [Google Scholar]
- Cornelissen G.; Krusa M. E.; Breedveld G. D.; Eek E.; Oen A. M. P.; Arp H. P. H.; Raymond C.; Samuelsson G.; Hedman J. E.; Stokland O.; Gunnarsson J. S. Remediation of Contaminated Marine Sediment Using Thin-Layer Capping with Activated Carbon-A Field Experiment in Trondheim Harbor, Norway. Environ. Sci. Technol. 2011, 45, 6110–6116. [DOI] [PubMed] [Google Scholar]
- Kitagawa W.; Suzuki A.; Hoaki T.; Masai E.; Fukuda M. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1 demonstrated by denaturing gradient gel electrophoresis. Biosci., Biotechnol. Biochem. 2001, 65, 1907–1911. [DOI] [PubMed] [Google Scholar]
- Kanaly R. A.; Harayama S. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 2000, 182, 2059–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuya W. Microorganisms relevant to bioremediation. Curr. Opin. Biotechnol. 2001, 12, 237–241. [DOI] [PubMed] [Google Scholar]
- Head I. M.; Jones D. M.; Roling W. F. M. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 2006, 4, 173–182. [DOI] [PubMed] [Google Scholar]
- Pérez-Pantoja D.; Donoso R.; Junca H.; González B.; Pieper D. H., Phylogenomics of aerobic bacterial degradation of aromatics. In Handbook of Hydrocarbon and Lipid Microbiology;Timmis K. N., Ed.; Springer Verlag: Heidelberg, 2010. [Google Scholar]
- Heitkamp M.; Franklin W.; Cerniglia C. Microbial metabolism of polycyclic aromatic hydrocarbons: Isolation and characterization of a pyrene-degrading bacterium. Appl. Environ. Microbiol. 1988, 54, 2549–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingley R. L.; Khan A. A.; Cerniglia C. E. Molecular characterization of a phenanthrene degradation pathway in Mycobacterium vanbaalenii PYR-1. Biochem. Biophys. Res. 2004, 322, 136–146. [DOI] [PubMed] [Google Scholar]
- Shimodaira J.; Miyauchi K.; Takeda H.; Kasai D.; Masai E.; Fukuda M. Regulatory mechanism of biphenyl/PCB-degradation gene transcription in Rhodococcus jostii RHA1. J. Biotechnol. 2010, 150, S246–S246. [Google Scholar]
- Arai H.; Kosono S.; Taguchi K.; Maeda M.; Song E.; Fuji F.; Chung S.; Kudo T. Two sets of biphenyl and PCB degradation genes on a linear plasmid in Rhodococcus erythropolis TA421. J. Ferment. Bioeng. 1998, 86, 595–599. [Google Scholar]
- Kesseler M.; Dabbs E.; Averhoff B.; Gottschalk G. Studies on the isopropylbenzene 2,3-dioxygenase and the 3-isopropylcatechol 2,3-dioxygenase genes encoded by the linear plasmid of Rhodococcus erythropolis BD2. Microbiology 1996, 142, 3241–3251. [DOI] [PubMed] [Google Scholar]
- HuiJie L.; CaiYun Y.; Yun T.; GuangHui L.; TianLing Z. Using population dynamics analysis by DGGE to design the bacterial consortium isolated from mangrove sediments for biodegradation of PAHs. Int. Biodeter. Biodegrad. 2010, 65, 269–275. [Google Scholar]
- Vinas M.; Sabate J.; Espuny M.; Solanas M. Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl. Environ. Microbiol. 2005, 71, 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.; Crandell D.; Singleton D. Stable-isotope probing of the polycyclic aromatic hydrocarbon-degrading bacterial guild in a contaminated soil. Environ. Microbiol. 2011, 13, 2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin B. R.; Nakatsu C. H.; Nies L. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl. Environ. Microbiol. 2003, 69, 3350–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cébron A.; Norini M.-P.; Beguiristain T.; Levyval C. Real-Time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 2008, 148–159. [DOI] [PubMed] [Google Scholar]
- Bamforth S. M.; Singleton I. Bioremediation of polycyclic aromatic hydrocarbons: current knowledge and future directions. J. Chem. Technol. Biotechnol. 2005, 80, 723–736. [Google Scholar]
- Parales R. E. The role of active-site residues in naphthalene dioxygenase. J. Ind. Microbiol. Biotechnol. 2003, 30, 271–278. [DOI] [PubMed] [Google Scholar]
- Laurie A. D.; Lloyd-Jones G. Quantification of phnAc and nahAc in contaminated New Zealand soils by competitive PCR. Appl. Environ. Microbiol. 2000, 66, 1814–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillis L.; Clipson N.; Doyle E. Quantification of catechol dioxygenase gene expression in soil during degradation of 2,4-dichlorophenol. FEMS Microbiol. Ecol. 2010, 73, 363–369. [DOI] [PubMed] [Google Scholar]
- Widada J.; Nojiri H.; Kasuga K.; Yoshida T.; Habe H.; Omori T. Quantification of the carbazole 1,9a-dioxygenase gene by real-time competitive PCR combined with co-extraction of internal standards. FEMS Microbiol. Lett. 2001, 202, 51–57. [DOI] [PubMed] [Google Scholar]
- Van Hamme J. D.; Singh A.; Ward O. P. Recent advances in petroleum microbiology. Microbiol. Mol. Biol. Rev. 2003, 67, 503–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry J. C. Direct methods and biomass estimation. Methods Microbiol. 1990, 22, 41–85. [Google Scholar]
- Hawthorne S. B.; Azzolina N. A.; Neuhauser E. F.; Kreitinger J. P. Predicting Bioavailability of sediment polycyclic aromatic hydrocarbons to Hyalella azteca using equilibrium partitioning, Supercritical fluid extraction, and pore water concentrations. Environ. Sci. Technol. 2007, 41, 6297–6304. [DOI] [PubMed] [Google Scholar]
- Cho Y. M.; Werner D.; Choi Y.; Luthy R. G. Long-term monitoring and modeling of the mass transfer of polychlorinated biphenyls in sediment after pilot-scale in-situ amendment with activated carbon. J. Contam. Hydrol. 2012, 129–130, 25–37. [DOI] [PubMed] [Google Scholar]
- Ahn S.; Werner D.; Luthy R. G. Physicochemical characterization of coke plant soil for the assessment of PAH availability and the feasibility of phytoremediation. Environ. Toxicol. Chem. 2005, 24, 2185–2196. [DOI] [PubMed] [Google Scholar]
- Hale S. E.; Werner D. Modeling the mass transfer of hydrophobic organic pollutants in briefly and continuously mixed sediment after amendment with activated carbon. Environ. Sci. Technol. 2010, 44, 3381–3387. [DOI] [PubMed] [Google Scholar]
- Wiedemeier T. H.; Rifai H. S.; Newell C. J.; Wilson J. T.. Natural Attenuation of Fuels and Chlorinated Solvents in the Subsurface; Wiley: New York, 1999; p 617. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.