Abstract
The mechanisms underlying evolutionary changes in protein length are poorly understood. Protein domains are lost and gained between species and must have arisen first as within-species polymorphisms. Here, we use Drosophila melanogaster population genomic data combined with between species divergence information to understand the evolutionary forces that generate and maintain polymorphisms causing changes in protein length in D. melanogaster. Specifically, we looked for protein length variations resulting from premature termination codons (PTCs) and stop codon losses (SCLs). We discovered that 438 genes contained polymorphisms resulting in truncation of the translated region (PTCs) and 119 genes contained polymorphisms predicted to lengthen the translated region (SCLs). Stop codon polymorphisms (SCPs) (especially PTCs) appear to be more deleterious than other polymorphisms, including protein amino acid changes. Genes harboring SCPs are in general less selectively constrained, more narrowly expressed, and enriched for dispensable biological functions. However, we also observed exceptional cases such as genes that have multiple independent SCPs, alleles that are shared between D. melanogaster and Drosophila simulans, and high-frequency alleles that cause extreme changes in gene length. SCPs likely have an important role in the evolution of these genes.
Keywords: nonsense mutation, selective constraint, population genomics, polymorphism, stop codon loss
Introduction
Genetic variation in natural populations has long been a source of interest to both population biologists and functional geneticists, and the genus Drosophila has been a system of choice for describing such variation (Timofeev-Ressovsky H and Timofeev-Ressovsky NW 1927; Timofeev-Ressovsky 1930; Dubinin et al. 1937; Ives 1945; Spencer 1947). Natural variations allow one to infer patterns of gene flow, migration, and selection. In addition, alleles discovered in natural populations have been used as tools to elucidate molecular mechanisms of specific phenotypes—for example, meiotic mutants from natural populations (Sandler et al. 1968). More recently, effort has focused on determining what specific genetic changes have led to adaptation between and within species. One hotly debated question is whether protein sequence, copy number, or gene regulation are more likely to be the genetic basis of adaptation (Prud'homme et al. 2007; Wray 2007; Emerson et al. 2008). However, the evolutionary role of genetic variants that lead to deviations from annotated gene models—such as the position of initiation codons, splicing junctions, and stop codons—has received relatively little attention considering the potential impact of such variants on gene function. Alleles containing premature termination codons (PTCs) are well characterized as the genetic cause of many human diseases including retinosis pigmentosa (Chang and Kan 1979; Rosenfeld et al. 1992) and beta-thalassemia and so have been of particular interest to the genetics of human disease, but the prevalence of such changes within and between populations is rarely studied.
We expect the functional consequences of stop codon polymorphisms (SCPs)—in particular PTCs—on the affected gene to be at least as severe as those caused by nonsynonymous mutations and more severe than those caused by synonymous mutations. This is because transcripts of genes carrying PTCs are expected to undergo nonsense-mediated decay (Chang et al. 2007), which results in loss of gene expression and function. In humans, stop codons occurring more than 50 bases prior to the final exon–exon junction are silenced by nonsense-mediated decay (Nagy and Maquat 1998). This process occurs in all organisms in which it has been studied (Chang et al. 2007), but the trigger for nonsense-mediated decay is not as clear in other organisms as it is in humans (Gatfield et al. 2003; Behm-Ansmant et al. 2007). If transcripts harboring PTCs are not targeted by nonsense-mediated decay, they will likely still be deleterious because of the loss of 3′ protein domains or dissociation from 3′ untranslated region regulatory elements. The stop codon of a transcript may also be lost (stop codon loss, SCL), leading to either downregulation of expression through the nonstop decay pathway (Vasudevan et al. 2002) or an expansion of the open reading frame. Nonstop decay results in posttranscriptional degradation of transcripts without an in-frame stop codon prior to the polyadenylation signal and is conserved throughout eukaryotes (Gatfield et al. 2003). SCLs that are not silenced could acquire novel downstream structural or regulatory sequence elements that might alter protein expression or function.
The length of the open reading frame of genes has clearly changed over evolutionary time (Yandell et al. 2006). It has been observed that divergence in the length of coding regions is disproportionately found at the beginnings and ends of genes (Bjorklund et al. 2005; Weiner et al. 2006), with the latter possibly caused by either loss or gain of the stop codon. The fact that we observe such changes between species implies that they must first arise as within-species polymorphisms. But where do these SCPs first arise within populations and genomes, and what is their evolutionary fate after they arise? Previous work has documented the number and frequency of polymorphisms causing changes in the position of termination codons in humans. Yamaguchi-Kabata et al. (2008) used human database single nucleotide polymorphism (SNP) data (Sherry et al. 1999) and found 1,183 SNPs resulting in PTCs, 581 of which were predicted to trigger nonsense-mediated decay and were thus annotated as null alleles. They also observed 119 polymorphisms causing SCLs, which typically led to short expansions of the open reading frames. SCPs were found at a lower density (polymorphic site per mutable site) than nonsynonymous amino acid changes, implying that SCPs are more likely to be deleterious than changes to amino acid sequence. In another study, Yngvadottir et al. (2009) genotyped a subset of the SNPs reported above in order to measure allele frequency of these SNPs and confirmed that PTCs were generally at low frequency and evenly distributed within the coding region of the proteins they were found in. Finally, the 1000 human genomes project has cataloged additional PTCs in the human population (Durbin et al. 2010).
The population genetics of null alleles has long been an area of interest in Drosophila (Voelker et al. 1980; Langley et al. 1981; Burkhart et al. 1984), and a number of individual SCPs have been described and characterized in detail (Begun and Lindfors 2005; Lazzaro 2005; Kelleher and Markov 2009). However, SCPs have not yet been described in Drosophila on a genome-wide scale. This analysis will provide a useful contrast to human data in an experimentally tractable organism, providing a unique opportunity to determine the functional importance and fitness impacts of SCPs observed in natural populations. With the advent of next-generation sequencing technology, we are now able to describe thousands of natural variants in Drosophila simultaneously. Recently, 37 whole genomes from a population of Drosophila melanogaster near Raleigh, North Carolina, USA (RAL), and 7 genomes from a population in Malawi, Africa (MW), have been resequenced as part of the Drosophila Population Genomics Project (DPGP) (www.dpgp.org; Langley et al. 2012). Additionally, six genomes of D. melanogaster’s close relative, D. simulans (Begun et al. 2007), and ten other species of Drosophila (Clark et al. 2007) have been sequenced and annotated, providing a wealth of data for inferring the evolutionary history of within-species variation. In contrast to previous surveys of natural variants (e.g., Sandler et al. 1968), the described alleles from these 44 D. melanogaster genomes are preserved in living stocks, which can be rapidly leveraged toward functional work. This represents an unparalleled resource for answering questions about the origin, maintenance, and functional impact of natural variants on a genomic scale.
Here, we describe one type of variation that was uncovered in the DPGP sequencing project: SNPs that cause changes in the position of the stop codon (SCPs). Our observations generally supported our a priori hypothesis that the sampled SCPs are, as a group, selected against, and generally more deleterious than other types of SNPs. However, we did find a number of alleles that were exceptions to this pattern, such as alleles that have been segregating since before the split between D. melanogaster and D. simulans, high frequency–derived alleles and alleles at high frequency despite causing large changes in the original gene model. We also found 56 genes carrying more than two alleles with different stop codon positions. The evolution of these genes may be strongly affected by changes in gene model. Furthermore, the alleles described in this study are available in living stocks, providing an opportunity to directly measure the phenotypic consequences of stop codon variation.
Materials and Methods
Characterizing SCPs within D. melanogaster
We used FlyBase version 5.16 for gene model annotations (Tweedie et al. 2009), giving a total of 14,072 annotated protein-coding genes. For each gene model, we searched through the 44 D. melanogaster genomes from DPGP (www.dpgp.org; Langley et al. 2012) and identified genes that varied across the 44 genomes with respect to the position of the stop codon. To avoid conflating other changes in gene structure with changes in the stop codon, we removed from consideration any alleles that had splice sites or initiation codons that differed from the reference annotation. For genes with more than one isoform, we determined which isoforms were affected. If the genomic position of a variant was the same for multiple isoforms, we only considered the isoform with the longest coding region when calculating statistics on the changes in gene model. For each allele, we counted the number of lines agreeing or disagreeing with the reference annotations for North American (Raleigh, “RAL”) and African (Malawi, “MW”) populations, respectively. It is worth noting that although all the major chromosomes of the 37 Raleigh strains were sequenced, only some chromosomes were sequenced from each of the nine Malawi strains. This resulted in seven first (X), six second (2L and 2R), and five third (3L and 3R) chromosomes in the Malawi population. We then defined each allele with respect to the allele frequency across both populations. If the minor allele was the shorter one, we considered the polymorphism to be a PTC, whereas if the minor allele was the longer of the two, we considered the polymorphism to be an SCL. We considered polarizing with respect to the ancestral state but were concerned that this would bias against alleles of rapidly evolving genes. The ancestral state of these genes is difficult to determine due to the poor alignment between distantly related species (D. yakuba/D. erecta) with fast-evolving sequences. Indeed, we found nearly half of our alleles dropped out of the analysis when we included D. yakuba and D. erecta as the outgroup lineage (see Results). There were nine alleles that would have a different definition if we had polarized with respect to the ancestral state (designated “Ancestor minor” in supplementary table 2, Supplementary Material online). The reference genome carried a minor PTC allele in 8 cases and a minor SCL allele in 13 cases (designated “Reference Minor” in supplementary table 2, Supplementary Material online), though the reference genome was not included as an allele in the population for our analyses.
For alleles with an SCL and an annotated 3′ untranslated region, the “expanded region” for a given allele was defined as the downstream transcribed sequence until one of three features was encountered: 1) an in-frame stop codon, 2) an uncalled (“N”) base that would have been an in-frame stop codon assuming the genome matched the reference genome, and 3) the end of the known transcribed region. If no stop codon was encountered before an annotated polyadenylation site, the gene was labeled as a target of nonstop decay and was not included in the expansion length analysis. In total, we predicted four alleles to undergo nonstop decay and could not confirm nonstop decay status for another 90 alleles because there was no 3′ untranslated region sequence data available or the region did not contain a polyadenylation site.
We estimated the density of SCPs following Yngvadottir et al. (2009). Density represents the proportion of sites in which an SNP resulted in a PTC or SCL. The density of PTCs is the number of observed PTCs divided by 2,387,149—the number of sites in the genome that can mutate directly to a stop codon (one mutable site for all codons in annotated genes that are one mutational step away from one of the three stop codons except TGG, which has two mutable sites). For SCLs, the density is the observed number of SCLs divided by the number of unique stop codons across all the isoforms of all genes (total 42,315 sites).
Allele frequency for each polymorphism was estimated as the proportion of the minor allele among all available alleles. Each DPGP assembly is missing data for some lines due to assembly problems and/or low sequencing quality. We controlled for the resulting variation in allelic coverage (the number of genomes at each site that have data) by removing sites with allelic coverage below 20 from the entire data set. We also used maximum likelihood methods to estimate the number of minor alleles if all alleles were available, assuming minor allele counts have a hypergeometric distribution. This method was only applied to Raleigh and all D. melanogaster samples. Our observations were consistent between the two methods and we only present the former. We contrasted the frequency of SCPs, with that of nonsynonymous and synonymous SNPs (fig. 1) from the DPGP data set. Among the annotated genes in version 5.16, there are 16 genes with PTCs in the annotated coding regions of the reference annotations and 12 genes that do not have a stop codon at the end of the reference annotated translated region. These 28 genes were excluded from our analysis (supplementary table 1, Supplementary Material online).
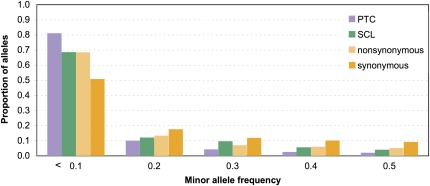
FIG. 1.—
The allele frequency spectra for SCPs are skewed toward rare alleles. PTC (violet) and SCL (green) polymorphisms are enriched for rare alleles. SCPs are more likely to be at low frequency than synonymous SNPs (dark orange). In addition, PTCs—but not SCLs—are more skewed than highly constrained nonsynonymous SNPs (light orange).
Error/Sequence Quality Control
Release 1.0 of the DPGP data consists of fastq files, where the quality score of a base is derived from the quality of the Illumina reads covering that base and the quality of the consensus assembly at that base. The scoring system is based on Phred quality scores where a score of Q50 indicates an estimated 1/100,000 error rate (Ewing et al. 1998). We used SNPs surrounding our SCPs to estimate the expected distribution of quality data for SNPs. The median of the distribution is Q50 with scores ranging from a minimum of Q30 to a maximum of Q74 (supplementary fig. 1, Supplementary Material online, orange).
We calculated above that 2,387,149 bp could mutate to become a stop codon. Because we are calling bases independently across an average of 44 genomes, this is a total of approximately 103 million bases that could become stop codons. If all of these bases had the median Phred quality score of Q50 (1/100,000 error rate), then 1,026 bases would be incorrectly called as PTCs. We used an empirical distribution of quality scores for polymorphic bases in the 44 genomes to determine how many total false-positive mutations would be expected. Given this distribution, we expect about 7,026 false-positive PTCs would be called, and most of them (5,095) would have quality scores below Q40. Given that we actually observed 2,104 PTCs across the genomes (across all quality scores), the error rate is likely lower than calculated above. We found that PTC SNPs had lower quality scores than other SNPs, with an apparent excess of SNPs with quality scores less than 40 (supplementary fig. 1A, Supplementary Material online). However, the experimental distribution of quality scores for observed SCPs is not consistent with the expectation if most of the SCPs called were errors (supplementary fig. 1B, Supplementary Material online), implying that most SCPs we have observed are not errors.
We went on to sequence 73 alleles chosen randomly from the different quality score classes and found that alleles with a quality score below 40 were usually false positives (3/12 alleles were validated). Conversely, alleles with a quality score of 40 or greater were validated the majority of the time (29/46), and alleles with quality above 60 were validated in 12/15 cases. We decided to pursue the remaining analyses having discarded all alleles with a quality score below 40, as these alleles were mostly erroneous. Although some of the remaining alleles are likely false positives, we are confident that the majority of them are true positive. We also did all analyses with a more conservative data set in which polymorphisms called in only a single line were removed (table 1, nonsingletons). As each SNP is called using an independent sequencing data set for each line, it is extremely unlikely that the same error would be found in more than a single line provided that error is random. Furthermore, we found that there was no difference in quality scores between nonsingleton and singleton alleles, implying no obvious bias in base calling across genomes. We used the same quality score cutoff (Q40) for other types of SNPs (nonsynonymous, synonymous, and noncoding) that were used to contrast with SCPs.
Table 1.
SCPs in Drosophila melanogaster
| All Alleles |
Nonsingletons Only |
||||||
| Malawi | Raleigh | Total | Malawi | Raleigh | Total | ||
| PTC | Genes | 146 | 353 | 438 | 88 | 147 | 157 |
| Alleles | 153 | 395 | 498 | 91 | 158 | 170 | |
| SCL | Genes | 62 | 93 | 119 | 21 | 59 | 65 |
| Alleles | 65 | 97 | 124 | 22 | 63 | 68 | |
Phylogenetic Analyses
For each SCP, we asked whether the sequenced genome of other Drosophila species in the melanogaster subgroup shared the major or minor allele from the D. melanogaster population. This allowed us to infer which of the extant D. melanogaster alleles are newly derived versus shared with D. simulans. We used the multispecies whole-genome alignment (D. melanogaster, D. simulans, D. yakuba, and D. erecta) created for the DPGP genome project (Langley et al. 2012). In order to infer the age of the origin for an SCP, we required that a base that is polymorphic in D. melanogaster has data available (not an “N” or deletion) in at least one D. simulans genome and either the D. yakuba or D. erecta genomes in the multispecies alignment. For analyses using an outgroup, we used the D. yakuba data if available, and if it was not available, we used the D. erecta data. Alleles without sufficient data were designated as “missing data” (fig. 4). If fixed, the D. simulans allele was required to be the same as the D. yakuba/D. erecta allele. Nineteen alleles that violated this rule were given the designation of “ambiguous history.” The age of the remaining alleles were annotated (with the assumption of only a single mutation leading to the allele) as having arisen in the ancestor of D. simulans and D. melanogaster, in the ancestor of the two D. melanogaster populations, or in one of the two D. melanogaster populations. We also asked whether the major allele currently found in D. melanogaster was the ancestral allele or the derived allele by comparing it to D. yakuba/D. erecta. Finally, we asked whether any genes were segregating with two or more alleles in one or both of the D. melanogaster populations and/or the D. simulans populations. A caveat of using D. yakuba and D. erecta as an outgroup is that fast-evolving genes have a higher chance of being misaligned in the multispecies genomic alignment and thus removed from the analysis. We performed above analyses and tests without using the yakuba–erecta outgroup to polarize the changes—that is, we only asked if an allele was specific to one or both D. melanogaster populations or if it was shared with D. simulans. Our observations were insensitive to whether or not we use the D. yakuba/D. erecta clade to polarize the direction of the changes. To contrast the age of SCPs to other SNPs, we used the same criteria to classify nonsynonymous and synonymous polymorphism into different age classes.
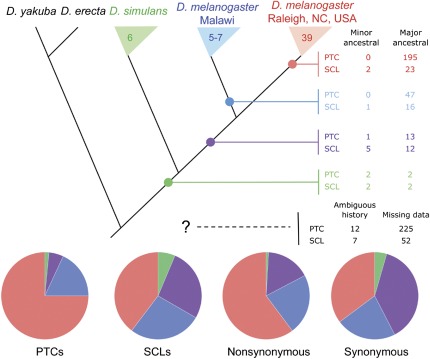
FIG. 4.—
PTCs are more derived than nonsynonymous polymorphisms. We classified each PTC and SCL allele as recently derived in the Raleigh, NC, population (red) or the Malawi population (blue); shared by the two Drosophila melanogaster populations (violet); or shared with Drosophila simulans (green). The number of alleles sampled is shown for each branch (branch lengths are not to scale). The outgroup alleles (D. yakuba and D. erecta) allowed us to determine whether the current D. melanogaster major or minor allele was likely ancestral (side panel minor/major). Almost half of the alleles could not be categorized due to a lack of sequencing/alignment data from one or more species (“Missing data”) or because it was unclear whether the minor or major allele was ancestral (“Ambiguous history”). Pie charts show the proportion of alleles in each described age category for PTCs, SCLs, nonsynonymous SNPs, and synonymous SNPs.
When comparing the number of SCPs unique to either MW or RAL populations, we made the following correction. It has been demonstrated that the expected number of polymorphic sites observed from a sample of size n is proportional to (Watterson 1975). Accordingly, in the chi-square test table, we applied this correction to the sample size of each population.
Population Genetics Analyses of Genes with SCPs
The GC content of each gene was estimated as the proportion of GC bases in the coding regions annotated in the reference D. melanogaster genome. The Codon bias index Fop (frequency of optimal codons) was estimated with CodonW (Peden 1999). We used PAML (version 4; Yang 1997) to estimate the lineage-specific substitution rate on the D. melanogaster and D. simulans lineage, using D. yakuba as the outgroup. For each gene, we used the two D. melanogaster and two D. simulans alleles with the highest allelic coverage per base pair (e.g., the proportion of bases that are not missing data) together with the D. yakuba allele, to estimate dN/dS on the D. melanogaster branch. This prevents within-species polymorphism from inflating the estimate of the substitution rate. dN/dS estimates tend to have larger variance when there is not enough information. We thus removed estimates for genes that have fewer than 100 sites (nonsynonymous plus synonymous sites) included in the PAML analysis or whose dS estimates are below 0.001. To account for the variation in allelic coverage in the D. melanogaster genomes, Tajima’s D (Tajima 1989) was calculated as the sum of Tajima’s D for each allelic coverage class normalized by the square root of the number of allelic classes. We also calculated Tajima’s D and estimated dN/dS for protein-coding genes without SCPs using the DPGP polymorphism data and multispecies alignment. All statistical analyses were done using R version 2.8 (www.r-project.org).
Gene Expression Analysis
In order to determine 1) whether genes having SCPs were likely to be expressed more or less broadly than other genes and 2) genes having SCPs were enriched in certain tissues, we used multiple tissue microarray data from FlyAtlas (www.flyatlas.org; Chintapalli et al. 2007). We downloaded the raw data from the FlyAtlas gene expression database then categorized each gene as 1) protein coding, 2) protein coding and harboring an SCL or PTC, and 3) nonprotein coding. For the rest of the analysis, we excluded nonprotein-coding genes.
To test for broadness of expression, we used the FlyAtlas “present” call data. The FlyAtlas data consist of four duplicate arrays for each tissue type tested. Each gene is called as either present or not on each array. Therefore, a given gene can have a present call score from 0/4 to 4/4. We declared a gene as expressed if it was called as present in 3/4 or 4/4 of the arrays in at least one of the probes for that gene. We first asked how many tissues a gene was called as present in and calculated the means and variances for PTCs, SCLs, and all protein-coding genes. Next, we used a contingency test (chi-square test) to ask whether the most broadly expressed category (i.e., present call in all tissues) or the least expressed category (i.e., present call in zero tissues) were enriched among PTCs and SCLs compared with the remaining protein-coding genes.
To determine if any single tissue was enriched among SCPs, we used the raw expression data from FlyAtlas to determine what the most highly expressed tissue was for each gene. We then asked whether there was an excess or paucity of SCPs expressed at their highest level in any given tissue compared with the total genes annotated as being expressed in at least one tissue (Fisher’s exact test). We then corrected for the number of tissue types tested (10) using the Bonferroni adjustment (Abdi 2007).
Gene Ontology Analysis
We used the online gene ontology (GO) functional annotation tool DAVID to determine if the genes were enriched for any biological, cellular, or molecular functional terms (Huang da et al. 2009). We used the FATGO annotation categories, which give extra weight to GO terms that are more specific (e.g., less weight is given to broad GO terms such as “cellular component” and more weight is given to specific terms such as “vesicle”). DAVID uses a modified Fisher’s exact test called the EASE score to test for enrichment (Dennis et al. 2003). We separately uploaded the list of genes found to contain PTCs and SCLs to DAVID’s servers, bulk-downloaded the resulting enriched GO categories, and then ranked the results by P value to obtain a list of the top enriched categories for each gene list.
Annotation with InterProScan
We used the program InterProScan (Quevillon et al. 2005) to annotate domains in coding regions lost due to PTCs or gained due to SCLs. InterProScan cannot annotate domains in peptides shorter than 20 amino acids. Accordingly, for both SCLs and PTCs, we excluded truncated or expanded sequences from PTCs and SCLs that were shorter than 60 bp. PTCs can lead to a truncated protein or silencing of the gene by nonsense-mediated decay. Studies using D. melanogaster Adh transgenes suggested that the decay process was triggered if there was more than 400 bp between the stop codon and the polyadenylation site (Behm-Ansmant et al. 2007). Because the average size of a 3′ untranslated region is 200 bp in D. melanogaster (Retelska et al. 2006), we looked for domains in coding sequences (CDS) truncations that were 200 bp or shorter, as these are predicted to avoid nonsense-mediated decay. For SCLs, we only used alleles from genes that were not predicted to trigger nonstop decay as described above. Extracted sequences were translated and sent in bulk to the InterProScan server (www.ebi.ac.uk/Tools/InterProScan/).
Results and Discussion
Hundreds of SCPs Are Present in D. melanogaster
We searched through the 44 D. melanogaster genomes generated by DPGP (www.dpgp.org; Langley et al. 2012) for alleles that varied in the position of the stop codon, using the annotations of the D. melanogaster reference genome (version 5.16). To confirm that observed SNPs are not sequencing or assembly errors, we used direct sequencing and found that polymorphisms with an assembly quality score Q40 or greater were correct 60% of the time, whereas alleles with quality below Q40 were errors 80% of the time. This false discovery rate implies the quality of a DPGP Q40 SNP is actually much higher than Phred Q40 (see Materials and Methods and supplementary fig. 1, Supplementary Material online). As a result, we used the DPGP genome assembly with a cutoff of quality score Q40 (bases with quality score lower than Q40 are treated as missing data). We defined the direction of the change for each allele with respect to the major allele in the population, which was not always the same as the annotated reference allele (supplementary table 2, Supplementary Material online). We considered the traditional approach of polarizing by ancestry but were unable to determine the ancestry of nearly half of the alleles (237 PTCs and 59 SCLs). This was often because of high levels of divergence between D. melanogaster and the D. yakuba/D. erecta clade.
In D. melanogaster, we observed 438 genes harboring 498 PTC alleles and 119 genes harboring 124 SCL alleles (table 1, supplementary table 2, Supplementary Material online). After quality screening and polarization, there were a total of 1,667 occurrences of all minor alleles across all genomes—this gives roughly 37.9 SCPs per genome analyzed. Although we are confident that most of the observed SCPs are not sequencing or assembly errors, we created a more conservative data set by removing alleles that were present only once across the two D. melanogaster populations (table 1, nonsingletons). We performed our analyses using both data sets but present only results using all alleles unless the observations are different between the two data sets.
We expect our data set to be biased toward polymorphisms with minor fitness effects because the sequenced DPGP genomes were prepared as inbred strains (RAL populations) or strains with targeted pairs of homozygous chromosomes (MW populations). In both cases, polymorphisms that are strongly deleterious in nature were likely removed from the strains prior to sequencing.
SCPs Are as a Group Selected Against
Due to the potential impact of SCPs on the function and expression of the genes harboring them, we expected a priori that most SCPs should be selected against more strongly than other types of variation. Four aspects of the data supported our hypothesis. First, the density (the number of polymorphic sites per mutable site across the genome) of SNPs resulting in PTCs and SCLs are 0.00021 and 0.0029, respectively, both of which are small when compared with a density of 0.0089 for nonsynonymous sites and 0.090 for synonymous sites. Second, we found that the allele frequency distributions of SCPs of both types are skewed toward rare variants when compared with synonymous polymorphisms (fig. 1, chi-square test, P < 10−16 [PTC], P = 0.03 [SCL]). The allele frequency distribution of PTCs is also more skewed than that observed for highly constrained nonsynonymous polymorphisms (fig. 1, chi-square test, P < 10−11), whereas the distribution for SCLs was neither more nor less skewed than nonsynonymous polymorphisms (fig. 1). We noted that several SCPs only affect some of the many isoforms of the genes they reside in. The fitness consequences of such polymorphisms are likely to be less extreme and may be less likely to be selected against. To test this hypothesis, we removed any SCPs that affected less than 50% of a gene’s isoforms (18.9% of PTCs and 23.4% of SCLs; supplementary table 2, Supplementary Material online “<50% Isoforms”), repeated the comparison, and observed an even stronger enrichment of rare alleles for PTCs but no change in the result for SCLs.
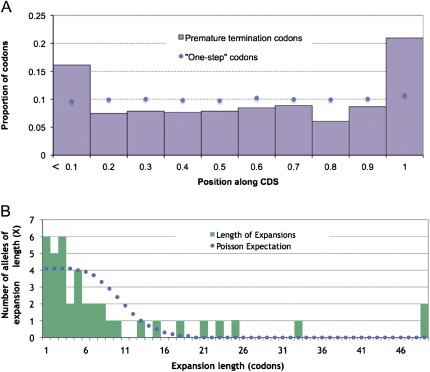
Third, we found that both PTCs and SCLs were enriched for alleles that cause less extreme changes of the coding region length (fig. 2), suggesting that extreme alleles are more strongly selected against and less likely to be sampled. Assuming mutations occur randomly, the positions of PTCs within coding regions should be uniformly distributed. However, we estimated the empirical distribution of codons that are one mutational step away from a stop codon (one-step codons) and found that such codons are “not” uniformly distributed within CDS, having a slightly higher proportion of one-step codons in the 3′ regions of genes (fig. 2A, blue dots). We found that the distribution of PTCs was significantly different from both the uniform distribution and the one-step codon distribution (chi-square test, both P < 10−4), with an excess of PTCs at the start and end of coding regions (fig. 2A). Our observation that an excess of PTCs are found near the start of CDS is intriguing. As expected, most of the alleles with highly truncated CDS are segregating at low frequency in the population with a few interesting exceptions (see below). The number of base pairs added after an SCL is expected to follow a “Poisson” process with parameter λ as the mean length, provided that stop codons are randomly distributed in the 3′ untranslated region. However, the absence of a stop codon can lead to gene silencing by nonstop decay (Vasudevan et al. 2002), which is triggered when there is no stop codon prior to the polyadenylation site. In order to measure the effect of length expansion on allele frequency, we considered only those alleles that are not predicted to undergo nonstop decay (see Materials and Methods). Among these, the mean number of codons added was 5.5, with the longest and shortest expansions being 1 and 49 codons, respectively. We found that the distribution of length change was significantly different from the Poisson process expectation, with an excess of both small and large length changes (Kolmogorov–Smirnov test, P < 10−7; fig. 2B). The excess of small changes may be due to selection against extreme changes in gene length, whereas the excess of longer changes is intriguing and could be due to nonrandom distribution of stop codons in some 3′ untranslated regions. Given these observations, we expected to see a negative correlation between the size of change caused by the SCP and its frequency in the population. However, we did not see this correlation for either PTCs or SCLs (Spearman’s rank ρ, P all > 0.05). Restricting to alleles influencing more than half of a gene’s isoforms yielded a similar insignificant result. It is possible that the realized allele frequency is more affected by the function of the gene in question than by the extremity of the allele.
FIG. 2.—
Extremely long truncations and expansions of genes are rarer than expected. The change in gene length was predicted for PTCs (A) and SCLs (B). (A) PTCs appearing earlier in the coding regions are expected to have a more extreme effect on gene function than those appearing near the ends of genes. There was a significant excess of short truncations compared with the distribution of one-step codons (blue dots). (B) For alleles with SCLs whose gene model has an annotated 3′ untranslated region, the number of codons added is shown (green bars) along with the expectation if the length expansions due to SCLs followed a Poisson process (blue dots). The distribution was significantly different from the Poisson process expectation, with an excess of both long and short alleles.
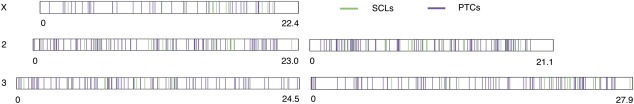
Finally, we observed that there is a deficiency of genes harboring PTCs on the X chromosome compared with autosomes, which likely results from stronger purifying selection against deleterious recessive alleles on the X chromosome in males (1.56% for X chromosomes and 3.29% of autosomes, Fisher’s exact test, P < 10−3; fig. 3). This pattern was only marginally significant for SCLs (0.37% of X-linked and 0.91% of autosomal genes, Fisher’s exact test, P = 0.02). We repeated the analysis with SCPs that affected more than 50% of a gene’s isoforms and found a significant paucity of SCLs on the X (0.18% of X-linked and 0.71% of autosomal genes, Fisher’s exact test, P = 0.007) and an even stronger X deficiency for PTCs (1.5% on the X and 3.4% on the autosomes, Fisher’s exact test, P < 10−4). This pattern was not significant when we only considered nonsingletons because such alleles are likely to be less deleterious due to their high population frequency.
FIG. 3.—
Fewer SCPs are found on the X chromosome. The genomic position of each PTC (violet) and SCL (green) are shown to scale on the chromosomes on which they are found (the length in megabase of each chromosome is shown). The X chromosome is underrepresented for PTCs and SCLs compared with the expectation given the number of genes on each chromosome.
PTCs that trigger nonsense-mediated decay and SCLs that trigger nonstop decay are expected to have greatly reduced expression and to be functional null alleles. Alleles that escape these surveillance processes are likely to have impaired gene function. A priori then, these polymorphisms should have equal likelihood to be deleterious. However, this conclusion must be taken with caution, as it is not known how universal these processes are. We observed that the change in length of protein sequence is more dramatic in alleles harboring PTCs than SCLs (fig. 2) and that most SCLs are not predicted to trigger nonstop decay. This is due to the fact that length expansion resulting from SCLs is constrained by the length of the 3′ untranslated region, which is in general shorter than the coding regions where PTCs could happen. Therefore, it seems likely that PTCs would be more deleterious than SCLs. Our observations are consistent with this scenario. We found that PTCs are present at a lower density (0.00021 for PTCs and 0.0029 for SCLs), their frequency spectrum is more skewed toward rare variants (chi-square test, P = 0.01), and a smaller proportion of PTCs are observed on the X chromosome compared with SCLs, though the difference was not statistically significant (8.43% for PTCs and 9.68% for SCLs, Fisher’s exact test, P > 0.05).
Our empirical results support our hypothesis that SCPs of both types are selected against. If the alleles were strictly neutral and at equilibrium between genetic drift and mutation, 23.5% in a sample of 44 alleles are expected to be found only once (Tajima 1989). We observed a much more skewed frequency spectrum for SCPs (60.1% singletons), consistent with the hypothesis that selection removes SCP alleles from the population. However, it is worth asking whether the SCPs we observed are mostly deleterious (Nes >> 1). In D. melanogaster (where Ne ∼ 106 [Kreitman 1983; Charlesworth 2009] and μ ∼ 10−9 per base per generation [Keightley et al. 2009]), the upper bound for the expected equilibrium frequency of a partially recessive deleterious allele under mutation–selection balance is μ/hs = 10−3 (this assumes the least deleterious case, where Nes ∼ 10, h ∼ 0.1; Simmons and Crow 1977; Charlesworth and Charlesworth 2010). With the assumption that segregating alleles are at equilibrium and conditioned on observing a segregating allele using binomial sampling, the probability of sampling a deleterious allele more than once among 44 chromosomes is around 2.1% (the probability of sampling nonsingletons divided by the probability of sampling a segregating allele at least once). This is much lower than the observed number of nonsingletons SCPs (39.9%). Accordingly, while we may have sampled a few SCPs with large fitness effects, an appreciable proportion of SCPs should be under weaker selection and are only weakly deleterious (Nes ∼ 1).
Most SCPs Are Newly Derived on the D. melanogaster Lineage
Given that SCPs are selected against as a group, we predicted that most SCPs should be newly derived. To infer whether SCPs are recently derived on the D. melanogaster lineage and to identify alleles with interesting evolutionary histories, we determined whether any of the six D. simulans genomes or the D. yakuba and D. erecta reference genomes shared each SCP with the D. melanogaster populations (fig. 4). Three hundred and fifteen SCPs within D. melanogaster were fixed in D. simulans for the allele in the D. yakuba/D. erecta outgroup, suggesting a recent origin of these SCPs on the D. melanogaster lineage. Conversely, we found 13 alleles that are polymorphic in both D. simulans and D. melanogaster (“Polymorphic in simulans,” supplementary table 2, Supplementary Material online), although only eight of these (four PTCs and four SCLs) have data available in the outgroup. These alleles likely have been segregating since before the species diverged approximately 5.4 Ma (Tamura et al. 2004) and are of substantial interest. We also observed that D. melanogaster population frequencies of SCL and PTC alleles that are shared between the two species are significantly higher than those that are specific to D. melanogaster, suggesting these shared polymorphisms may have been present for long periods of time (chi-square test, P < 10−5 for PTCs and 0.03 for SCLs). However, we cannot exclude the alternative possibility that our observations were the result of independent mutations arising in the two lineages.
We next asked whether the age distribution of PTCs or SCLs differed from nonsynonymous or synonymous polymorphisms using chi-square tests (fig. 4). We found that PTCs had an excess of Raleigh- and Malawi-specific alleles compared with either nonsynonymous polymorphisms (P < 10−6) or synonymous polymorphisms (P < 10−16). The age distribution of SCLs was not different from the observations for synonymous polymorphisms (P > 0.05) but was different from either PTCs (P < 10−8) or nonsynonymous polymorphisms (P < 10−5), having an excess of alleles shared between the two D. melanogaster populations and with D. simulans. These results show that PTCs are even more likely to be new mutations than nonsynonymous polymorphisms, whereas SCLs show a very different pattern, with a similar age distribution as synonymous polymorphisms. This corroborates the pattern in our data that suggested PTCs are more strongly selected against than SCLs.
Among the D. melanogaster-specific alleles, 31 alleles are polymorphic in both D. melanogaster populations, 64 are segregating in only the Malawi population and 220 are segregating only in the Raleigh population (fig. 4, inset table). Previous research suggests that the Malawi population has higher overall polymorphism than non-African populations (Begun and Aquadro 1993; Haddrill et al. 2005; Hutter et al. 2007). However, after correcting for the effect of sample size (see Materials and Methods), we found no significant excess of alleles in the Malawi population (Fisher’s exact test, P > 0.05). This may be explained by the recent demographic history of non-African D. melanogaster populations (Stephan and Li 2007), which could result in less effective selection against deleterious SCPs. Finally, we found nine alleles where the major allele in the population is derived with respect to the inferred ancestral state (supplementary table 2, Supplementary Material online, Ancestor minor). These alleles have recently increased in frequency and are good candidates to be targets of recent positive selection (see below).
Mutation Contributes to the Appearance of New SCPs
Our observations supported the hypothesis that most SCPs are likely to be either deleterious or weakly deleterious. Population frequencies of SCPs should thus be determined by the intensity of selection removing SCP alleles and the rate of new mutations increasing their frequency. Accordingly, we expected that genes with larger mutational targets and/or weaker selective constraint would be more likely to harbor SCPs. The mutational targets of PTCs are any codons that can mutate directly to a stop codon (one-step codons). Hence, genes containing a larger number of one-step codons should be more likely to harbor PTCs. We would expect the pattern to be even stronger when considering the proportion of codons that are one mutational step away from two stop codons (2-fold one-step codons, TAC, TAT, TCA, TTA, TGG). Indeed, although we did not find an excess of one-step codons in genes carrying PTCs, these genes had a significantly larger number of 2-fold one-step codons (32.1 vs. 28.2 Mann–Whitney U test, P = 0.001). Additionally, the three stop codons of Drosophila are AT rich, and we observed higher AT content among PTC genes than other genes (49.5% vs. 46.4%, Mann–Whitney U test, P < 10−16). However, it is worth noting that most of the unpreferred codons in D. melanogaster are also AT-rich (Akashi 1994). Highly expressed slowly evolving genes have stronger codon bias and higher GC content (Duret and Mouchiroud 1999; Marais et al. 2004; Subramanian and Kumar 2004; Lemos et al. 2005; Larracuente et al. 2008), and we also found that genes carrying PTCs have weaker codon bias than other genes (Fop 0.44 vs. 0.51, Mann–Whitney U test, P < 10−14). Accordingly, it is difficult to tease apart whether mutation or indirect selective forces are the underlying cause of our observation that PTCs have larger numbers of 2-fold one-step codons.
The mutational target of SCLs is the original stop codon. We would predict that TGA and TAG stop codons should more likely to be lost than TAA codons because two possible mutations from the TAA retain a stop codon, whereas only one mutation from TGA or TAG is silent. Supporting this idea, we observed that TAA stop codons are more likely to harbor silent polymorphisms (minor allele has an alternative stop codon at the same position) than TAG or TGA stop codons (Fisher’s exact test, P = 0.02). However, we did not see the predicted paucity of TAA nonsilent changes among SCLs compared with TGA and TAG changes (Fisher’s exact test, P > 0.05). Therefore, we cannot conclude that mutational bias has a strong role in the origin of new SCLs. Yet, this was a weak test as TAA codons have only a marginally lower chance of being lost than TAG or TGA codons (two of nine mutations are silent rather than one of nine).
Genes Harboring SCPs Exhibit Lower Evolutionary Constraint than Other Genes
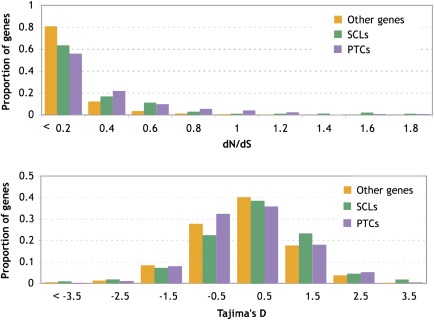
Given our a priori expectations of fitness impacts of SCPs, the intensity of selection against an SCP depends both on how severely the SCP allele affects gene function and how essential the affected gene is. We can test the hypothesis that genes harboring SCPs are less evolutionarily constrained by comparing the dN/dS estimates between genes with and without SCPs. High dN/dS estimates can be interpreted as either elevated rates of adaptive evolution (positive selection) or as weaker selective constraint (reduced purifying selection). Here, we used the dN/dS ratio as a proxy for selective constraint, as most of the genes have a ratio well below one and so are not likely to be under positive selection. Our results are consistent whether or not we include genes showing evidence of adaptive protein evolution (dN/dS > 1). We found that genes harboring SCPs have significantly higher dN/dS ratios than other genes (Mann–Whitney U, P < 10−6 for both PTCs and SCLs; fig. 5A). We also used Tajima’s D to address this question. Tajima’s D summarizes the frequency spectrum of the within-population polymorphism, and strong purifying or directional selection usually leads to highly negative Tajima’s D estimates. We found no significant difference in Tajima’s D between genes with and without SCPs (fig. 5B).
FIG. 5.—
The dN/dS ratios for genes harboring SCPs are higher than typical genes. dN/dS (A) and Tajima’s D (B) were calculated across the CDS for genes harboring SCLs (green), genes harboring premature stop codons (violet), and all other genes (orange). PTCs and SCLs had a significantly elevated dN/dS ratio compared with all genes. There was no statistical difference in Tajima’s D between the different gene types.
We might predict certain groups of SCPs are particularly likely to be under weak constraint or even affected by positive selection. For example, alleles that have increased in frequency recently could be under positive selection or could have drifted to fixation as nearly neutral alleles. We found that the nine genes harboring alleles in which the major allele is derived relative to the ancestral state (supplementary table 2, Supplementary Material online, Ancestor minor) had less negative (closer to zero) Tajima’s D statistics and larger (but still on average < 1) dN/dS estimates than other genes or genes harboring other SCPs (Mann–Whitney U tests, P < 0.05 all tests). The genes carrying the 13 SCPs segregating in both D. melanogaster and D. simulans also showed a less negative Tajima’s D than other genes and than other SCP genes (Mann–Whitney U test, P < 0.05 for both tests) and a larger, though insignificant, dN/dS ratio. Together, these observations are consistent with the hypothesis that genes carrying these subsets of SCPs are under weaker selective constraint than other genes. It is worth noting that our overall observation (SCP genes have higher dN/dS than other genes) was not driven by these special groups, as removal of these genes still yielded significant differences (Mann–Whitney U, P < 10−4 for both PTCs and SCLs).
Genes Harboring SCPs Are More Narrowly Expressed than Other Genes
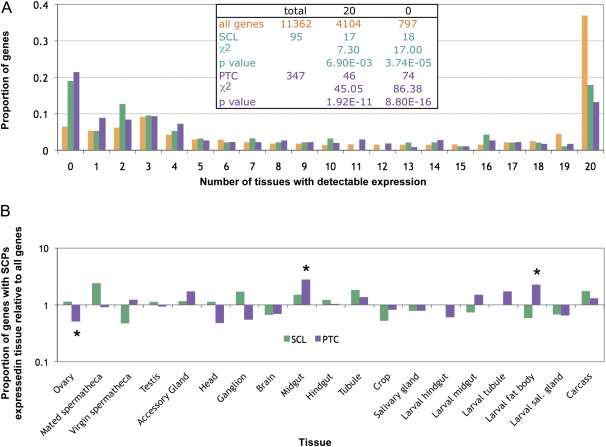
We found above that genes harboring SCPs are likely to be under weak functional constraint. The expression pattern of a gene is one of the most important indicators of gene function. It has been shown that genes expressed broadly and at a high level are more likely to be under strong selective constraint, whereas narrowly and weakly expressed genes are more likely to evolve with less selective constraint (Subramanian and Kumar 2004; Larracuente et al. 2008) or frequent directional selection (Begun and Lindfors 2005; Schully and Hellberg 2006). Given our observation that SCLs have elevated dN/dS ratios, we expected to see an excess of narrowly expressed genes. We used microarray expression data from FlyAtlas (Chintapalli et al. 2007) to determine whether genes harboring SCPs had different expression patterns than other genes. We asked whether genes harboring SCPs were more likely than other genes to have no detectable expression, and whether they were less likely to be expressed broadly (see Materials and Methods). Consistent with our predictions, PTCs and SCLs were significantly more likely than other protein-coding genes to be expressed in none of the tissues tested and significantly less likely to be expressed in all 20 tissues (fig. 6A, inset, chi-square test, P < 0.05).
FIG. 6.—
SCPs are expressed in fewer tissues than other protein-coding genes. (A) All protein-coding genes (orange) were far more likely to be expressed in all 20 tissues tested than genes harboring either SCLs (green) or PTCs (violet) and far less likely than either group of SCPs to be expressed in none of the tissues tested (inset chi-square test, P < 0.01 for all cases). (B) Gene harboring PTCs were more likely than other genes to be expressed at their highest level in the larval fat body and midgut but less likely in the ovary. None of the assayed tissues are significantly enriched for genes harboring SCLs compared with other genes.
We also asked whether there was an enrichment of genes expressed in particular tissues among either PTCs or SCLs. For each gene, we asked what the most highly expressed tissue was and determined whether each tissue was enriched or depleted among either type of SCP compared with all genes (fig. 6B). PTCs were more likely than expected to occur in genes expressed at their highest level in the larval fat body (Fisher’s exact test, P = 0.011) and the adult midgut (Fisher’s exact test, P = 0.004), and they are less likely to occur than expected in genes expressed at their highest level in the ovary (Fisher’s exact test, P = 0.011). SCLs had no significant enrichment or depletion in any tissue. Genes expressed in the ovary include maternally deposited developmental genes, many of which are essential. This may explain why few ovary-specific genes carry PTCs. Conversely, the larval fat body is a common place for immunity genes to be expressed. Several SCPs in immunity genes have previously been observed (Jiggins and Kang-Wook 2005; Lazzaro 2005). Furthermore, immunity genes are known to show unusually rapid copy number evolution (Sackton et al. 2007), including changes in copy number due to duplication, deletion, and pseudogenization. As acquisition of stop codons can lead to pseudogenization, we may be witnessing the early stages of copy number evolution.
GO Analysis Shows SCPs Are Enriched for Chemoreceptors
We can also infer levels of functional constraint using the functional annotation of a gene. Loss-of-function alleles in genes with dispensable functions are less likely to be strongly selected against than similar alleles in essential genes. We used the GO annotation tool DAVID (see Materials and Methods) to determine whether genes with SCPs were enriched for specific functions. We found that genes with PTCs and SCLs are equally likely to be associated with at least one GO category as other genes (all genes 63.5%, PTCs 66.7%, SCLs 67.2%, chi-square tests, P > 0.05), indicating that genes with SCPs are not strongly biased toward unannotated genes. We found that PTCs were enriched for GO terms associated with proteolytic activity and that both PTCs and SCLs were enriched for GO terms associated with the sensation of chemical stimuli and the plasma membrane (table 2). However, nonsingleton PTCs and SCLs did not show enrichment for these chemoreceptory GO terms. For both PTCs and SCLs, the enrichment of chemical sensation and plasma membrane GO terms appears to be driven by the fact that many gustatory receptors (GRs), odorant receptors (ORs), and other chemoreceptors (IRs) harbor SCPs. Most chemoreceptors are dispensable (i.e., null mutations do not cause lethality or sterility), and both GRs and ORs are known to evolve rapidly between species (Matsuo et al. 2007; McBride 2007; McBride et al. 2007; Dworkin and Jones 2009). Therefore, it is unsurprising we find SCP alleles present in these genes in D. melanogaster.
Table 2.
Enriched GO Terms among Genes with SCPs
| Allele Type | GO Term | Description | Number of Genes | P value |
| PTC | GO:0006508 | Proteolysis | 37 | 3.7 × 10−06 |
| PTC | GO:0007606 | Sensory perception of chemical stimulus | 17 | 6.1 × 10−06 |
| PTC | GO:0008233 | Peptidase activity | 37 | 4.1 × 10−05 |
| PTC | GO:0070011 | L-amino acid peptidase activity | 35 | 6.9 × 10−05 |
| PTC | GO:0044421 | Extracellular region component | 14 | 8.0 × 10−05 |
| PTC | GO:0050909 | Sensory perception of taste | 9 | 8.9 × 10−05 |
| PTC | GO:0005576 | Extracellular region | 28 | 9.9 × 10−05 |
| PTC | GO:0008527 | Taste receptor activity | 9 | 1.2 × 10−04 |
| PTC | GO:0007600 | Sensory perception | 18 | 1.4 × 10−04 |
| PTC | GO:0007186 | G-protein–coupled receptor signaling | 19 | 2.1 × 10−04 |
| SCL | GO:0007186 | G-protein–coupled receptor signaling | 8 | 2.6 × 10−03 |
| SCL | GO:0005615 | Extracellular space | 4 | 1.5 × 10−02 |
| SCL | GO:0050890 | Cognition | 7 | 1.8 × 10−02 |
| SCL | GO:0007600 | Sensory perception | 6 | 2.5 × 10−02 |
| SCL | GO:0033043 | Regulation of organelle organization | 4 | 2.8 × 10−02 |
| SCL | GO:0004965 | Gamma-aminobutyric acid–B receptor activity | 2 | 3.2 × 10−02 |
| SCL | GO:0007166 | Cell surface receptor signal transduction | 9 | 3.3 × 10−02 |
| SCL | GO:0016021 | Integral to membrane | 15 | 3.4 × 10−02 |
| SCL | GO:0051493 | Regulation of cytoskeleton organization | 3 | 3.6 × 10−02 |
| SCL | GO:0031224 | Intrinsic to membrane | 15 | 3.9 × 10−02 |
Genes with Unusual Evolutionary Histories
The pattern of variation observed among SCPs is consistent with our expectations if SCPs are as a group selected against. However, we were also interested in investigating genes that may not be following this overall pattern. First, we noted that 56 genes harbored more than two SCPs in D. melanogaster (supplementary table 2, Supplementary Material online, “Multiple alleles”). These genes may be evolving under weak selective constraint but could also be selected for multiple variants (diversifying or balancing selection). Named genes in this group included Acp26Aa, which is one of the most rapidly evolving genes in the D. melanogaster genome (Schully and Hellberg 2006; Wong et al. 2006), att-ORFB, two GRs (Gr59f and Gr36a), and one predicted chemosensory protein (CheA86a). Acps were observed to undergo rapid loss-and-gain in the melanogaster species subgroup (Begun et al. 2006), and length variations of Acp26Aa in this species subgroup have been described (Aguade 1998). Several loss-of-function and PTC alleles of other Acps were also documented in a survey of natural variation (Begun and Lindfors 2005). Consistent with this, we observed that Acp26Aa harbors one SCL allele that expands the open reading frame by one codon and one PTC allele that shortens it by seven codons, along with the major allele that matches the D. melanogaster reference annotation. It is possible that rapid diversifying selection of Acp26Aa includes the acquisition of SCPs among other types of polymorphism. The observation of att-ORFB, one of the several transcripts from a bicistronic messenger RNA (mRNA) expressed in adult testes (Madigan et al. 1996), is unsurprising given that many genes related to male reproduction are rapidly evolving in Drosophila (Zhang et al. 2004; Schully and Hellberg 2006; Richards et al. 2005). Similarly, chemoreceptors are also known to rapidly evolve between species (Matsuo et al. 2007; McBride 2007; McBride et al. 2007; Dworkin and Jones 2009). Genes that carry many SCPs warrant further study due to the possibility that diversifying selection may drive these genes to carry many alleles. On the other hand, not all of these genes have positive evidence for protein-coding ability, raising the possibility that their open reading frames are less constrained because they are mRNA-like noncoding RNA genes that are misannotated as protein-coding genes (Rymarquis et al. 2008). Such genes would be expected to tolerate SCPs because they are not translated.
Another interesting group is nine genes (one PTC and eight SCLs) whose major alleles are derived relative to the ancestral state, possibly resulting from recent increases in allele frequencies (supplementary table 2, Supplementary Material online, Ancestor minor). However, only small protein length differences were generated by these SCP alleles. Most of these are unnamed genes and none have known functions. Two interesting cases are CG15531, a predicted stearoyl-CoA 9-desaturase, and att-ORFB, a testis-expressed gene that also harbors multiple SCP alleles (see above).
We noted that SCPs are enriched with alleles causing small as well as large protein length changes (see above). Among the PTC alleles causing extreme changes (truncation of more than half of the CDS), ten alleles have population frequency above 25%, and three named genes (gfA, Flo-2, and dpr2) are among this list. However, PTCs in these named genes influenced only a few isoforms out of the many isoforms of the genes, suggesting their influence on D. melanogaster fitness may be less severe than predicted by change of coding region alone. All the SCL alleles with extreme number of codons added or predicted under nonstop decay have low population frequency.
Finally, we observed 13 D. melanogaster SCP alleles that are also segregating in D. simulans and 4 of them (PTCs) are in named genes (Sucb, dpr2, Vha100-1, and Fak56D). Although large truncations of protein sequences (from 16% to 97% of CDS) were caused by PTCs in these named genes, only one isoform was affected. These results are generally consistent with our finding that purifying selection is removing mutations in essential genes or essential parts of genes and mutations causing extreme changes in protein length. These alleles usually affect only unnamed genes or a few isoforms of named genes. Thus, the unusual alleles we found may be explained by the overall pattern we have observed—SCPs are as a group selected against and affect weakly constrained genes. However, we also found an enrichment of genes previously known to be rapidly evolving within Drosophila or to harbor nonsense alleles (e.g., chemoreceptors and male-specific genes), indicating that SCPs might be important to the evolution of these genes.
SCPs Lead to the Loss and Gain of Protein Regions
Previous studies have shown that domains of proteins can be lost and gained through evolutionary time and that these mutations are biased toward the 5′ and 3′ ends of proteins (Bjorklund et al. 2005; Weiner et al. 2006). Although we observed that there is a bias toward SCPs causing small changes in protein length, we wanted to know whether protein sequence features might be added or lost in the SCP alleles. We used the annotation tool InterProScan (Quevillon et al. 2005) to determine if truncated parts of PTC alleles that are not targeted by nonsense-mediated decay or expanded parts of SCP alleles that are not targeted by nonstop decay contained any known sequence features or domains.
We found that 23 of 71 alleles causing truncations that are expected to escape nonsense-mediated decay had lost characterized sequence features including signal peptides, protein-binding domains, DNA-binding domains, and catalytic domains. However, one caveat is that the exact trigger for nonsense-mediated decay is not well understood on a genome-wide scale (Behm-Ansmant et al. 2007; Hansen et al. 2009). Exactly which PTC alleles will lead to domain loss and which will lead to silencing will vary depending on how much the mechanism of nonsense-mediated decay differs between genes, which has not yet been established in Drosophila.
Among SCLs expected to avoid nonstop decay (the same set as used for gene expansion analysis above), we found one gene with an SCL allele resulting in the acquisition of an apparently novel sequence features. The SCL allele of muscleblind, which codes for a zinc-finger protein with roles in apoptosis, muscle development (Begemann et al. 1997), and sexual behavior (Juni and Yamamoto 2009), acquired a 20 amino acid signal peptide. This allele has a population frequency of 0.19, which is among the highest frequency SCLs. The idea that a protein might expand into its 3′ untranslated region and acquire a novel peptide is intriguing and certainly warrants further functional study.
Conclusion
Natural mutations causing null alleles of genes have long been of interest to geneticists. Many of the first disease causing alleles characterized in humans carried PTCs (Chang and Kan 1979; Rosenfeld et al. 1992), and null alleles of allozymes in Drosophila were some of the earliest natural variants to be characterized (Voelker et al. 1980; Langley et al. 1981; Burkhart et al. 1984). Until recently, it has been unclear how common null alleles caused by variation in the position of stop codons are, as study has been restricted primarily to alleles defined by lack of function. Furthermore, we do not understand how stop codon variants first arise within populations, leading to changes in gene models over evolutionary time.
Here, we used newly available D. melanogaster genomes from North American and African populations and performed a genome-wide survey for alleles causing changes in the position of the stop codon. We found several hundred such polymorphisms segregating in the D. melanogaster genome, and these alleles are a mixture of deleterious and slightly deleterious mutations. SCPs had more extreme allele frequency spectra than other types of polymorphisms, were enriched for small changes in protein length, and were found less often on the X chromosome, indicating purifying selection is acting to reduce the frequency of such polymorphisms. An appreciable number of SCPs in more than one genome were also observed, suggesting some of the observed SCPs are subject to both the effects of selection and genetic drift. We also found evidence that both mutational pressure and selective constraint are important in determining the likelihood a gene harbors SCPs. We described several exceptional SCPs, which include alleles that are shared between D. melanogaster and D. simulans, alleles with high population frequency despite causing dramatically altered protein lengths, and alleles that arose and quickly became the major allele in D. melanogaster. Additionally, there are 56 genes that carry more than two alleles with different stop codon positions in D. melanogaster. These include rapidly evolving genes such chemoreceptors and male-expressed genes. Finally, one SCL gene, muscleblind, appears to have gained 3′ sequence with similarity to a signal peptide. This implies the possibility for genes to gain domains as well as lose them.
Parallel resequencing projects have uncovered SCPs in humans (Yamaguchi-Kabata et al. 2008; Yngvadottir et al. 2009; Durbin et al. 2010), providing an opportunity to contrast findings across species. The human and Drosophila data differ in some important ways—human genomes were sequenced in a heterozygous state, whereas the DPGP project sequenced homozygous flies. It is therefore expected that the human data would contain more alleles—especially deleterious alleles—than does the fly data. Indeed, the reported density of PTCs and number of observed PTCs per genome in humans is much higher than in Drosophila (PTC density: 0.00021 [fly] vs. 0.00085 [human]; Yamaguchi-Kabata et al. 2008; PTC per individual: 37.9 [fly] vs. 80–100 [human]; Durbin et al. 2010). Furthermore, it was reported that PTCs are distributed evenly across the coding regions in humans (Yngvadottir et al. 2009), which are in contrast to our observation that PTC alleles are enriched for those causing small changes. These differences may also be explained by the much smaller effective population size of human compared with Drosophila (Charlesworth 2009), which results in less effective selection. Furthermore, 59% of human nonsense alleles were found to be present in the homozygous state in some individuals (Yngvadottir et al. 2009). If this frequency was similar in Drosophila, a sampling of alleles in the heterozygous state should uncover many more SCPs than we were able to find in this study. Yet, we must be cautious when comparing these data sets because there may be different (and unknown) biases resulting from the fundamental differences in sequencing technology and SNPs-calling methods between the human and Drosophila data. Finally, while both the Drosophila and human data showed that selection is acting to reduce population frequency of nonsense SNPs as a whole, some SCPs violating this pattern were identified. Yngvadottir et al. (2009) reported MAGEE2, which appeared to have increased in frequency in Asian human populations despite causing a 77% truncation of the open reading frame. Likewise, we identified several SCPs that have increased in frequency (Ancestor minor alleles) and several genes that carry many SCPs. Intriguingly, GO enrichment analysis in both species found chemosensory receptors are enriched with nonsense SNPs, consistent with the idea that dispensable, rapidly evolving genes are more likely to harbor strong-effect mutations.
In sum, our study provides the first comprehensive description of the variation in stop codon position in Drosophila, and we show that polymorphisms changing the position of the stop codon were as a group selected against. However, a number of genes that broke this pattern in various ways were identified and warrant further analysis. Because the study system was Drosophila, this analysis also provides a list of D. melanogaster and D. simulans stocks harboring a variety of natural nonsense polymorphisms (supplementary table 2, Supplementary Material online), which can be readily applied to studies of the functional consequences of these natural variants.
Supplementary Material
Supplementary tables 1 and 2 and figure 1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank DPGP for providing access to the Drosophila melanogaster and Drosophila simulans genome sequences and multispecies genomic alignment. We thank FlyAtlas for fly expression data. Thanks to D. J. Begun, C. D. Jones, C. H. Langley, A. Kopp, E. J. Earley, and N. D. White for ideas, helpful discussion, and critical reading of the manuscript. We are grateful to P. L. Ralph for providing statistical help, P. Saelao for experimental assistance and fly stocks, and C. M. Cardeno for providing DNA samples. We also thank E. Betran and three anonymous reviewers for constructive comments that greatly improved this manuscript. This work was partially supported by the Center for Population Biology of UC Davis and NSF Grant MCB 0920196. Authors’ contributions: J.A.R. conceived the study, performed DNA sequencing, carried out evolutionary genetic analyses, and wrote the manuscript. Y.C.G.L. carried out SCP detection, performed evolutionary genetic and statistical analyses, and wrote the manuscript.
References
- Abdi H. Bonferroni and Sidak corrections for multiple comparisons. In: Salkind N, editor. Encyclopedia of measurement and statistics. Thousand Oaks (CA): Sage; 2007. pp. 103–107. [Google Scholar]
- Aguade M. Different forces drive the evolution of the Acep26Aa and Acep 26Ab accessory gland genes in the Drosophila melanogaster species complex. Genetics. 1998;150(3):1079–1089. doi: 10.1093/genetics/150.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, et al. Muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development. 1997;124:4321–4331. doi: 10.1242/dev.124.21.4321. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature. 1993;365:548–550. doi: 10.1038/365548a0. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA. Rapid evolution of genomic Acp complement in the melanogaster subgroup of Drosophila. Mol Biol Evol. 2005;22:2010–2021. doi: 10.1093/molbev/msi201. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA, Thompson ME, Alisha KH. Recently evolved genes identified from Drosophila yakuba and D. erecta accessory gland expressed sequences. Genetics. 2006;172(3):1675–1681. doi: 10.1534/genetics.105.050336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund AK, Ekman D, Light S, Frey-Skott J, Elofsson A. Domain rearrangements in protein evolution. J Mol Biol. 2005;353:911–923. doi: 10.1016/j.jmb.2005.08.067. [DOI] [PubMed] [Google Scholar]
- Burkhart BD, Montgomery E, Langley CH, Voelker RA. Characterization of allozyme null and low activity alleles from two natural populations of Drosophila melanogaster. Genetics. 1984;107(2):295–306. doi: 10.1093/genetics/107.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, Kan YW. beta 0 thalassemia, a nonsense mutation in man. Proc Natl Acad Sci U S A. 1979;76:2886–2889. doi: 10.1073/pnas.76.6.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. Elements of evolutionary genetics. 2010 Greenwood Village (CO): Roberts and Company. [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dubinin N, Romashov DD, Heptner MA, Demidova ZA. Aberrant polymorphism in Drosophila fasciata Meig. Biol Zh. 1937;6:311–354. [Google Scholar]
- Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin I, Jones CD. Genetic changes accompanying the evolution of host specialization in Drosophila sechellia. Genetics. 2009;181:721–736. doi: 10.1534/genetics.108.093419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Thronton KR, Charlesworth B, Andolfatto P. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 2005;15:790–799. doi: 10.1101/gr.3541005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, et al. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hutter S, Haipeng L, Beisswanger S, De Lorenzo D, Stephan W. Distinctly different sex ratios in African and European populations of Drosophila melanogaster inferred from chromosome wide single nucleotide polymorphism data. Genetics. 2007;177(1):469–480. doi: 10.1534/genetics.107.074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives PT. The genetic structure of American populations of Drosophila melanogaster. Genetics. 1945;30:167–196. doi: 10.1093/genetics/30.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, Kang-Wook K. The evolution of antifungal peptides in Drosophila. Genetics. 2005;171(4):1847–1859. doi: 10.1534/genetics.105.045435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juni N, Yamamoto D. Genetic analysis of chaste, a new mutation of Drosophila melanogaster characterized by extremely low female sexual receptivity. J Neurogenet. 2009;23:329–340. doi: 10.1080/01677060802471601. [DOI] [PubMed] [Google Scholar]
- Keightley PD, et al. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;19(7):1195–1201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Markov TA. Duplication selection and gene conversion in a Drosophila mojavensis female reproductive protein family. Genetics. 2009;181:1451–1465. doi: 10.1534/genetics.108.099044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature. 1983;304:412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- Langley CH, et al. Null allele frequencies and allozyme loci in natural populations of Drosophila melanogaster. Genetics. 1981;99(1):151–156. doi: 10.1093/genetics/99.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, et al. Genomic variation in natural populations of Drosophila melanogaster. Genetics. Forthcoming 2012 doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, et al. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP. Elevated polymorphism and divergence in the class C scavenger receptors of Drosophila melanogaster and D. simulans. Genetics. 2005;169:2023–2034. doi: 10.1534/genetics.104.034249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol. 2005;22:1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- Madigan SJ, Edeen P, Esnayra J, McKeown M. att, a target for regulation by tra2 in the testes of Drosophila melanogaster, encodes alternative RNAs and alternative proteins. Mol Cell Biol. 1996;16(8):4222–4230. doi: 10.1128/mcb.16.8.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G, Domazet-Loso T, Tautz D, Charlesworth B. Correlated evolution of synonymous and nonsynonymous sites in Drosophila. J Mol Evol. 2004;59:771–779. doi: 10.1007/s00239-004-2671-2. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 2007;5:985–996. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc Natl Acad Sci U S A. 2007;104:4996–5001. doi: 10.1073/pnas.0608424104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS, Arguello JR, O'Meara BC. Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics. 2007;177:1395–1416. doi: 10.1534/genetics.107.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Peden J. Analysis of codon usage [PhD thesis] [Nottingham (UK)]: University of Nottingham; 1999. [Google Scholar]
- Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci U S A. 2007;104(Suppl 1):8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retelska D, Iseli C, Bucher P, Jongeneel CV, Naef F. Similarities and differences of polyadenylation signals in human and fly. BMC Genomics. 2006;7:176. doi: 10.1186/1471-2164-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld PJ, et al. A null mutation in the rhodopsin gene causes rod photoreceptor dysfunction and autosomal recessive retinitis pigmentosa. Nat Genet. 1992;1:209–213. doi: 10.1038/ng0692-209. [DOI] [PubMed] [Google Scholar]
- Rymarquis LA, Kastenmayer JP, Huttenhofer AG, Green PJ. Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 2008;13:329–334. doi: 10.1016/j.tplants.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Sackton TB, et al. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Sandler L, Lindsley DL, Nicoletti B, Trippa G. Mutants affecting meiosis in natural populations of Drosophila melanogaster. Genetics. 1968;60:525–558. doi: 10.1093/genetics/60.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schully SD, Hellberg ME. Positive selection on nucleotide substitutions and indels in accessory gland proteins of the Drosophila pseudoobscura subgroup. J Mol Evol. 2006;62:793–802. doi: 10.1007/s00239-005-0239-4. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- Simmons MJ, Crow JF. Mutations affecting fitness in Drosophila populations. Annu Rev Genet. 1977;11:49–78. doi: 10.1146/annurev.ge.11.120177.000405. [DOI] [PubMed] [Google Scholar]
- Spencer WP. Mutations in wild populations in Drosophila. Adv Genet. 1947;1:359–402. doi: 10.1016/s0065-2660(08)60489-7. [DOI] [PubMed] [Google Scholar]
- Stephan W, Li H. The recent demographic and adaptive history of Drosophila melanogaster. Heredity. 2007;98:65–68. doi: 10.1038/sj.hdy.6800901. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Timofeev-Ressovsky H, Timofeev-Ressovsky NW. Genetische analyse einer freilebenden Drosophila melanogaster population. Arch Entw Mech Org. 1927;109:70–109. doi: 10.1007/BF02079096. [DOI] [PubMed] [Google Scholar]
- Timofeev-Ressovsky N. Das Genovariieren in verschiedenen Richtungen bei Drosophila melanogaster unter dem Einfluss der Rontgenbestrahlung. Naturwiss. 1930;18:434–437. [Google Scholar]
- Tweedie S, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Peltz SW, Wilusz CJ. Non-stop decay–a new mRNA surveillance pathway. Bioessays. 2002;24:785–788. doi: 10.1002/bies.10153. [DOI] [PubMed] [Google Scholar]
- Voelker RA, Schaffer HE, Mukai T. Spontaneous allozyme mutation in Drosophila melanogaster: rate of occurrence and nature of the mutants. Genetics. 1980;94(4):961–968. doi: 10.1093/genetics/94.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Weiner J, 3rd, Beaussart F, Bornberg-Bauer E. Domain deletions and substitutions in the modular protein evolution. FEBS J. 2006;273:2037–2047. doi: 10.1111/j.1742-4658.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- Wong A, Albright SN, Wolfner MF. Evidence for structural constraint on ovulin, a rapidly evolving Drosophila melanogaster seminal protein. Proc Natl Acad Sci U S A. 2006;103:18644–18649. doi: 10.1073/pnas.0601849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Kabata Y, et al. Distribution and effects of nonsense polymorphisms in human genes. PLoS One. 2008;3:e3393. doi: 10.1371/journal.pone.0003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M, et al. Large-scale trends in the evolution of gene structures within 11 animal genomes. PLoS Comput Biol. 2006;2:e15. doi: 10.1371/journal.pcbi.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yngvadottir B, et al. A genome-wide survey of the prevalence and evolutionary forces acting on human nonsense SNPs. Am J Hum Genet. 2009;84:224–234. doi: 10.1016/j.ajhg.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hambuch TM, Parsch J. Molecular evolution of sex-biased genes in Drosophila. Mol Biol Evol. 2004;21(11):2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]