Abstract
Core histone proteins are essential for packaging the genomic DNA into chromatin in all eukaryotes. Since multiple genes encode these histone proteins, there is potential for generating more histones than what is required for chromatin assembly. The positively charged histones have a very high affinity for negatively charged molecules such as DNA, and any excess of histone proteins results in deleterious effects on genomic stability and cell viability. Hence, histone levels are known to be tightly regulated via transcriptional, posttranscriptional and posttranslational mechanisms. We have previously elucidated the posttranslational regulation of histone protein levels by the ubiquitin-proteasome pathway involving the E2 ubiquitin conjugating enzymes Ubc4/5 and the HECT (Homologous to E6-AP C-Terminus) domain containing E3 ligase Tom1 in the budding yeast. Here we report the identification of four additional E3 ligases containing the RING (Really Interesting New Gene) finger domains that are involved in the ubiquitylation and subsequent degradation of excess histones in yeast. These E3 ligases are Pep5, Snt2 as well as two previously uncharacterized Open Reading Frames (ORFs) YKR017C and YDR266C that we have named Hel1 and Hel2 (for Histone E3 Ligases) respectively. Mutants lacking these E3 ligases are sensitive to histone overexpression as they fail to degrade excess histones and accumulate high levels of endogenous histones on histone chaperones. Co-immunoprecipitation assays showed that these E3 ligases interact with the major E2 enzyme Ubc4 that is involved in the degradation related ubiquitylation of histones. Using mutagenesis we further demonstrate that the RING domains of Hel1, Hel2 and Snt2 are required for histone regulation. Lastly, mutants corresponding to Hel1, Hel2 and Pep5 are sensitive to replication inhibitors. Overall, our results highlight the importance of posttranslational histone regulatory mechanisms that employ multiple E3 ubiquitin ligases to ensure excess histone degradation and thus contribute to the maintenance of genomic stability.
Introduction
Histones are essential basic proteins that package the genomic DNA of all eukaryotes into nucleosomes to form chromatin [1], [2], [3]. In doing so, histones also regulate access to the genetic information contained within the DNA and thereby affect all aspects of DNA metabolism. Most histones are encoded by multiple genes in all eukaryotes [4] that have the potential for generating histones in excess of the requirement for chromatin assembly [5]. Any excess of the positively charged histones can allow them to potentially associate non-specifically with negatively charged molecules such as DNA in the cell, resulting in deleterious effects on cell viability and genomic stability [6], [7], [8]. As such, in organisms such as the Xenopus that undergo rapid embryonic cell cycles in the absence of transcription, large quantities of histones are stored bound to chaperone proteins such as Nucleoplasmin in their oocytes to provide adequate quantities of histones for chromatin assembly during DNA synthesis [9], [10]. A related strategy has also been reported in Drosophila and other fly embryos wherein histones appear to be stored in lipid droplets [11]. In the budding yeast and mammalian cells, histone chaperones such as Asf1 (Anti-Silencing Function 1), CAF-1 (Chromatin Assembly Factor-1), Spt16 (Suppressor of Ty 16) and Nap1 (Nucleosome Assembly Protein 1) can bind to and buffer against only a limited excess of histone proteins [6], [12], [13], [14], [15], [16]. Therefore, to avoid the problems associated with excess histone accumulation, most eukaryotic cells rely largely on the strict regulation of their histone protein levels via transcriptional, posttranscriptional, translational and posttranslational mechanisms [5], [17].
Proteins are the structural and functional workhorses of all cells. In order to respond effectively to constantly changing environmental cues, protein levels must be regulated by eliminating the pools that have either been damaged or are simply no longer required for normal cell function. Therefore, cells have evolved regulated proteolysis to fine-tune their metabolic processes [18], [19]. This regulated proteolysis involves a fairly well defined choreography known as the ubiquitin-proteasome pathway. Ubiquitin is a short 76-amino acid peptide that is universally conserved among eukaryotes (hence the name ubiquitin). Ubiquitin is attached to substrate proteins to mark them for destruction or transduce other signals in the cell. Several ubiquitin molecules (at least four) need to be covalently attached to the same lysine residue in the target protein for it to be recognized and degraded by the multifunctional protease known as the 26S proteasome [20]. The conjugation of ubiquitin to proteins requires the sequential action of three enzymes. The ubiquitin-activating enzyme (Enzyme 1 or simply E1; Uba1 in the budding yeast) hydrolyzes ATP to convert ubiquitin into an activated form, which is covalently linked at its carboxyl terminus to a cysteine (Cys) residue of the E1 enzyme via a high-energy thiol-ester linkage [21]. The activated ubiquitin molecule is then transferred to the second enzyme of this pathway, an ubiquitin conjugating enzyme (Ubc or E2; Ubc1–Ubc13 in the budding yeast), and the activated form is maintained through the formation of a thiol-ester linkage with a Cys residue in the E2 enzyme. The third enzyme in the process, the ubiquitin ligase (E3; approximately 80 E3 ligases in yeast), supports the transfer of ubiquitin to substrates and generally provides the specificity [20], [22], [23]. E3 ubiquitin ligases have been broadly classified into three main categories, namely RING (Really Interesting New Gene) finger domain containing E3 ligases [24], the U-box domain containing E3 ligases that carry a modified RING domain [25] and HECT (homologous to E6-AP C-terminus) domain containing E3 ligases [26]. The RING domain is a Cys rich sequence motif that can bind two zinc atoms and the majority of the E3 ligases belong to this class [24]. The consensus RING motif has been defined as a unique pattern of Cys and histidine (His) residues at defined positions in a peptide sequence that is Cys-X2-Cys-X9–39-Cys-X1–3-His-X2–3-Cys-X2-Cys-X4–48-Cys-X2-Cys (and is often abbreviated to Cys3-His-Cys4), where X can be any amino acid. The RING domain has been shown to facilitate the interaction between the E2 and the substrate being ubiquitylated without ever being covalently attached to ubiquitin. U-box E3 ligases contain a U-box motif which is similar to RING motif except that it lacks the canonical Cys residues for zinc coordination [22], [25]. The consensus 75-amino acid U-box domain is poorly conserved and is characterized by the same sequence of -helices, ß-sheets and unstructured loops found in RING fingers. The HECT E3 ligases form a relatively small family of conserved E3 ligases that are present from yeast to humans [26], [27]. These ligases are characterized by the HECT domain, a C-terminal region of approximately 350 amino acids with significant similarity to the C-terminus of E6-AP. Unlike RING domain and U-box E3 ligases, the HECT E3 ligases interact with the cognate E2 ubiquitin conjugating enzymes leading to covalent attachment of the ubiquitin moiety by a thiol-ester bond to a conserved Cys residue in HECT domain. Subsequently this ubiquitin is transferred directly from the E3 ligase to the substrate protein. Elongation of the initial ubiquitin into polyubiquitin chains is carried out by the concerted actions of E2 and E3 enzymes. Even though some E2s might provide the chain-type specificity, most of the substrate specificity is achieved by the E3 ligases. Not surprisingly, cells have a large number of proteins in their proteome (1–2% of the total number of proteins) that are putative E3 ligases [23]. The importance of E3 ligases is underscored by the fact that a number of human diseases including cancer, neurodegenerative processes, metabolic disorders, familial Parkinson's disease and Angelman's syndrome have been found to have defects in this proteolytic pathway [28], [29], [30], [31]. Further, some of the most important cell cycle regulators are under very tight regulation and this is achieved by employing multiple E3s for this task. The most studied protein to fall in this class is the tumor suppressor p53, which is known to be ubiquitylated by at least 11 different E3 ligases (Mdm2, Pirh2, Cop1, TOPORS, Synoviolin, CHIP, UBC13, CARP1, CARP2, p300/CBP and ARFBP1) [32]. Even though these E3 ligases have other targets, they all contribute individually in regulating p53 levels, which is vital for proper cell cycle progression.
We have been engaged for the past decade in trying to understand the posttranslational regulation of histone protein levels using the budding yeast Saccharomyces cerevisiae as a model system. Histones are generally considered to be extremely stable proteins with half-lives in the order of several months in mammalian cells [33]. However, these half-lives are likely to mainly reflect the contribution of chromatin bound histones, as we have previously shown that non-chromatin bound histones are rapidly degraded with a half-life of 30–40 minutes in yeast cells [6], [7]. We went on to demonstrate that the phosphorylation, ubiquitylation and subsequent degradation of excess histones in yeast occurs via the ubiquitin-proteasome pathway. We identified the DNA damage checkpoint kinase Rad53 as the master regulator of this pathway which also employs the E2 ubiquitin conjugating enzyme Ubc4/5 and the HECT domain E3 ligase Tom1 for the degradation related polyubiquitylation of excess histones. Interestingly HUWE1 (HECT, UBA and WWE domain containing 1), the human homolog of Tom1, has been reported to ubiquitylate all four core histones in vitro [34] as well as control the levels or activities of important cellular regulators such as Cdc6 [35], Mcl-1 [36], Myc [37] and p53 [38]. tom1 deletion strains are defective in efficient degradation of ectopically expressed histone H3 [7]. Subsequent experiments carried out by us strongly suggested that additional E3 ligases maybe involved in the degradation of excess histones in budding yeast. Here we report the identification and characterization of four additional histone E3 ligases, namely Pep5, Snt2 and two novel E3 ligases that correspond to previously uncharacterized ORFs. These ORFs (YKR017c on chromosome XI and YDR266c on chromosome IV) encode RING finger domain containing proteins. Based on their involvement in ubiquitylating histones, we propose the name HEL1 and HEL2 respectively (for Histone E3 Ligase) for these two ORFs. Using a combination of genetic and biochemical assays we show that these genes encode genuine ubiquitin E3 ligases and histones serve as at least one of their major substrates. As such, these E3 ligases play an important role in regulating histone protein levels and are likely to contribute to the maintenance of genomic stability in the budding yeast.
Results
Identification of four RING finger containing predicted E3 ligases that are sensitive to histone overexpression
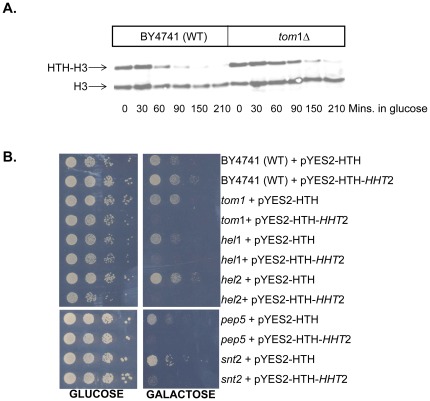
Although we had previously demonstrated that tom1 mutants were defective in the degradation of exogenous histones [7], when followed for longer time points, the tom1 mutants appeared to degrade the exogenously expressed histones, albeit with much slower kinetics compared to the wild type cells (Figure 1A). Additionally, the tom1 mutant was not as sensitive to genotoxic agents as the ubc4 ubc5 double deletion strain that lacks both the E2 enzymes responsible for the degradation related ubiquitylation of histones in yeast [7]. These data strongly suggested that additional E3 ligases may be involved in the degradation of excess histones in budding yeast. The yeast proteome has approximately 80 E3 ligases [23] and we used a simple screen based on the sensitivity of yeast mutants defective in the histone degradation pathway to histone overexpression as described previously [7] to identify additional histone E3 ligases. For this, we acquired all the non-essential deletions strains corresponding to the predicted E3 ligases from the yeast genome deletion collection (Open Biosystems). Additionally, we acquired temperature sensitive mutants for a few of the essential E3 ligases as well. All the mutant strains along with the isogenic wild type strain were transformed with either the empty vector (pYES2-HTH) or a galactose inducible HIS10-TEV-HA tagged histone H3 overexpression construct (pYES2-HTH-HHT2) described previously [7]. The transformants were grown overnight on a minimal liquid media with raffinose as the sole source of carbon, but without uracil to maintain selection for the plasmid. Then 10-fold serial dilutions corresponding to each strain were plated on glucose or galactose containing plates lacking uracil and incubated at 30°C (semi-permissive temperature for the temperature sensitive mutants) for 2–3 days prior to being photographed. The growth of each strain was compared between galactose (in the presence of histone overexpression) and glucose plates (no histone overexpression). In addition to the tom1 deletion strain that served as a positive control in our screen, four putative E3 deletion strains were sensitive to histone overexpression to varying degrees (Figure 1B). Two of the strains corresponded to deletions of previously characterized genes PEP5 and SNT2, while the remaining two strains carried deletions of previously uncharacterized ORFs and we named them hel1 and hel2 for Histone E3 Ligase. Since histone overexpression causes cytotoxicity via multiple mechanisms in yeast cells [8] we needed to further characterize the four genes identified in our initial screen to determine if they encoded bona fide histone E3 ligases.
Figure 1. Identification of strains lacking specific E3 ligases that confer sensitivity to histone overexpression.
(A) The tom1 deletion strain can degrade exogenous histones with much slower kinetics than the wild type strain. Histone degradation assay was carried out exactly as described previously [7] whereby a galactose inducible, HIS10-TEV-HA (HTH) tagged histone H3 was expressed in G1 arrested cells for 90 minutes, following which cells were switched to glucose media and harvested at the indicated time points. Whole cell extracts were prepared and resolved on 18% polyacrylamide gels and subjected to Western blotting with the H3-C antibody described previously [6] that recognizes both the endogenous chromatin associated histone H3 as well as the overexpressed H3. The entire experiment was carried out in G1 arrested cells to prevent incorporation of exogenous histones into chromatin that occurs in replicating cells. (B.) Strains lacking Hel1, Hel2, Pep5 and Snt2 E3 ligases are sensitive to histone overexpression. The histone overexpression sensitivity screen was carried out exactly as described previously [7]. Briefly, wild type (WT) BY4741 and deletion strains corresponding to the indicated E3 ligases were transformed with either an empty vector (pYES2-HTH) or a galactose inducible HTH tagged histone H3 expressing plasmid (pYES2-HTT-HHT2). Then 10-fold serial dilutions of the resulting transformants were plated on glucose (no histone overexpression) or galactose (allows histone overexpression) containing media without uracil to select for the plasmid. Plates were photographed after 3 days of incubation at 30°C.
hel1, hel2, pep5 and snt2 mutants are defective in histone degradation
If hel1, hel2, pep5 and snt2 mutants were sensitive to histone overexpression due to a defect in histone degradation, then this defect could be detected using our histone degradation assays described previously [6], [7]. For this assay, G1 arrested cells were induced with galactose for 90 minutes to express HA tagged histone H3, after which cells were switched to glucose media to stop the synthesis of the tagged H3. Samples were harvested at 30-minute or indicated intervals and processed for Western blotting using the H3-C antibody as described previously [6], [7], except that the signals were detected using fluorescently labeled secondary antibodies on a Li-COR Odyssey imager. Compared to the wild type BY4741 strain, the hel1, hel2, pep5 and snt2 deletion strains were clearly defective in degrading ectopically expressed histone H3 to varying degrees, with pep5 being the most defective and nearly indistinguishable from rad53 mutants (Figure 2A). The levels of endogenous chromatin bound histone H3 do not change during the course of the experiment and serve as an excellent internal loading control. Next, we investigated if the RING domains of the putative histone E3 ligases were required for the degradation of excess histones. For this we used site-directed mutagenesis to replace the critical Cys and His residues in the RING domains of Hel1, Hel2 and Snt2 with Alanine (Ala) residues as described in Materials and Methods. These hel1-r1, hel1-r2, hel1-r1r2, hel2-r and snt2-r2 mutants carrying point mutations in their respective RING domains were as defective in excess histone degradation as the hel1, hel2 and snt2 deletion strains (Figure 2B), strongly suggesting that their RING domains are important for histone protein regulation.
Figure 2. Strains lacking putative E3 ligases Hel1, Hel2, Pep5 and Snt2 are defective in degrading excess histones.
(A) Strains carrying deletions of hel1, hel2, pep5 and snt2 are deficient in degrading exogenously expressed histones. The exogenous histone degradation assay was carried out in the indicated strains carrying the pYES2-HTH-HHT2 plasmid as described in Figure 1A. The rad53 deletion strain serves as a positive control in this assay as we have previously shown that it is defective in the degradation of exogenously expressed histones [6]. The endogenous chromatin bound histone H3 is not degraded in this assay and serves as a loading control. Glu = glucose. (B) The RING domains of Hel1, Hel2 and Snt2 are required for efficient degradation of excess histones. Strains carrying mutations in the RING (r) domains of Hel1, Hel2 and Snt2 were generated as described in the Materials and Methods section. These mutant strains were then transformed with the pYES6/CT-HA-HHT2 plasmid carrying a Blasticidin resistance marker and a galactose inducible, HA-tagged histone H3 gene (HA-H3). The excess histone degradation assay was carried out as described in (A) except that the duration of the experiment was limited to 90 minutes and the exogenous HA-H3 was detected using HA.11 antibodies, while the endogenous H3 was detected using the H3-C antibody described previously [6], [7].
hel1, hel2, pep5 and snt2 mutants accumulate excess histones on the histone chaperone Asf1
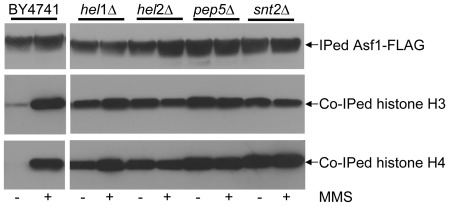
We have previously shown that yeast strains defective in the histone degradation pathway such as rad53, ubc4, ubc5 and tom1 accumulate excess endogenous histones on histone chaperones such as Asf1 which typically binds small amounts of histones under normal conditions but becomes saturated with histones upon replication arrest or DNA damage [6], [7]. If hel1, hel2, pep5 and snt2 are indeed defective in degrading endogenous histones, they should also exhibit histone chaperones overloaded with endogenous histones under normal conditions. Wild-type and E3-mutant strains carrying FLAG3 tagged Asf1 (Asf1-FLAG) were treated with or without methyl methane sulfonate (MMS) to induce alkylation damage, following which Asf1-FLAG was immunoprecipitated using FLAG-M2 affinity resin. As expected, in the absence of MMS, small amounts of H3 were associated with Asf1–FLAG in wild-type cells, which increased dramatically upon MMS treatment (Figure 3). In contrast, hel1, hel2, pep5 and snt2 deletion strains showed high levels of H3 associated with Asf1–FLAG both in the absence and presence of MMS-induced DNA damage. The near saturation of Asf1 with histones in each of these mutants even in the absence of DNA damage strongly suggests that the corresponding E3 ligases contribute independently of each other in the posttranslational regulation of endogenous histone protein levels.
Figure 3. hel1, hel2, pep5 and snt2 deletion strains accumulate excessive amounts of endogenous histones on the histone chaperone Asf1.
The chromosomal copy of the gene encoding Asf1 was tagged with a 3×FLAG epitope at the C-terminus in the indicated strains. Whole cell extracts were prepared from 0.5 L cell cultures treated with or without DNA damaging agent Methylmethane Sulfonate (MMS; 0.033%) as described previously [7]. Then, Asf1-FLAG was immunoprecipitated (IPed) using FLAG-M2 antibodies and the co-immunoprecipitated (Co-IPed) histones were analyzed by Western blotting using the H3-C and H4 polyclonal antibodies described previously [6].
Hel1, Hel2, Pep5 and Snt2 interact with Ubc4 and histones
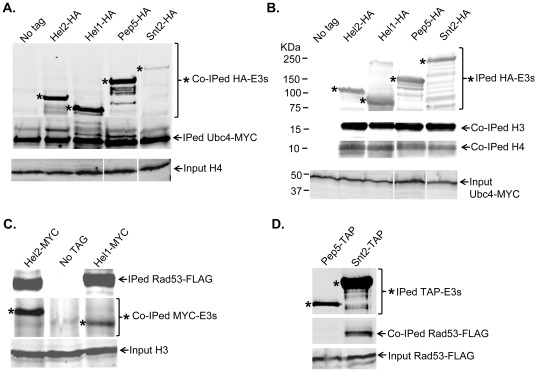
All the new histone E3 ligases identified in our screen are RING finger domain containing proteins. RING domains are known to directly interact with Ubc (E2) enzymes as well as the substrate, thereby helping the Ubc enzymes transfer the ubiquitin to the substrate proteins [24], [39]. Hence, if the putative RING finger histone E3 ligases identified by us are bona fide E3 ligases, they should interact physically with their substrate histones as well as the E2 enzymes involved in the degradation related ubiquitylation of histones. We tested this idea directly by checking if Hel1, Hel2, Pep5 and Snt2 interact with the major histone ubiquitin conjugating enzyme Ubc4 and histones. For this we transformed a strain expressing MYC13-tagged Ubc4 [7] with constructs expressing HIS6-HA-Protein A (HIS6-HA-PrA) tagged E3 ligases. Then we prepared whole cell extracts and immunoprecipitated (IPed) either Ubc4-MYC using anti-MYC EZ View beads (Sigma) (Figure 4A) or HA-tagged E3 ligases using magnetic beads conjugated to HA.11 antibodies (Figure 4B). All IPed material was resolved on precast gradient gels. Western blotting with MYC and HA antibodies clearly shows that all the four E3 ligases co-immunoprecipitated (co-IPed) with Ubc4-MYC (Figure 4A). Similar results were obtained using reciprocal immunoprecipitation experiments (data not shown). Further, both histones H3 and H4 co-IPed with all the four E3 ligases (Figure 4B). Overall, these interactions demonstrate that the new RING finger containing histone E3 ligases can potentially mediate the ubiquitin transfer from the E2 enzyme Ubc4 to the substrate histones by acting as a scaffold to bring these proteins together.
Figure 4. Hel1, Hel2, Pep5 and Snt2 interact with the E2 enzyme Ubc4 as well as histones.
(A) Hel1, Hel2, Pep5 and Snt2 interact with Ubc4. Whole cell extracts prepared from 0.5 liter cultures of the indicated strains were used to IP Ubc4-MYC essentially as described for Asf1-FLAG IPs in Figure 3, but using anti-MYC EZ view beads (Sigma). Co-IPed HA-tagged E3 enzymes were detected using HA.11 antibodies. Apart from the predominant bands corresponding to the full length proteins (indicated by the asterisks), additional bands of weaker intensity are detected and these are likely to be either degradation products (faster migrating) or ubiquitylated (slower migrating) forms of these proteins. No co-IPed proteins were detected in the parental untagged strain carrying Ubc4-MYC but lacking HA-tagged proteins. 1% of the whole cell extracts were loaded on a separate gel to determine that roughly equal amounts of proteins were present in the input fraction by measuring the amount of histone H4. (B) Hel1, Hel2, Pep5 and Snt2 interact with histones. The HA-tagged E3 enzymes were IPed from 1 liter cultures of the indicated strains using HA.11 antibodies. Co-IPed endogenous histones H3 and H4 was detected using the polyclonal antibodies described previously [6]. No co-IPed histones were detected in the “no tag” control. The amount of Ubc4-MYC was measured in 1% of the input fraction to demonstrate that roughly equal amounts of cell extracts were used for the IP reactions. (C) Hel1 and Hel2 interact with Rad53. MYC tagged Hel1 or Hel2 were IPed using MYC antibody beads from 3 liter cultures of strains carrying FLAG-tagged Rad53. IPed and Co-IPed proteins were detected by Western Blotting using appropriate antibodies. The relative amount of histone H3 present in each of the “input” extracts used for the IP is shown to demonstrate that same amount of material was used for each IP. (D) Snt2 interacts with Rad53 whereas Pep5 does not. Tandem Affinity Purification (TAP) tagged Pep5 or Snt2 was IPed from 3 liter cultures of strains carrying FLAG-tagged Rad53. Any co-IPed Rad53 was detected using FLAG antibodies. The similar levels of Rad53-FLAG present in the extracts confirm that the same amount of extracts was used for each IP. No evidence for any interaction between Rad53 and Pep5 was observed even upon scaling up the Pep5-TAP IP by 3-fold (data not shown).
Hel1, Hel2, Pep5 and Snt2 promote ubiquitylation of histones in vitro
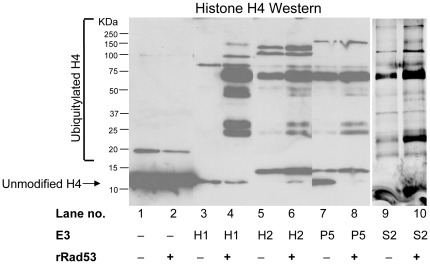
To further demonstrate that the E3 ligases identified by us were bona fide histone E3 ligases, next we tested if they can polyubiquitylate histones in vitro. For this we set up in vitro histone ubiquitylation reactions using recombinant E1, E2, ubiquitin and histone H4 along with purified TAP-tagged E3 ligases as described previously [7]. Reactions were stopped by the addition of Sodium Dodecyl Sulfate (SDS) loading buffer and boiling prior to analysis by Western blotting using histone H4 antibodies. The formation of high-molecular weight species clearly demonstrates that histone H4 is efficiently ubiquitylated in vitro using Hel1, Hel2, Pep5 and Snt2 E3 ligases (Figure 5). Further, the inclusion of Rad53 significantly stimulated the activity of all these E3 ligases in the in vitro ubiquitylation reactions, especially in the case of Hel1 (Figure 5, compare lanes 3 to 4, 5 to 6 and 7 to 8), suggesting that the newly identified E3 ligases may recognize the same Rad53-dependent phosphorylation as degradation signals on histones as the Tom1 ligase identified previously [7].
Figure 5. Hel1, Hel2, Pep5 and Snt2 can efficiently ubiquitylate histone H4 in vitro in a reaction that is stimulated by Rad53.
Recombinant and purified components were used to reconstitute the ubiquitylation of histone H4 in vitro exactly as described previously [7]. The reaction products were resolved on an 18% polyacrylamide gel and processed for Western blotting with H4 antibodies. Addition of commercially available recombinant yeast Uba1 (E1), human UbcH5A (E2; homolog of yeast Ubc4) and HIS6-tagged ubiquitin to recombinant human histone H4 with or without Rad53 did not result in appreciable histone modifications, apart from some monoubiquitylation (lanes 2 and 1 respectively). Only the addition of the E3 ligases Hel1 (H1), Hel2 (H2), Pep5 (P5) and Snt2 (S2) purified from yeast extracts via TAP epitope tags resulted in considerable high molecular weight histone modifications, which were further stimulated to varying degrees by the addition of recombinant Rad53 to the reaction mixture.
Hel1, Hel2, Pep5 and Snt2 do not regulate the levels of the E2 enzyme Ubc4
Although the in vivo and in vitro data presented here so far strongly suggest that the newly identified E3 ligases are likely to be directly involved in histone ubiquitylation and degradation, it is also formally possible that one or more of these ligases may be indirectly affecting histone levels by controlling the abundance of Ubc4, the major E2 enzyme involved in the ubiquitylation mediated degradation of histones. To investigate this possibility, we measured the levels of full-length MYC13-tagged Ubc4, as well as its ubiquitylated forms and degradation products in wild type and E3 mutant strains. Although we readily detected multiple bands migrating slower than full-length Ubc4-MYC, we did not observe any differences in the levels of the full-length or ubiquitylated forms, as well as in the degradation products of this protein upon the deletion of any of the newly identified E3 ligases (Figure 6). This data strongly suggests that these E3 ligases do not regulate Ubc4 protein levels, although it is still formally possible that these E3 ligases may regulate the levels of Tom1 or each other, thereby indirectly influencing histone levels.
Figure 6. The E3 ligases Hel1, Hel2, Pep5, Snt2 and Tom1 do not regulate the protein levels of Ubc4, the major E2 enyme involved in excess histone degradation.
Whole cell lysates were prepared as described previously [69] from wild type and the indicated E3 ligase deletion strains carrying MYC-tagged Ubc4. The lysates were resolved on a 12% polyacrylamide gel and processed for Western blotting using MYC antibodies. No significant differences are observed in the levels of full-length Ubc4-MYC, or in its slower migrating modified forms and faster migrating degradation products.
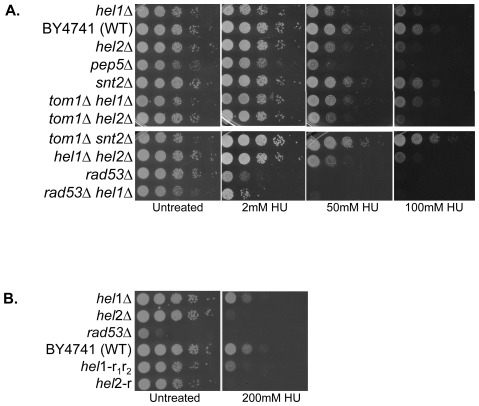
Histone E3 ligases exhibit varying degrees of sensitivity to genotoxic agents such as the replication inhibitor hydroxyurea
DNA damage in eukaryotes occurs in context of chromatin. Therefore, histone proteins and chromatin structure is likely to influence DNA damage and repair. DNA repair enzymes need to gain access to the damaged DNA in order to repair it. We have previously shown that yeast strains lacking factors involved in the regulation of histone protein levels such as the E3 ligase Tom1 are sensitive to genotoxic agents to varying degrees [7]. To assess if the new histone E3 ligases identified in our screen were also sensitive to genotoxic agents, we plated 10-fold serial dilutions of mutant and wild type strains on plates containing the replication inhibitor hydroxyurea (HU). We found the pep5 deletion strain to be very sensitive to HU, though not as sensitive as the rad53 deletion strain, while the hel1 and hel2 deletion mutants were sensitive to HU to a lesser extent (Figure 7A). The snt2Δ mutant was not sensitive at all to HU, suggesting the possibility that different histone E3 ligases may be employed for the degradation of excess histones in different contexts. Combining the tom1 deletion mutant which is slightly sensitive to HU [7] with the hel1, hel2 and snt2 deletion mutants individually did not further exacerbate the HU sensitivity of any of these E3 ligases (Figure 7A), suggesting that they may be working in the same pathway. Similar results were obtained for the hel1 hel2 and the rad53 hel1 double mutants, as the HU sensitivity of the hel2 and rad53 single mutants was not enhanced any further in these double mutants (Figure 7A). However, all our attempts to generate the tom1Δ pep5Δ double mutant have failed so far, suggesting that this combination may be synthetically lethal (data not shown) and that these two E3 ligases may work in parallel pathways that are redundant with each other. We also investigated if the RING domains of Hel1 and Hel2 played any role in resistance of yeast cells to HU. Mutation of the critical residues within the RING finger domains of Hel1 and Hel2 rendered them as sensitive to HU as the complete loss of the proteins (Figure 7B), once again suggesting that the RING finger domains are essential for the function of these E3 ligases.
Figure 7. Histone E3 ligase mutants exhibit varying degrees of sensitivity to the replication inhibitor hydroxyurea.
(A) hel1, hel2 and pep5 mutants are sensitive to hydroxyurea. 10-fold serial dilutions of the indicated strains were plated on media containing glucose with or without the indicated concentrations of hydroxyurea (HU) and incubated at 30°C for 3 days before being photographed. (B) Mutations in the RING finger domains of Hel1 and Hel2 render them sensitive to HU. 10-fold serial dilutions of the indicated strains were plated on media containing galactose with or without 200 mM HU and incubated at 30°C for 3 days before being photographed. The hel1-r1r2 mutant carries inactivating mutations in both r finger domains of Hel1, while the hel2-r mutant carries inactivating mutations in the single r finger domain of Hel2. The rad53 deletion strain was included as a positive control.
Discussion
In this study we have identified and characterized four putative E3 ligases that contribute to the posttranslational regulation of histone protein levels in the budding yeast. As opposed to the previously identified HECT-domain containing histone E3 ligase Tom1 [7], all the four new histone E3 ligases carry RING finger domains that enable them to polyubiquitylate excess histones for subsequent degradation. Non-degradation related polyubiquitylation of histones H2B has also been reported to occur in the budding yeast [40]. As such, although we have focused here on the involvement of these new E3 ligases in the degradation related ubiquitylation of histones, we acknowledge the possibility that they may also be involved in histone ubiquitylation that is not related to degradation. The five histone E3 ligases identified so far in the posttranslational regulation of histone proteins represent roughly 7% of the 80 predicted E3 ligases in the budding yeast [23]. However, it is possible that this list may expand further upon the analysis of the remaining essential predicted E3 ligases that we have not screened so far. This involvement of multiple E3 ligases in the regulation of histone protein levels is reminiscent of the multiple E3 ligases that independently contribute to the regulation of p53 tumor suppressor protein levels in mammalian cells [32]. Overall, the identification of multiple E3 ligases, each of which appears to contribute independently to the regulation of histone protein levels, highlights the importance of the posttranslational degradation of histones in yeast cells.
Two of the E3 ligases identified in our screen, Hel1 and Hel2, correspond to previously uncharacterized budding yeast ORFs YKR017C and YDR266C respectively. Hel1 is only one of two yeast E3 ligases that belong to the RBR (RING-in between-RING) or TRIAD sub-class of E3 ligases that are characterized by the presence of two RING finger domains with a novel Cys-rich Cys6-His-Cys in-between-RING (IBR) domain present in between the two RING domains [41], [42]. The other RBR E3 ligase in the budding yeast, Itt1, is not sensitive to histone overexpression and as such it is unlikely to be involved in the regulation of histone protein levels (data not shown). The RBR family of E3 ligases include human E3 ligases such as the human homolog of Drosophila Ariadne (HHARI; also known as ARIH1) that shares homology with Hel1, as well as TRIAD1 (or ARIH2) and Parkin (or PARK2), the latter of which has been implicated in a subset of Parkinson's disease [43]. It remains to be determined if these human RBR E3 ligases that share similarities with Hel1 can ubiquitylate histones. Recent mechanistic insight into these RBR E3 ligases suggest that they may function as hybrid RING/HECT E3 ligases whereby the first RING domain interacts with the E2 enzyme while the second RING domain participates in the transfer of the ubiquitin moiety to the substrate [44]. Not surprisingly, the second RING domain of HHARI appears to have a very different structure compared to canonical RING domains [45]. Mutations in any one of the two RING domains of Hel1 result in similar defects in histone degradation (Figure 2B) and confer the same degree of DNA damage sensitivity as the null mutant (Figure 7B), further confirming that both the RING domains are required for the function of RBR E3 ligases. Overall, our studies on Hel1 constitute the first characterization of an RBR E3 ligase in yeast and suggest that histones are a potential substrate.
Hel2 is a canonical RING domain containing E3 ligase that has been previously shown to be phosphorylated by the DNA damage checkpoint kinase and histone protein regulator Rad53 [46]. Not surprisingly, in vitro ubiquitylation of histone H4 in the presence of Hel2 is significantly stimulated by the presence of Rad53 (Figure 5). Further, hel2 mutants are sensitive to the replication inhibitor HU and this maybe in part due to its role in regulating histone protein levels, although it is quite likely that there are additional substrates that are ubiquitylated in vivo by Hel2 in response to replication arrest and we are currently investigating this. The human protein with the highest similarity to Hel2 is an uncharacterized zinc finger protein 598 (ZNF598), although known E3 ligases such as KPC1 (Kip1 Ubiquitination-Promoting Complex 1; also known as RNF123) also show limited similarity to Hel2 [47]. It is unclear if these human E3 enzymes are capable of ubiquitylating histones.
Snt2 is a poorly characterized protein. It was initially identified and named for the presence of the DNA binding SANT (SWI3, ADA2, N-CoR and TFIIIB) domain and has been predicted to function as a transcriptional regulator based largely on in silico analyses, although this prediction is yet to be verified experimentally [48], [49]. Snt2 also has three PHD (Plant Homeo-Domain) fingers consisting of the Cys4-His-Cys3 motif which is typically involved in binding methylated lysine residues on histones, although the Snt2 PHD fingers show weak or nonspecific binding to methylated histone H3 peptides at best [50]. The PHD finger is very similar to the Cys3-His-Cys4 containing RING finger domain and two of the PHD fingers of Snt2 are also RING fingers potentially capable of E3 ligase activity. Our data clearly show that the snt2 deletion strain is sensitive to histone overexpression (Figure 1B), defective in histone degradation (Figure 2A) and accumulates excess endogenous histones (Figure 3). Further, point mutations in the second RING domain of Snt2 result in a defect in excess histone degradation (Figure 2B). More importantly, Snt2 interacts with Ubc4, histones and Rad53 (Figure 4) and exhibits robust histone ubiquitylation activity in vitro that is further stimulated by Rad53 (Figure 5). Hence, based on these data we conclude that Snt2 is a genuine histone E3 ligase and contributes to the regulation of histone protein levels.
Pep5 (also known as Vps11) was initially isolated as a protease deficient mutant [51]. Pep5 was subsequently shown to be involved in protein trafficking and vacuole biogenesis pathways [52], although it has not been studied as a RING E3 ligase. In our in vitro ubiquitylation assay, Pep5 shows robust activity in ubiquitylating histone H4 (Figure 5), indicating that histones may serve as its substrates in vivo as well. Of the four histone E3 ligases identified in this study, the pep5 deletion strain exhibited the most pronounced defects in the degradation of exogenous histones (Figure 2A) and accumulated high amounts of excess endogenous histones on the histone chaperone Asf1 (Figure 3). Interestingly, pep5 mutants have been reported to exhibit a synthetic growth defect when combined with hir1, hir2 or asf1 mutations, and are synthetically lethal when combined with rad53 mutations [53]. Hir1 and Hir2 are part of the four-subunit HIR complex that serves as a negative transcriptional regulator of histone gene expression outside of S-phase in yeast [54], apart from functioning as a histone chaperone [55]. Asf1 also contributes to the transcriptional regulation of yeast histone genes [56], while Rad53 is known to regulate histone protein levels [6], [7]. Taken together, our data suggests that Pep5 is likely to be a major player in the regulation of histone proteins, which would explain the synthetic phenotypes observed upon the simultaneous loss of pep5 along with known regulators of histone levels. Loss of the histone transcriptional regulator complex HIR may result in an increased production of endogenous histones, thereby increasing the burden on the remaining histone regulators, including Pep5. Similarly, loss of Asf1 would result in a reduction in the capacity of the cells to sequester excess endogenous histones on the remaining histone chaperones, thereby channeling more histones for degradation via Pep5. It will be interesting to determine if Vps11, which appears to be a clear human homolog for Pep5 [57], is also involved in histone regulation.
We have previously provided evidence that the kinase activity of Rad53 is required for the efficient polyubiquitylation of histones both in vitro and in vivo [7]. We further identified the tyrosine 99 residue of histone H3 as a crucial determinant for the degradation of this histone in vivo, presumably due to phosphorylation at this residue. Based on the phosphorylation-ubiquitylation mediated degradation of well-known proteins [58], [59], we proposed a similar model for the degradation of excess histones whereby these histones were first detected and phosphorylated by Rad53, leading to their efficient polyubiquitylation and subsequent degradation by the proteasome. Consistent with this model, our current in vitro ubiquitylation reactions demonstrate that the activities of all the identified histone E3 ligases in ubiquitylating histone H4 were further stimulated by the addition of Rad53, but to varying degrees (Figure 5), suggesting that these ligases may also recognize phosphorylation dependent degradation signals on histones similar to Tom1. Further, just like Tom1 [7], Hel1 shows limited H4 ubiquitylation activity in the absence of Rad53, while Hel2, Snt2 and Pep5 show significant H4 ubiquitylation under the same conditions (Figure 5). Upon the addition of Rad53, the H4 ubiquitylation activity of Hel1 was stimulated strongly, while the activities of Hel2, Snt2 and Pep5 showed a more modest increase. The varying degree of Rad53-dependent stimulation of the in vitro histone ubiquitylation reactions by these E3 ligases may be indicative of the requirement for additional cellular regulatory factors, such as accessory proteins and possibly other kinases that are missing from our in vitro reactions, which may be crucial for the full range of regulated activities of these ligases in vivo. Since deletion of pep5 confers a synthetic lethal phenotype when combined with the rad53 deletion [53] (and data not shown), it is possible that at least this and possibly all of these E3 ligases may carry out significant histone ubiquitylation independent of Rad53 in vivo by relying on other kinases to phosphorylate excess histones and mark them for degradation. This idea would be consistent with the strong defects in histone protein regulation in the pep5 (Figure 2A and 3) and rad53 [6], [7] single mutants, the synthetic lethality of the rad53 pep5 [53] and possibly the tom1 pep5 double mutants (data not shown), as well as the mutually exclusive localization of Pep5 and Rad53 in the cytoplasm and the nucleus respectively.
Our previous studies have shown that excess histones are degraded by the proteasomes following polyubiquitylation [7]. Since the proteasomes are primarily localized in the nucleus in budding yeast [60], [61], [62], following the initial identification of the nuclear histone E3 ligase Tom1 [7], we had suggested that excess histone degradation was likely to be occurring in the nucleus. However, the cytoplasmic localization of the histone E3 ligases such as Hel2 and Pep5, as well as the nucleocytoplasmic localization of Snt2 [63], suggest the possibility that some excess histone degradation may occur in the cytoplasm (the localization of Hel1 has not been determined yet). Due to the predominant nuclear localization of proteasomes in budding yeast [60], [61], [62], cytoplasmic degradation of excess histones may even occur in a proteasome independent manner, perhaps in the vacuoles. A very recent report suggests that histones may be targeted for degradation via the Chaperone Mediated Autophagy (CMA) pathway in mammalian cells [64]. Although CMA has not been reported to occur in yeast, a related autophagy process known as Vacuolar Import and Degradation (VID) has indeed been described in yeast [65] and this pathway requires components of the ubiquitylation pathway [66], [67]. Hence, it is possible that the known function of Pep5 in vacuolar sorting [52] is somehow involved in such a proteasome independent cytoplasmic degradation of excess histones. Based on their localization, another possibility is that Tom1 targets excess histones in the nucleus for degradation, while Hel2 and Pep5 are involved in the cytoplasmic degradation of excess histones, whereas Snt2 contributes to histone degradation in both cellular compartments. Future studies will determine if this is indeed the case.
Materials and Methods
All yeast strains used are listed in Table S1. These strains were either obtained from Open Biosystems budding yeast genome deletion collection or were generated in the lab as needed using standard yeast manipulations [68]. MYC13-tagged Ubc4 and the plasmid pRS416-RAD53-FLAG for the expression of FLAG-tagged Rad53 have been described previously [7]. TAP-tagged strains were obtained from Open Biosystems. Plasmids pYES2-HTH and pYES2-HTH-HHT2 have been described previously [7]. Plasmid BG1805 based constructs expressing C-terminal HIS6-HA-Protein A (HIS6-HA-PrA) tagged Hel1, Hel2, Pep5 and Snt2 under the control of a galactose inducible promoter were also obtained from Open Biosystems. Mutagenesis of the HEL1, HEL2 and SNT2 RING finger domains was carried out on the plasmid BG1805 based constructs expressing Hel1, Hel2 and Snt2 using Stratagene's QuikChange Multi Site-Directed Mutagenesis kit following the manufacturer's instructions. Briefly, the crucial Cys and His residues in the RING (r) finger domains were mutated to Ala residues. The hel1-r1 mutant carries the Cys-195, His-197, Cys-200→Ala mutations in r1, while the hel1-r2 of carries the His-356, Cys-359, Cys-362→Ala mutations in r2. The hel1-r1r2 mutant carries both the r1 and r2 mutations simultaneously. The hel2-r mutant carries the Cys-79, His-81, Cys-87→Ala mutations in its r finger domain. The snt2-r2 carries the Cys1061, His1066, Cys1069→Ala mutations in its second r2 finger domain. All mutations were confirmed by sequencing the entire mutant gene. The plasmids bearing the r finger mutants of the E3 ligases were transformed into cells lacking the endogenous gene for the respective E3 ligase to perform the experiments shown in figures 2B and 7B. All other methods used such as the histone degradation assay, histone overexpression sensitivity assay, Western blotting, co-immunoprecipitation assay and in vitro ubiquitylation assay have been described extensively in our previous publications [6], [7].
Supporting Information
List of strains used in this study.
(PDF)
Acknowledgments
We wish to thank Alain Verreault and Yanchang Wang for reagents. We thank Dr. Johanna Paik for critical reading of this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: These authors have no support or funding to report.
References
- 1.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 2.van Holde KE. New York: Springer-Verlag; 1988. Chromatin; van Holde KE, editor. [Google Scholar]
- 3.Wolffe AP. San Diego: Academic Press; 1995. Chromatin: Structure and Function.299 [Google Scholar]
- 4.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 5.Singh RK, Paik J, Gunjan A. Generation and management of excess histones during the cell cycle. Front Biosci. 2009;14:3145–3158. doi: 10.2741/3441. [DOI] [PubMed] [Google Scholar]
- 6.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 7.Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh RK, Liang D, Gajjalaiahvari UR, Kabbaj MH, Paik J, et al. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 2010;9:4236–4244. doi: 10.4161/cc.9.20.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodland HR, Adamson ED. The synthesis and storage of histones during the oogenesis of Xenopus laevis. Developmental Biology. 1977;57:118–135. doi: 10.1016/0012-1606(77)90359-1. [DOI] [PubMed] [Google Scholar]
- 10.Dingwall C, Laskey RA. Nucleoplasmin: the archetypal molecular chaperone. Seminars in Cell Biology. 1990;1:11–17. [PubMed] [Google Scholar]
- 11.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Current Biology. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 12.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Molecular Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groth A, Corpet A, Cook AJ, Roche D, Bartek J, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 14.Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, et al. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Morillo-Huesca M, Maya D, Munoz-Centeno MC, Singh RK, Oreal V, et al. FACT prevents the accumulation of free histones evicted from transcribed chromatin and a subsequent cell cycle delay in G1. PLoS Genet. 2010;6:e1000964. doi: 10.1371/journal.pgen.1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang D, Burkhart SL, Kabbaj MH, Gunjan A. Nucleic Acids Res (In Press); 2012. Histone dosage regulates DNA damage sensitivity in a checkpoint independent manner via the homologous recombination pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 19.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 20.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. Journal of Clinical Oncology. 2005;23:4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 21.Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. Journal of Biological Chemistry. 1982;257:2543–2548. [PubMed] [Google Scholar]
- 22.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays in Biochemistry. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual Review of Biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 25.Hatakeyama S, Nakayama KI. U-box proteins as a new family of ubiquitin ligases. Biochemical and Biophysical Research Communications. 2003;302:635–645. doi: 10.1016/s0006-291x(03)00245-6. [DOI] [PubMed] [Google Scholar]
- 26.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 27.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5249. doi: 10.1073/pnas.92.11.5249-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciechanover A, Schwartz AL. Ubiquitin-mediated degradation of cellular proteins in health and disease. Hepatology. 2002;35:3–6. doi: 10.1053/jhep.2002.30316. [DOI] [PubMed] [Google Scholar]
- 29.Ciechanover A. The ubiquitin proteolytic system and pathogenesis of human diseases: a novel platform for mechanism-based drug targeting. Biochemical Society Transactions. 2003;31:474–481. doi: 10.1042/bst0310474. [DOI] [PubMed] [Google Scholar]
- 30.Scheffner M, Staub O. HECT E3s and human disease. BMC Biochem. 2007;8(Suppl 1):S6. doi: 10.1186/1471-2091-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ardley HC, Robinson PA. The role of ubiquitin-protein ligases in neurodegenerative disease. Neurodegener Dis. 2004;1:71–87. doi: 10.1159/000080048. [DOI] [PubMed] [Google Scholar]
- 32.Brooks CL, Gu W. p53 regulation by ubiquitin. FEBS Letters. 2011;585:2803–2809. doi: 10.1016/j.febslet.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Commerford SL, Carsten AL, Cronkite EP. Histone turnover within nonproliferating cells. Proc Natl Acad Sci U S A. 1982;79:1163–1165. doi: 10.1073/pnas.79.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Oughtred R, Wing SS. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol Cell Biol. 2005;25:2819–2831. doi: 10.1128/MCB.25.7.2819-2831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall JR, Kow E, Nevis KR, Lu CK, Luce KS, et al. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Molecular Biology of the Cell. 2007;18:3340–3350. doi: 10.1091/mbc.E07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Chen D, Kon N, Li M, Zhang W, Qin J, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 39.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 40.Geng F, Tansey WP. Polyubiquitylation of histone H2B. Mol Biol Cell. 2008;19:3616–3624. doi: 10.1091/mbc.E08-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Reijden BA, Erpelinck-Verschueren CA, Lowenberg B, Jansen JH. TRIADs: a new class of proteins with a novel cysteine-rich signature. Protein Science. 1999;8:1557–1561. doi: 10.1110/ps.8.7.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marin I, Ferrus A. Comparative genomics of the RBR family, including the Parkinson's disease-related gene parkin and the genes of the ariadne subfamily. Molecular Biology and Evolution. 2002;19:2039–2050. doi: 10.1093/oxfordjournals.molbev.a004029. [DOI] [PubMed] [Google Scholar]
- 43.Marin I, Lucas JI, Gradilla AC, Ferrus A. Parkin and relatives: the RBR family of ubiquitin ligases. Physiol Genomics. 2004;17:253–263. doi: 10.1152/physiolgenomics.00226.2003. [DOI] [PubMed] [Google Scholar]
- 44.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Capili AD, Edghill EL, Wu K, Borden KL. Structure of the C-terminal RING finger from a RING-IBR-RING/TRIAD motif reveals a novel zinc-binding domain distinct from a RING. Journal of Molecular Biology. 2004;340:1117–1129. doi: 10.1016/j.jmb.2004.05.035. [DOI] [PubMed] [Google Scholar]
- 46.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 47.Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, et al. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol. 2004;6:1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, Zhang Z, Li Y, Zhu XG, Liu Q. Identifying cooperative transcription factors by combining ChIP-chip data and knockout data. Cell Research. 2010;20:1276–1278. doi: 10.1038/cr.2010.146. [DOI] [PubMed] [Google Scholar]
- 49.Ward LD, Bussemaker HJ. Predicting functional transcription factor binding through alignment-free and affinity-based analysis of orthologous promoter sequences. Bioinformatics. 2008;24:i165–171. doi: 10.1093/bioinformatics/btn154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. Journal of Biological Chemistry. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones EW. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977;85:23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srivastava A, Woolford CA, Jones EW. Pep3p/Pep5p complex: a putative docking factor at multiple steps of vesicular transport to the vacuole of Saccharomyces cerevisiae. Genetics. 2000;156:105–122. doi: 10.1093/genetics/156.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan X, Ye P, Yuan DS, Wang X, Bader JS, et al. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 54.Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 55.Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, et al. Replication-independent histone deposition by the HIR complex and Asf1. Current Biology. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sutton A, Bucaria J, Osley MA, Sternglanz R. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huizing M, Didier A, Walenta J, Anikster Y, Gahl WA, et al. Molecular cloning and characterization of human VPS18, VPS 11, VPS16, and VPS33. Gene. 2001;264:241–247. doi: 10.1016/s0378-1119(01)00333-x. [DOI] [PubMed] [Google Scholar]
- 58.Deshaies RJ, Ferrell JE Multisite phosphorylation and the countdown to S phase. Cell. 2001;107:819–822. doi: 10.1016/s0092-8674(01)00620-1. [DOI] [PubMed] [Google Scholar]
- 59.Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, et al. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 60.Russell SJ, Steger KA, Johnston SA. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. Journal of Biological Chemistry. 1999;274:21943–21952. doi: 10.1074/jbc.274.31.21943. [DOI] [PubMed] [Google Scholar]
- 61.Enenkel C, Lehmann A, Kloetzel PM. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO Journal. 1998;17:6144–6154. doi: 10.1093/emboj/17.21.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laporte D, Salin B, Daignan-Fornier B, Sagot I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. Journal of Cell Biology. 2008;181:737–745. doi: 10.1083/jcb.200711154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 64.Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3–H4 in the histone supply chain. Molecular Cell. 2011;44:918–927. doi: 10.1016/j.molcel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 65.Alibhoy AA, Chiang HL. Vacuole import and degradation pathway: Insights into a specialized autophagy pathway. World J Biol Chem. 2011;2:239–245. doi: 10.4331/wjbc.v2.i11.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shieh HL, Chen Y, Brown CR, Chiang HL. Biochemical analysis of fructose-1,6-bisphosphatase import into vacuole import and degradation vesicles reveals a role for UBC1 in vesicle biogenesis. Journal of Biological Chemistry. 2001;276:10398–10406. doi: 10.1074/jbc.M001767200. [DOI] [PubMed] [Google Scholar]
- 67.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 68.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 69.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of strains used in this study.
(PDF)