Abstract
We have cloned a yeast gene, SKO1, which in high copy number suppresses lethal overexpression of cAMP-dependent protein kinase. SKO1 encodes a bZIP protein that binds to the CRE motif, TGACGTCA. We found that SKO1 also binds to a CRE-like site in SUC2, a yeast gene encoding invertase which is under positive control by cAMP. A disruption of the SKO1 gene causes a partial derepression of SUC2, indicating that SKO1 is a negative regulator of the SUC2 gene. SKO1 interacts positively with MIG1, a zinc finger protein that mediates glucose repression of SUC2. A kinetic analysis revealed a complex regulation of the SUC2 mRNA in response to glucose. First, MIG1 mediates a rapid and strong repression of SUC2, which is complete within 10 minutes. Second, a MIG1-independent process causes a further slow reduction in the mRNA. Third, in the absence of MIG1, there is also a rapid but transient glucose induction of the SUC2 mRNA. This induction is correlated with a transient loss of SKO1-dependent repression.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argüelles J. C., Mbonyi K., Van Aelst L., Vanhalewyn M., Jans A. W., Thevelein J. M. Absence of glucose-induced cAMP signaling in the Saccharomyces cerevisiae mutants cat1 and cat3 which are deficient in derepression of glucose-repressible proteins. Arch Microbiol. 1990;154(2):199–205. doi: 10.1007/BF00423333. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Deschenes R. J. The function of ras genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- Cameron S., Levin L., Zoller M., Wigler M. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell. 1988 May 20;53(4):555–566. doi: 10.1016/0092-8674(88)90572-7. [DOI] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986 Sep 12;233(4769):1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Cherry J. R., Johnson T. R., Dollard C., Shuster J. R., Denis C. L. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell. 1989 Feb 10;56(3):409–419. doi: 10.1016/0092-8674(89)90244-4. [DOI] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Audino D. C. The CCR1 (SNF1) and SCH9 protein kinases act independently of cAMP-dependent protein kinase and the transcriptional activator ADR1 in controlling yeast ADH2 expression. Mol Gen Genet. 1991 Oct;229(3):395–399. doi: 10.1007/BF00267461. [DOI] [PubMed] [Google Scholar]
- Eraso P., Gancedo J. M. Catabolite repression in yeasts is not associated with low levels of cAMP. Eur J Biochem. 1984 May 15;141(1):195–198. doi: 10.1111/j.1432-1033.1984.tb08174.x. [DOI] [PubMed] [Google Scholar]
- Fassler J. S., Winston F. The Saccharomyces cerevisiae SPT13/GAL11 gene has both positive and negative regulatory roles in transcription. Mol Cell Biol. 1989 Dec;9(12):5602–5609. doi: 10.1128/mcb.9.12.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor-Chaiken M., Deschenes R. J., Broach J. R. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990 Apr 20;61(2):329–340. doi: 10.1016/0092-8674(90)90813-t. [DOI] [PubMed] [Google Scholar]
- Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992 Mar 20;68(6):1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Gonzalez G. A., Montminy M. R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989 Nov 17;59(4):675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Heitman J., Movva N. R., Hiestand P. C., Hall M. N. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1948–1952. doi: 10.1073/pnas.88.5.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelfarb H. J., Pearlberg J., Last D. H., Ptashne M. GAL11P: a yeast mutation that potentiates the effect of weak GAL4-derived activators. Cell. 1990 Dec 21;63(6):1299–1309. doi: 10.1016/0092-8674(90)90425-e. [DOI] [PubMed] [Google Scholar]
- Hoeffler J. P., Meyer T. E., Yun Y., Jameson J. L., Habener J. F. Cyclic AMP-responsive DNA-binding protein: structure based on a cloned placental cDNA. Science. 1988 Dec 9;242(4884):1430–1433. doi: 10.1126/science.2974179. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hubbard E. J., Yang X. L., Carlson M. Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein kinase and invertase expression in Saccharomyces cerevisiae. Genetics. 1992 Jan;130(1):71–80. doi: 10.1093/genetics/130.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. H., Jones N. C. Mammalian cAMP-responsive element can activate transcription in yeast and binds a yeast factor(s) that resembles the mammalian transcription factor ANF. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2176–2180. doi: 10.1073/pnas.86.7.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992 Feb 21;68(4):709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Krebs E. G., Beavo J. A. Phosphorylation-dephosphorylation of enzymes. Annu Rev Biochem. 1979;48:923–959. doi: 10.1146/annurev.bi.48.070179.004423. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Green M. R. Identification and purification of a Saccharomyces cerevisiae protein with the DNA binding specificity of mammalian activating transcription factor. Proc Natl Acad Sci U S A. 1989 Jan;86(1):109–113. doi: 10.1073/pnas.86.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A., Cereghino G. P., Scheffler I. E. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Jul;12(7):2941–2948. doi: 10.1128/mcb.12.7.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Sakura H., Kanei-Ishii C., Sudo T., Yoshimura T., Fujisawa J., Yoshida M., Ishii S. Leucine zipper structure of the protein CRE-BP1 binding to the cyclic AMP response element in brain. EMBO J. 1989 Jul;8(7):2023–2028. doi: 10.1002/j.1460-2075.1989.tb03610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T. Control of cell division in Saccharomyces cerevisiae mutants defective in adenylate cyclase and cAMP-dependent protein kinase. Exp Cell Res. 1983 Jun;146(1):151–161. doi: 10.1016/0014-4827(83)90333-6. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T., Oshima Y. Cyclic AMP may not be involved in catabolite repression in Saccharomyces cerevisiae: evidence from mutants unable to synthesize it. J Bacteriol. 1983 Nov;156(2):898–900. doi: 10.1128/jb.156.2.898-900.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T. Regulation of repressible acid phosphatase by cyclic AMP in Saccharomyces cerevisiae. Genetics. 1984 Sep;108(1):53–66. doi: 10.1093/genetics/108.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Oshima Y., Ishikawa T. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2355–2359. doi: 10.1073/pnas.79.7.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye-Rowley W. S., Harshman K. D., Parker C. S. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 1989 Mar;3(3):283–292. doi: 10.1101/gad.3.3.283. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A repetitive DNA sequence that confers cell-cycle START (CDC28)-dependent transcription of the HO gene in yeast. Cell. 1985 Aug;42(1):225–235. doi: 10.1016/s0092-8674(85)80118-5. [DOI] [PubMed] [Google Scholar]
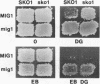
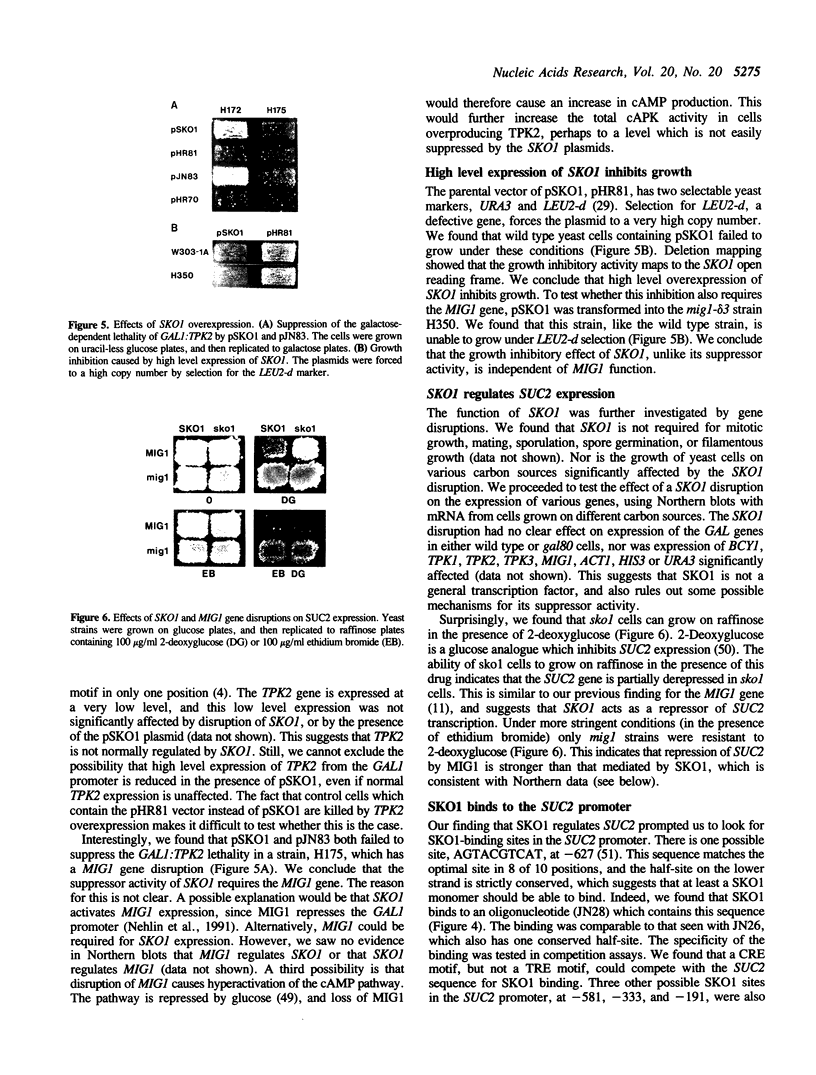
- Nehlin J. O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991 Nov;10(11):3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Yeast galactose permease is related to yeast and mammalian glucose transporters. Gene. 1989 Dec 28;85(2):313–319. doi: 10.1016/0378-1119(89)90423-x. [DOI] [PubMed] [Google Scholar]
- Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990 Sep;9(9):2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M., Suzuki Y., Nogi Y., Matsumoto K., Fukasawa T. Yeast Gal11 protein mediates the transcriptional activation signal of two different transacting factors, Gal4 and general regulatory factor I/repressor/activator site binding protein 1/translation upstream factor. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5373–5377. doi: 10.1073/pnas.87.14.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
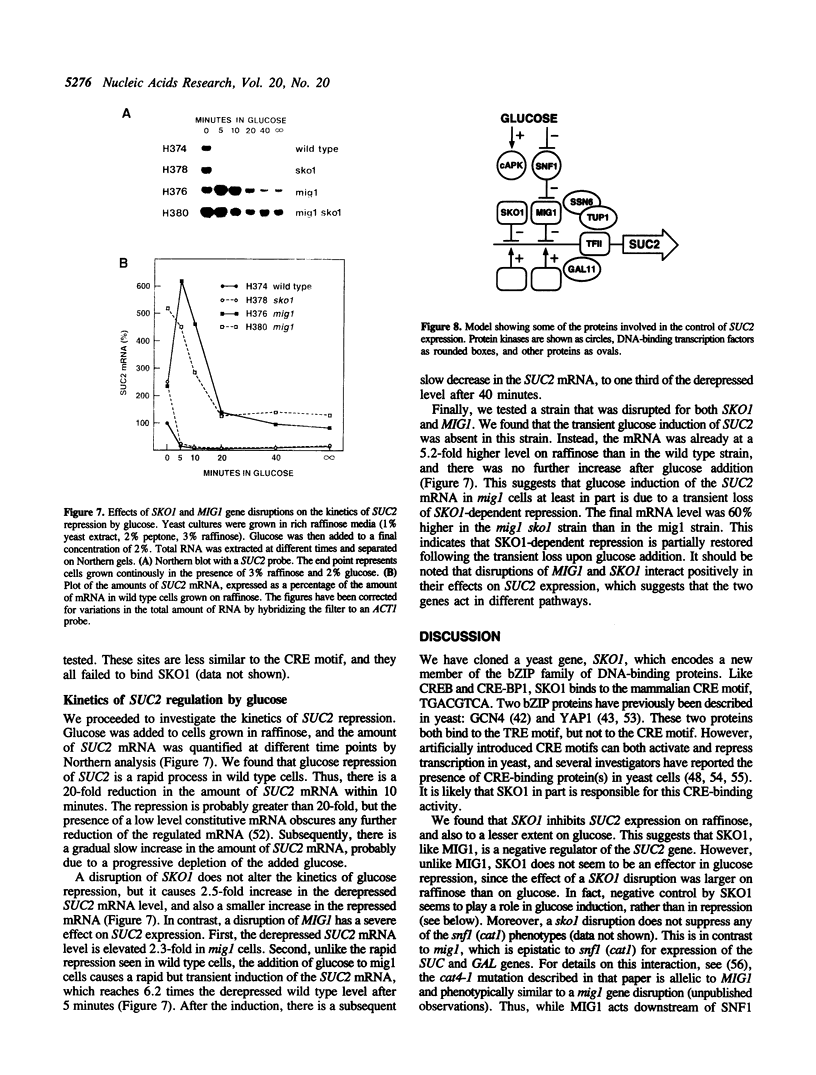
- Nogi Y., Shimada H., Matsuzaki Y., Hashimoto H., Fukasawa T. Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. II. The isolation and dosage effect of the regulatory gene GAL80. Mol Gen Genet. 1984;195(1-2):29–34. doi: 10.1007/BF00332719. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell. 1982 Sep;30(2):517–528. doi: 10.1016/0092-8674(82)90249-5. [DOI] [PubMed] [Google Scholar]
- Perkins K. K., Admon A., Patel N., Tjian R. The Drosophila Fos-related AP-1 protein is a developmentally regulated transcription factor. Genes Dev. 1990 May;4(5):822–834. doi: 10.1101/gad.4.5.822. [DOI] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Sakai A., Shimizu Y., Kondou S., Chibazakura T., Hishinuma F. Structure and molecular analysis of RGR1, a gene required for glucose repression of Saccharomyces cerevisiae. Mol Cell Biol. 1990 Aug;10(8):4130–4138. doi: 10.1128/mcb.10.8.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarokin L., Carlson M. Upstream region required for regulated expression of the glucose-repressible SUC2 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2750–2757. doi: 10.1128/mcb.4.12.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell N., Krems B., Entian K. D. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr Genet. 1992 Apr;21(4-5):269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- Schüller H. J., Entian K. D. Extragenic suppressors of yeast glucose derepression mutants leading to constitutive synthesis of several glucose-repressible enzymes. J Bacteriol. 1991 Mar;173(6):2045–2052. doi: 10.1128/jb.173.6.2045-2052.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller H. J., Entian K. D. Isolation and expression analysis of two yeast regulatory genes involved in the derepression of glucose-repressible enzymes. Mol Gen Genet. 1987 Sep;209(2):366–373. doi: 10.1007/BF00329667. [DOI] [PubMed] [Google Scholar]
- Sellers J. W., Vincent A. C., Struhl K. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol Cell Biol. 1990 Oct;10(10):5077–5086. doi: 10.1128/mcb.10.10.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver B. J., Bokar J. A., Virgin J. B., Vallen E. A., Milsted A., Nilson J. H. Cyclic AMP regulation of the human glycoprotein hormone alpha-subunit gene is mediated by an 18-base-pair element. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2198–2202. doi: 10.1073/pnas.84.8.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986 Nov;6(11):3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J. M. Fermentable sugars and intracellular acidification as specific activators of the RAS-adenylate cyclase signalling pathway in yeast: the relationship to nutrient-induced cell cycle control. Mol Microbiol. 1991 Jun;5(6):1301–1307. doi: 10.1111/j.1365-2958.1991.tb00776.x. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Thompson-Jaeger S., François J., Gaughran J. P., Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991 Nov;129(3):697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987 Jul 17;50(2):277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Trumbly R. J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992 Jan;6(1):15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- Vallier L. G., Carlson M. New SNF genes, GAL11 and GRR1 affect SUC2 expression in Saccharomyces cerevisiae. Genetics. 1991 Nov;129(3):675–684. doi: 10.1093/genetics/129.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt P. K., Bos T. J., Doolittle R. F. Homology between the DNA-binding domain of the GCN4 regulatory protein of yeast and the carboxyl-terminal region of a protein coded for by the oncogene jun. Proc Natl Acad Sci U S A. 1987 May;84(10):3316–3319. doi: 10.1073/pnas.84.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills C. Regulation of sugar and ethanol metabolism in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1990;25(4):245–280. doi: 10.3109/10409239009090611. [DOI] [PubMed] [Google Scholar]
- Zerial M., Toschi L., Ryseck R. P., Schuermann M., Müller R., Bravo R. The product of a novel growth factor activated gene, fos B, interacts with JUN proteins enhancing their DNA binding activity. EMBO J. 1989 Mar;8(3):805–813. doi: 10.1002/j.1460-2075.1989.tb03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann F. K., Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol Gen Genet. 1977 Jul 7;154(1):75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]
- van Aelst L., Jans A. W., Thevelein J. M. Involvement of the CDC25 gene product in the signal transmission pathway of the glucose-induced RAS-mediated cAMP signal in the yeast Saccharomyces cerevisiae. J Gen Microbiol. 1991 Feb;137(2):341–349. doi: 10.1099/00221287-137-2-341. [DOI] [PubMed] [Google Scholar]
- van der Plaat J. B. Cyclic 3',5'-adenosine monophosphate stimulates trehalose degradation in baker's yeast. Biochem Biophys Res Commun. 1974 Feb 4;56(3):580–587. doi: 10.1016/0006-291x(74)90643-3. [DOI] [PubMed] [Google Scholar]