Abstract
In Saccharomyces cerevisiae, the telomerase complex binds to chromosome ends and is activated in late S-phase through a process coupled to the progression of the replication fork. Here, we show that the single-stranded DNA-binding protein RPA (replication protein A) binds to the two daughter telomeres during telomere replication but only its binding to the leading-strand telomere depends on the Mre11/Rad50/Xrs2 (MRX) complex. We further demonstrate that RPA specifically co-precipitates with yKu, Cdc13 and telomerase. The interaction of RPA with telomerase appears to be mediated by both yKu and the telomerase subunit Est1. Moreover, a mutation in Rfa1 that affects both the interaction with yKu and telomerase reduces the dramatic increase in telomere length of a rif1Δ, rif2Δ double mutant. Finally, we show that the RPA/telomerase association and function are conserved in Schizosaccharomyces pombe. Our results indicate that in both yeasts, RPA directly facilitates telomerase activity at chromosome ends.
Keywords: replication, RPA, telomerase, telomeres, yKu
Introduction
Telomeres are structures present at the ends of linear chromosomes that are composed of duplex telomeric DNA and various telomere-associated proteins. Most of the telomeric DNA is duplex, but the 3′ end of telomeric DNA consists of single-stranded 3′ overhangs. In budding yeast Saccharomyces cerevisiae, telomeres contain a 250–300 bp of TG1−3 repeats and a 12–14 nt long 3′ single-stranded overhang that is elongated during S-phase (Wellinger et al, 1993; Larrivee et al, 2004). The essential repressor/activator protein Rap1 is specifically associated with telomeric duplex DNA repeats, while the single-stranded 3′ overhang is bound by Cdc13 that is itself associated with Stn1–Ten1 to form the CST complex. CST acts as a platform involved in telomere end protection, telomere elongation and synthesis of the complementary C-strand (Lendvay et al, 1996; Lin and Zakian, 1996; Nugent et al, 1996; Grandin et al, 2000; Chandra et al, 2001; Grossi et al, 2004; Petreaca et al, 2006; Puglisi et al, 2008).
Telomere length maintenance depends on a specialized reverse transcriptase called telomerase that uses its RNA template to elongate the telomere terminal 3′ overhangs. Yeast telomerase consists of the catalytic subunit Est2 and TLC1 RNA template (Singer and Gottschling, 1994; Lingner et al, 1997). The telomerase RNA template TLC1 is a low abundance RNA (<1 TLC1/telomere) (Mozdy et al, 2008). TLC1 RNA secondary structure model (Dandjinou et al, 2004; Zappulla and Cech, 2004) and protein-binding experiments indicate that TLC1 is made of a central core, which contains the RNA template and the Est2-binding site, from which three RNA arms emanate and interact with Est1p, the Ku heterodimer and the Sm heteroheptamer, respectively (Peterson et al, 2001; Seto et al, 2002; Stellwagen et al, 2003; Zappulla et al, 2005). The TLC1-binding proteins Est1 and Est3 are subunits of the S. cerevisiae telomerase holoenzyme that are essential for telomerase activity in vivo (Lendvay et al, 1996; Hughes et al, 2000; Evans and Lundblad, 2002).
In vivo, the timing of telomere elongation by telomerase in late S-phase correlates with a telomerase-independent lengthening of G-tails whose length increases to about 50 nt during late S/G2 phase (Wellinger et al, 1993; Dionne and Wellinger, 1998). The Mre11/Rad50/Xrs2 (MRX) complex is required for G-tail formation from blunt ends of leading-strand telomeres (Larrivee et al, 2004; Bonetti et al, 2009; Faure et al, 2010). Whether a 5′ resection activity is also needed for the formation of the G-rich single-strand DNA at the lagging-strand telomere remains to be determined (Gilson and Geli, 2007; Faure et al, 2010). The generation of extended 3′ overhangs is positively regulated by Cdk1 (Cdc28) activity (Frank et al, 2006; Vodenicharov and Wellinger, 2006) and negatively regulated by long telomeric tracts that inhibit the binding of Mre11 (Negrini et al, 2007). Rap1 is able to interact via its C-terminus with two negative regulators of telomerase activity called Rif1 and Rif2 (Wotton and Shore, 1997; Levy and Blackburn, 2004). The lack of either protein causes telomere lengthening, a phenotype dramatically increased when both Rif1 and Rif2 are absent. It has been shown that cycling cells devoid of Rif2 display Mre11-dependent accumulation of ssDNA at native telomeres, indicating that Rif2 inhibits nucleolytic degradation at telomeres by preventing MRX recruitment at chromosome ends (Bonetti et al, 2010) in agreement with other results showing that Rif2 attenuates Tel1 binding to DNA ends (Hirano et al, 2009; McGee et al, 2010). Also, Rif1 has been very recently shown to have a synthetic interaction with the CST complex (Anbalagan et al, 2011). It was proposed that Rif1 helps the CST complex to fill the ssDNA at telomeres by a yet unknown mechanism.
In G1, Est2 is sequestered in an inactive form on the telomeres via an interaction of TLC1 RNA with yKu (Peterson et al, 2001; Taggart et al, 2002; Stellwagen et al, 2003; Fisher et al, 2004) and is then activated in late S-phase through a process coupled to the replication-dependent resection of the C-rich strand (Wellinger et al, 1996; Dionne and Wellinger, 1998). Binding of Cdc13 to the 3′ telomeric overhangs has been proposed to mediate the recruitment of telomerase through an interaction between Est1 and Cdc13 made at the expense of the interaction with Stn1–Ten1 (Evans and Lundblad, 1999; Chandra et al, 2001; Bianchi et al, 2004; Li et al, 2009).
Previous models proposed that the MRX-dependent recruitment of Tel1 to short telomeres (Arneric and Lingner, 2007; Bianchi and Shore, 2007; Hector et al, 2007; Sabourin et al, 2007) lead to the phosphorylation of Cdc13 that in turn helps telomerase recruitment to telomeres (Tseng et al, 2006; Bianchi and Shore, 2008). However, this model has been recently challenged by the finding that mutating every Tel1-potential consensus phosphorylation site of Cdc13 does not affect telomere length (Gao et al, 2010). In addition, alleles of YKU80 that disrupt the interaction with TLC1 were also found to decrease telomerase recruitment to telomeres and to result in telomere shortening phenotypes, indicating that the yKu70/80 heterodimer also contributes to the telomerase loading onto telomeres in late S-phase (Fisher et al, 2004; Chan et al, 2008). Moreover, the role of Est1 is not restricted to telomerase recruitment since mutant Est1 proteins that retain association with the telomerase enzyme were found to affect telomere length (Evans and Lundblad, 2002). In addition, recent data indicate that in vitro Est1 also favours telomerase-mediated DNA extension through a direct contact with Est2 (Dezwaan and Freeman, 2009). Taken together, these results suggest that the recruitment/activation of the telomerase holoenzyme is mediated by two pathways, one involving Cdc13 and the other yKu (Dezwaan and Freeman, 2010).
Replication protein A (RPA) is a highly conserved heterotrimeric single-stranded DNA-binding protein involved in DNA replication, recombination and repair (Binz et al, 2004). RPA has been also identified as an additional telomeric factor in S. cerevisiae. RPA was found to bind at telomeres in late S-phase and several alleles of RPA result in a telomere shortening phenotype in budding and fission yeasts (Smith et al, 2000; Mallory et al, 2003; Ono et al, 2003; Schramke et al, 2004; Kibe et al, 2007). In S. cerevisiae, a specific allele introducing a truncation of the N-terminal region of Rfa2 results in severe telomere shortening and a decreased association of Est1 (Schramke et al, 2004). However, the exact role of RPA in telomere maintenance remains poorly understood.
In this study, we provide insights showing that RPA is part of a transient complex comprising RPA, yKu, Cdc13 and the telomerase holoenzyme in late S-phase. We show in budding and fission yeasts that RPA facilitates telomere elongation by telomerase.
Results
Mre11 favours the binding of RPA to the leading-strand telomere
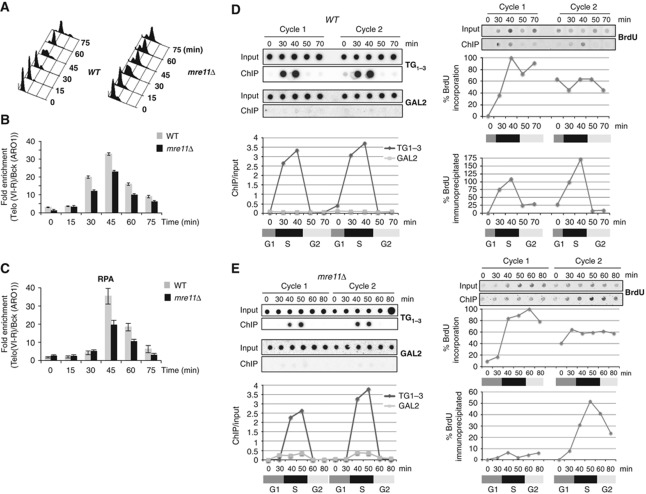
Deletion of MRE11 has been shown to reduce the telomere binding of the telomerase holoenzyme but to have only a modest effect on the binding of Cdc13 to telomeres (Goudsouzian et al, 2006; Faure et al, 2010). This prompted us to examine the role of MRE11 in the binding of RPA to telomeric DNA. For this purpose, we analysed the kinetics of association of RPA to telomeres in wild-type (WT) and mre11Δ strains. As an internal control for our experiments, we used strains expressing Flag-tagged Cdc13 and analysed the binding of RPA and Cdc13 in the same samples. Cells were synchronized with α-factor and released from the G1-arrest. Samples were collected every 15 min after the G1-release (Figure 1A) and chromatin immunoprecipitation (ChIP) experiments were performed with either anti-Rfa1 or Flag–M2 antibodies (Figure 1B and C). In WT cells, the binding of both Cdc13 and RPA increases in S-phase reaching maximum in late S-phase at the same time point (45 min) after the G1-release (Figure 1B and C). With the time resolution of this experiment, Cdc13 and RPA bind to telomere with similar kinetics. In the mre11Δ strain, Cdc13 and RPA binding to telomeres followed essentially the same kinetic profile although their binding was slightly reduced (Figure 1B). However, the lower enrichment of telomere fragments in the RPA ChIP could be partially due to the reduced telomere length of mre11Δ cells (see below).
Figure 1.
RPA binds to both the leading and lagging telomeres, but only the binding to the leading telomere is Mre11-dependent. WT and mre11Δ cells expressing CDC13–Flag were harvested from YPD cultures at the indicated times after release from an α-factor block. Cell-cycle progression was followed by FACS analysis (A). Binding of Cdc13–Flag (B) and RPA (C) to telomeres was analysed by ChIP. The relative enrichment of bound telomeric DNA (TelVI-R) over background (ARO1) is represented. PL9T163 (RFA1–MYC) (D) and PL9T163 (RFA1–MYC) mre11∷HIS3cells (E) were arrested in G1 with α-factor. BrdU was added to the medium (SD-LEU) 30 min before the cells were released from the α-factor block. α-factor was added again to obtain synchronized cells for the second cell cycle. Cells were washed and released into a new cell cycle in the absence of BrdU. ChIPs were performed with anti-myc (9E10) antibody at the indicated time points for the two successive cell cycles. The DNAs associated with the 9E10-ChIPs were spotted onto Hybond-N+ membrane and hybridize with a TG1−3 or a GAL2 probe as indicated in the Supplementary data (left panel). The signals were quantified with ‘Image Gauge’ software, the ratios ChIP/input signal are represented. The phases of cell cycle, monitored by FACS (data not shown), are represented by boxes. For BrdU detection (right panel), the DNAs associated to the 9E10-ChIPs were spotted onto a nitrocellulose membrane and immunodetection of BrdU was performed using a monoclonal antibody against BrdU. The fluorescence signal of each dot was quantified as indicated in the Supplementary data. BrdU incorporation was plotted as the signal of each dot normalized to the highest BrdU signal. The BrdU immunoprecipitated with the ChIP at each time point is normalized to the corresponding input signal.
We recently reported that lagging-strand and leading-strand synthesis of chromosomes leads to two structurally distinct ends at the telomeres. We have shown that Cdc13 and the telomerase subunits Est1 and Est2 can bind to the two daughter telomeres, but Mre11 promotes only their binding to the leading-strand telomere (Faure et al, 2010). To test the binding of RPA to the two daughter telomeres, we performed the assay described in Faure et al (2010). We label synchronized cells with BrdU during one cell cycle, chase BrdU and follow the cells for an additional cell cycle. We take samples throughout the two cell cycles and analyse the telomeric binding of RPA and the amount of BrdU in the immunoprecipitated DNA. Because S. cerevisiae telomere repeats are C1-3A/TG1–3, we are able to distinguish to which of the two daughter telomeres a given protein binds to by analysing the presence of BrdU in the ChIP. Cells that have the ability to incorporate BrdU (PL9T163 and PL9T163 mre11Δ) were harvested at different time points after the G1-release from the first and the second cell cycle, and divided into three samples. FACS analysis, ChIP experiments and BrdU incorporations were performed on each sample (Figure 1D and E; Supplementary Figure S1). For both strains PL9T163 and PL9T163 mre11Δ, the telomere association of RPA analysed by ChIP followed by qPCR showed a peak of telomere association in the late S-phase of the two successive cell cycles (Supplementary Figure S1). Consistent with Figure 1C, RPA telomere binding is reduced in late S-phase in mre11Δ cells. To determine whether the lower telomere binding of RPA could be at least in part due to the reduced telomere length of mre11Δ cells, the same ChIPs were then analysed by hybridization with a TG1−3 probe and a genomic control GAL2 probe (Figure 1D and E). We found that binding of RPA to telomeres in late S-phase was similar in WT and mre11Δ cells when the RPA ChIPs were normalized to the input DNAs. No hybridization signal was observed at any of the time points when the membrane was hybridized with the GAL2 probe. Probably, the time of residence of replicating RPA is too short at the GAL2 locus to give a detectable signal. We concluded that a large fraction of telomere-bound RPA detected by ChIP binds to telomeres independently of Mre11.
For each time point, we then analysed the incorporated BrdU in the RPA ChIP by a spot assay (Figure 1D and E). In WT cells (PL9T163), BrdU signals were mainly obtained for the time points when RPA binds to telomeres. BrdU was detected for the two consecutive cell cycles. In contrast in PL9T163 mre11Δ cells, BrdU signals were strongly reduced during the first cell cycle but not during the second cell cycle. Therefore, in mre11Δ cells, RPA telomere binding at the leading strand is decreased. Note that some BrdU was associated to the RPA ChIP at 60 and 80 min time points (Figure 1E), suggesting that in mre11Δ cells not all the BrdU signals correspond to telomeric DNA. These results support the notion that RPA binds to telomeric ssDNA during the lagging-strand telomere replication as well as the leading-strand telomere, but the presence of RPA to the leading-strand telomere is dependent on Mre11. Given the role of Mre11 in 5′ telomeric resection, the most likely interpretation of our results is that binding of RPA to the leading-strand telomere results from 5′ telomeric resection. However, one cannot exclude that another activity of Mre11 not related to resection could be required for RPA binding to the leading telomere.
RPA co-precipitates with yKu and TLC1
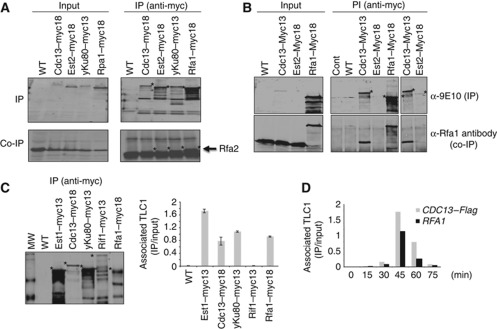
To further document the association of RPA with telomeres, we tested its interaction with several telomeric proteins. We immunoprecipitated Rfa1–myc, yKu80–myc, Cdc13–myc and Est2–myc with anti-Myc antibodies and tested for the presence of Rfa2 (the middle subunit of RPA) with a polyclonal antibody against Rfa2. As shown in Figure 2A, Rfa2 is efficiently co-immunoprecipitated with Rfa1 (our positive control) and with yKu80. Moreover, a very faint band corresponding to Rfa2 was observed in Cdc13 and Est2 immunoprecipitates that is completely absent when the immunoprecipitation (IP) was performed on cell extracts from an untagged strain. This result suggests that RPA interacts with Cdc13 and Est2. To further document this point, we immunoprecipitated Myc-tagged versions of Est2 and Cdc13 in late S-phase and analysed the presence of Rfa1 in the Cdc13 and Est2 IPs. As shown in Figure 2B, Rfa1 is efficiently co-immunoprecipitated with Cdc13 and weakly co-immunoprecipitated with Est2. The lower amount of RPA in the Est2 immunoprecipitates may reflect the low efficiency of the Est2 IP. These results indicate that RPA shows an association with yKu (see further), Cdc13 and Est2 suggesting the existence of a transient complex comprising RPA, yKu, Cdc13 and the telomerase holoenzyme.
Figure 2.
RPA interacts with yKu80, Cdc13 and TLC1 in vivo. (A) IPs were carried out using anti-Myc antibodies from cell extracts prepared from cells expressing the indicated proteins. The co-IP of RPA (co-IP) is monitored with anti-Rfa2 antibodies. The asterisks indicate the position of the immunoprecipitated Myc-tagged proteins. (B) WT, Cdc13–Myc13, Est2–Myc18 and Rfa1–Myc18 strains were synchronized in G1 with α-factor and released into the cell cycle. Cells were collected in late S-phase for IP using anti-Myc antibodies. The co-IP was revealed with an anti-Rfa1 antibody (lower panel). The asterisks indicate the co-immunoprecipitated Rfa1. No protein extract (cont) and untagged strain (WT) were used as controls to monitor the specificity of the IP. Inputs represent 2% of the IP. (C) (Left panel) IPs were carried out using anti-Myc antibodies and protein extracts prepared from cells expressing the indicated proteins. The first lane is untagged strain control. (Right panel) RNAs associated to the indicated IPs were extracted and analysed by reverse transcriptase assays followed by qPCR with primers specific for TLC1 as described by Fisher et al (2004). (D) At each indicated time point after release from a G1-arrest, IPs were carried out using anti-Flag (M2) or anti-Rfa1 antibodies on protein extracts prepared from the same cultures. RNAs recovered by the IPs were extracted and analysed by RT–PCR with primers specific for TLC1 and quantified by qPCR. Asterisks represent background bands.
The Zakian's laboratory showed by IP followed by RT–PCR that yKu interacts with TLC1 (Fisher et al, 2004) thereby providing a sensitive assay to monitor the yKu80/TLC1 interaction since this association could not be observed by IP followed by northern analysed with a TLC1 probe (Stellwagen et al, 2003). To confirm the RPA/telomerase interaction, we immunoprecipitated Rfa1–Myc and performed RT–PCR experiments using primers specific for TLC1 following the protocol described by Fisher (Figure 2C). As controls, we did the same IP experiments in cells expressing a myc-tagged version of yKu80, Est1 or Cdc13 that have been all shown to interact directly or indirectly with TLC1 (Hughes et al, 2000; Stellwagen et al, 2003; Fisher et al, 2004; Zappulla and Cech, 2004; Li et al, 2009) and in cells expressing Rif1–Myc that interacts with Rap1 and that is bound to telomeric chromatin (Wotton and Shore, 1997; Bourns et al, 1998). TLC1 was enriched in the immunoprecipitates of extracts from cells expressing Rfa1–myc, Est1–myc, Cdc13–myc and yKu80–myc but was not detected in Rif1–myc immunoprecipitates (Figure 2C). As additional controls, TLC1 was not found in the anti-Myc immunoprecipitates from the cells expressing an untagged version of Rfa1. Furthermore, the U1 snRNA was not found associated with Rfa1 (Supplementary Figure S2A). Finally, the association of RPA with TLC1 was sensitive to the addition of RNase in the immunoprecipitates but insensitive to the addition of DNase (Supplementary Figure S2B).
We next measured the association of RPA with TLC1 along the cell cycle. IPs of Cdc13–Flag and Rfa1 followed by RT–PCR were done on the same cell samples, those that were used to perform the ChIPs shown in Figure 1B and C. The amount of TLC1 associated with Rfa1 and Cdc13 was measured at each time point (Figure 2D). The interaction of TLC1 with either Rfa1 or Cdc13 was not detectable in G1 and peaked at 45 min after the G1-release when binding of both RPA and Cdc13 at telomeres is at its maximum. From these experiments, we conclude that RPA associates with TLC1 by the time RPA and Cdc13 telomere binding are at their maximum.
RPA/yKu interaction is maximum in late S-phase
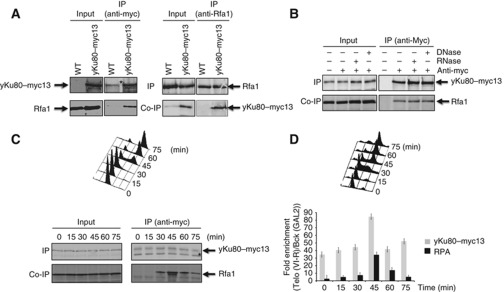
To characterize the interaction between RPA and yKu, we first checked that yKu80 and Rfa1 could be reciprocally co-immunoprecipitated (Figure 3A). In addition, we demonstrated that this interaction still occurs in the presence of DNase and RNase, indicating that the interaction RPA/yKu is mediated neither by DNA nor by TLC1 (Figure 3B). The association of RPA with yKu appears to be cell cycle regulated since this interaction is absent in G1 and early S-phase, is detectable in S-phase and peaks in late S-phase (Figure 3C). We next asked whether the interaction of RPA with yKu coincides with the time by which RPA and yKu telomere binding are at their maximum. As above, to obtain an internal control we used a strain carrying a Myc-tagged yKu80 and analysed in the same samples the binding of RPA and yKu80 to telomeres. We performed ChIP experiments in WT cells using either anti-Rfa1 or anti-Myc antibodies (Figure 3D). The results indicate that yKu80, which is constitutively bound to telomeres, peaks in late S-phase at the same time as RPA (Figure 3D). Taken together, these results indicate that RPA and yKu interact with each other in S-phase and G2, but their interaction is maximum at the end of S-phase, when both proteins are present at telomeres.
Figure 3.
RPA interacts with yKu80. (A) RPA co-precipitates with yku80–myc. IPs were carried out with either anti-myc (left panel) or Rfa1 antibodies (right panel) from cells expressing yKu80–myc. Co-precipitation was analysed by western blot with either anti-Rfa1 (left panel) or anti-myc antibodies (right panel). (B) The RPA/yKu80–myc interaction is DNase- and RNase-independent. yKu80–myc was immunoprecipitated with anti-myc antibodies in the presence of RNase or DNase. The co-precipitation of Rfa1 was analysed by western blot with anti-Rfa1 antibodies. (C) RPA/yKu80–myc interaction is cell cycle regulated. Interaction of yKu80–myc with RPA was analysed as described above on cells harvested at the indicated times after release from an α-factor block. Cell-cycle progression (upper panel), co-IP (lower panel). Asterisks represent background bands. (D) Binding of yKu80–Myc and RPA to telomeres was analysed by ChIP.
yKu and the telomerase subunit Est1 contribute to the interaction of RPA with telomerase
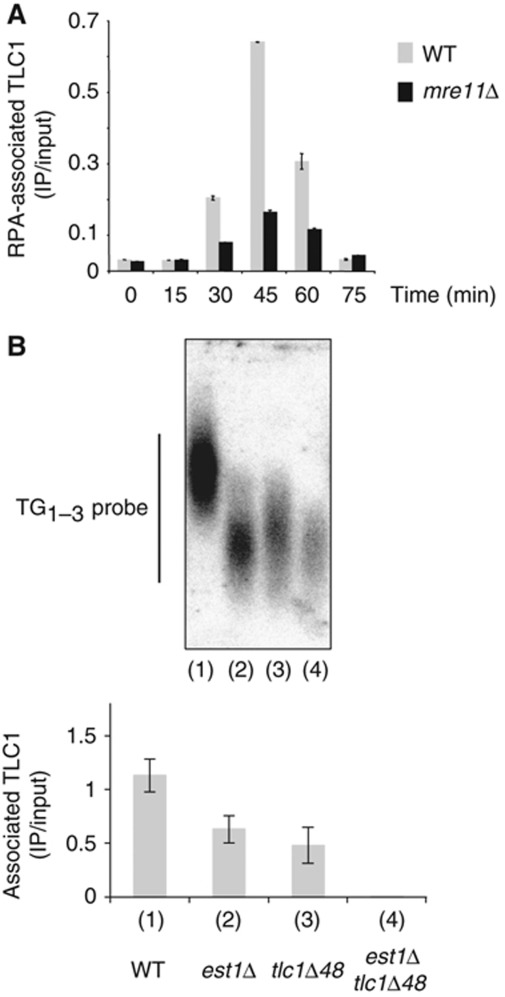
To understand how RPA interacts with telomerase, we first analysed whether the deletion of MRE11 interferes with the association between RPA and TLC1. We analysed the presence of TLC1 in the Rfa1 immunoprecipitates from WT and mre11Δ cells (Figure 4A). We found that association between RPA and TLC1 was severely reduced in mre11Δ cells (Figure 4A), indicating that impairing telomerase binding to telomeres almost abolished the interaction between RPA and TLC1.
Figure 4.
The interaction between RPA and telomerase is mediated by yKu and Est1. (A) The RPA/TLC1 co-precipitation requires Mre11.At each time point, IPs with anti-Rfa1 antibodies were carried out on protein extracts prepared from the same cultures than those used for the ChIP of Figure 1. RNAs associated to the IPs were extracted and analysed by RT–PCR with primers specific for TLC1. (B) MATa/α TEL∷URA3-VII-L tlc1Δ48/TLC1 est1Δ∷HIS3/EST1 diploid cells (Chan et al, 2008) were sporulated to generate spores of the indicated genotypes. (Top) Telomere length of the resulting cells were analysed after about 25 generations by Southern blot with a TG1−3 probe (bottom). For each spore, the RPA/TLC1 interaction was measured by IP directed against Rfa1 followed by RT–qPCR with primers specific for TLC1.
We further asked whether the binding of RPA to TLC1 could be mediated either by yKu (this work) or by Est1 that has been shown to genetically interact with RPA (Schramke et al, 2004) and to bind in vitro with RPA (Wu and Zakian, 2011). To test whether the interaction of RPA with TLC1 is bridged by either yKu80 and/or Est1, we used the tlc1Δ48 mutation that eliminates the specific interaction between yKu80 and TLC1 (Peterson et al, 2001) and a deletion of EST1 (est1Δ). Importantly, Est2 is completely absent from the telomere when the tlc1Δ48 allele is combined with the deletion of EST1 (Chan et al, 2008). We sporulated a diploid strain heterozygous for est1Δ/EST1 and for tlc1Δ48/TLC1 (Chan et al, 2008) to generate tetratype tetrads carrying the following mutant spores (WT, tlc1Δ48, est1Δ, tlc1Δ48 est1Δ). The telomere lengths of the mutant cells are consistent with their genotypes (Figure 4B, top). For each spore, we analysed the RPA/TLC1 association by IP of RPA followed by RT–PCR. In the single mutants (tlc1Δ48 and est1Δ), the RPA/TLC1 interaction decreased by about two-fold (Figure 4B, bottom) while the RPA/TLC1 interaction was not detectable in the tlc1Δ48 est1Δ double mutant. We concluded that both yKu and Est1 contribute to the interaction between RPA and TLC1. Taken together, these results indicate that the suppression of the telomerase recruiting pathways abolishes the interaction between RPA and TLC1.
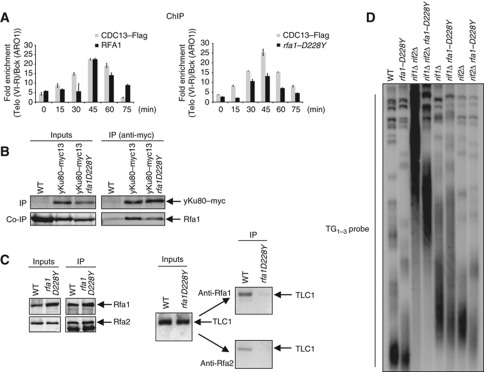
The rfa1–D228Y allele exhibits reduced interaction with telomeres, yKu and TLC1
The Rfa1–D228Y mutation was isolated in a screen aimed to find suppressors that would stimulate Rad52-independent pathways for direct-repeat recombination (Smith et al, 1995). It was proposed that in the absence of WT Rad52, the interaction of RPA with single-stranded DNA inhibits strand annealing, and that this inhibition is overcome by the rfa1–D228Y mutation (Smith and Rothstein, 1995, 1999). Moreover, a synergistic reduction in telomere length was observed when the rfa1–D228Y allele was combined with a null mutation of yKU70 (Smith et al, 2000). As shown in Supplementary Figure S3, the D228 residue lies within a conserved region, that is different from the canonical OB-fold region involved in the binding of the ssDNA, and which is likely to be involved in an interaction with another partner than ssDNA.
We analysed the binding of Rfa1–D228Y to telomeres and tested its co-precipitation with yKu and TLC1. We used a strain carrying a Flag-tagged Cdc13 and analysed in parallel the binding of either Rfa1 or Rfa1–D228Y with the one of Cdc13. Cells were harvested at different times after the G1-release. ChIP experiments were performed on the same cell samples with either anti-Rfa1 or Flag antibodies (Figure 5A). While Cdc13 binding remained unchanged in rfa1–D228Y cells, the binding of Rfa1–D228Y to telomeres was reduced about two-fold when compared with the WT Rfa1 (at 45 min after the G1-release) (Figure 5A). We then tested whether the Rfa1–D228Y retained the ability to interact with yKu. As exemplified in Figure 5B, we reproducibly observed that Rfa1–D228Y has a weaker interaction with yKu80. Finally, we immunoprecipitated RPA with antibodies directed against either the large subunit Rfa1 or the middle subunit Rfa2, in asynchronous WT and rfa1–D228Y cell cultures and analysed the presence of TLC1 in the immunoprecipitates as described above (Figure 5C). The amounts of Rfa1 (or Rfa1–D228Y) and Rfa2 immunoprecipitated from either WT or rfa1–D228Y cell extracts were checked by western blot (Figure 5C, left). We found that TLC1 was barely detectable in Rfa1 or Rfa2 immunoprecipitates obtained from the rfa1–D228Y cell extracts (Figure 5C, right). The fact that Rfa1–D228Y does not interact with TLC1 suggests that the D228Y mutation affects both pathways of RPA interaction with telomerase. Because Rfa1 and Rfa1–D228Y binds similarly to resected DNA after an HO-cut (unpublished observation), the lower telomeric binding of Rfa1–D228Y is likely to be due to defective protein/protein interactions.
Figure 5.
The Rfa1–D228Y mutation impairs the interaction with TLC1, yKu80 and affects telomere length. (A) Telomeric DNA binding of Cdc13–flag and Rfa1 was monitored by ChIP performed with anti-Flag (M2) or anti-rfa1 antibodies (ChIP; upper panels) from CDC13–flag and CDC13–flag rfa1–D228Y cells harvested at the indicated times after release from an α-factor block. The relative enrichment of bound telomeric DNA (TelVI-R) over background (ARO1) is represented. (B) IPs were carried out using anti-Myc antibodies from extracts prepared from WT (untagged), yku80–myc and yku80–myc rfa1–D228Y cells. The co-IP was revealed with an anti-Rfa1 antibody. (C) (Left panel) The Rfa1–D228Y mutation impairs the interaction between RPA and TLC1. Protein extracts from asynchronous WT and rfa1–D228Y cells were subjected to IP using either anti-Rfa1 or anti-Rfa2 antibodies. The presence of Rfa1 (or Rfa1–D228Y) and Rfa2 in the IPs was analysed by western blot. (Right panel) The presence of TLC1 in the Rfa1 and Rfa2 immunoprecipitates from the WT and the rfa1–D228Y strains was monitored as described above. (D) Southern blot of XhoI-digested genomic DNA hybridized with a TG1−3 probe from the indicated mutant strains. After tetrad dissection of the diploid strains, the indicated segregants were restreaked >10 times on YPD plates prior DNA preparation.
The rfa1–D228Y allele counteracts the rif1Δ rif2Δ overelongation of telomeres
To understand the effect of the rfa1–D228Y allele in telomere length regulation, we analysed the effect of the rfa1–D228Y allele in cells lacking Rif1 and Rif2 whose combined absence causes a telomerase-dependent dramatic telomere lengthening (Wotton and Shore, 1997; Levy and Blackburn, 2004). We crossed a rif1Δ rif2Δ double mutant with the rfa1–D228Y strain and sporulated the resulting diploid. Telomere length of the generated mutant spores were analysed after >10 restreaks to equilibrate telomere size. As expected, the lack of either Rif1 or Rif2 causes telomere lengthening, which is dramatically increased when both proteins are absent (Figure 5D). When the rfa1–D228Y allele was combined with each single rif mutant, we observed a moderate decrease of telomere size of the double mutants while the rfa1–D228Y allele strongly reduced the telomere length of the rif1Δ rif2Δ double mutant (Figure 5D). We then analysed the effect of the rfa1–D228Y allele combined with tel1Δ. We crossed tel1Δ with the rfa1–D228Y mutant. The resulting diploids were sporulated and telomere length of the spores was analysed as above. For different tetratypes, we observed that the single tel1Δ and double tel1Δ rfa1–D228Y mutants had similar telomere lengths (Supplementary Figure S4), suggesting that the rfa1–D228Y allele and tel1Δ affects the same pathway as was the case for the rfa2Δ40 allele (see further) that we previously described (Schramke et al, 2004).
We finally analysed the binding of RPA and Cdc13 in the absence of Rif1 and Rif2 (Supplementary Figure S5). Binding of RPA and Cdc13 were analysed by ChIP followed by hybridization with a TG1−3 probe since telomeres are overelongated in a rif1Δ rif2Δ double mutant. The results show that the total amount of Cdc13 bound to telomeres is similar in WT and rif1Δ rif2Δ cells (Supplementary Figure S5B) but when the level of telomere-bound Cdc13 is normalized to the total amount of TG1−3 sequences, the level of Cdc13 drops by a factor 4 (Supplementary Figure S5C). RPA telomere binding increases in the rif1Δ rif2Δ double mutant (Supplementary Figure S5B) but proportionately to the total TG1−3, there is less telomere DNA in the RPA ChIP in the rif1Δ rif2Δ mutant than in a WT strain (Supplementary Figure S5C). These results suggest that Cdc13 is mainly present at the end of the telomere, whereas RPA may be present at both, the replicating telomere and the telomere end. We then measured the interaction between RPA and telomerase in the rif1Δ rif2Δ double mutant. We observed a modest increase (30%) of the association between RPA and TLC1 in rif1Δ rif2Δ cells that may reflect an increased association between RPA and telomerase.
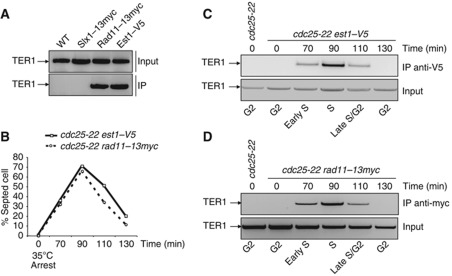
RPA is associated with TER1 in Schizosaccharomyces pombe
The RNA component of the telomerase was recently identified in S. pombe (Leonardi et al, 2008; Webb and Zakian, 2008). This telomerase RNA (TER1) was found to be over 1200 nt similar in size to TLC1 and to interact with Est1 and Trt1. This led us to test whether the interaction between RPA and telomerase RNA was conserved in fission yeast. To this purpose, we used a strain expressing a 13myc-tagged version of the large subunit of S. pombe RPA (spRPA) called Rad11 (Ono et al, 2003). As positive and negative controls, we used strains expressing a V5-tagged Est1 (PM Dehé and J Cooper, unpublished results) and the 13myc-tagged structure-specific endonuclease Slx1, respectively. As shown in Figure 6A, Rad11 and Est1 were found to both interact with TER1 while no interaction could be detected with Slx1, suggesting that the interaction between RPA and telomerase RNA is conserved in S. pombe. We next asked whether the association of RPA with TER1 is cell cycle regulated as we have shown for S. cerevisiae. We synchronized S. pombe cells expressing either Rad11–Myc or the Est1–V5 by using the cdc25-22 mutant, which block cells at the G2/M boundary at the restrictive temperature (Figure 6B–D). Cells were released from the G2/M block and the binding of TER1 to Rad11–13myc and Est1–V5 was analysed at each time point. The presence of TER1 was detected in both the Rad11–13myc and Est1–V5 immunoprecipitates 90 min after release when the number of septed cells is maximum (Figure 6B–D). Our results indicate that the interaction between RPA and TER1 is cell cycle regulated and coincides with the association of Est1 with TER1.
Figure 6.
The spRPA/TER1 interaction is conserved in fission yeast and coincides with Est1/TER1 association. (A) TER1 co-immunoprecipitates with Rad11 in asynchronous cells. Co-IP of Rad11 and TER1 has been performed as described by Webb (Webb and Zakian, 2008). IPs were carried out using anti-Myc or anti-V5 antibodies with cell extracts prepared from WT and Slx1–myc, Rad11–myc and Est1–V5 expressing strains. RT–PCR has been performed with TER1-specific oligonucleotides. The signal corresponds to the amplification of an internal fragment of TER1. (B) cdc25-22 rad11–myc and cdc25 est1–V5 strains were grown at 25°C until OD=0.4 and then shifted to 36°C for 3.5 h to synchronize them. Cells were then released at 25°C from the G2 block and aliquots were collected at the indicated time points. The septation index is an indicator of the cell-cycle phase. Est1–V5 (C) and Rad11–myc (D) have been immunoprecipitated from extracts of the corresponding strains and the presence of TER1 was analysed by RT–PCR as above.
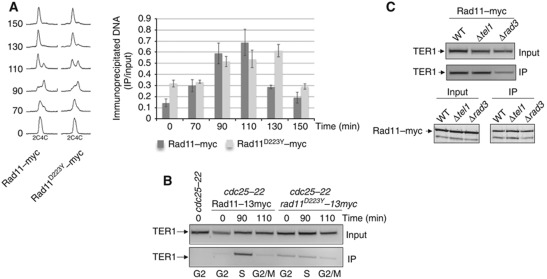
The Rad11–D223Y mutation affects the interaction between spRPA and TER1
To further characterize the interaction between spRPA and TER1, we analysed whether this interaction is affected by the point mutation Rad11–D223Y (the corresponding mutation of Rfa1–D228Y, a generous gift from M Ueno) (Ono et al, 2003). We then used a strain expressing a 13myc-tagged version of the Rad11–D223Y whose expression is identical to Rad11 (Kibe et al, 2007). We synchronized S. pombe cells expressing either Rad11–Myc or Rad11–D223Y–Myc by using the cdc25-22 mutant. Cells were released from the G2/M block and harvested at different time points after the shift to permissive temperature. Cell-cycle progression was followed by FACS. We first analysed by ChIP the binding of Rad11 and Rad11–D223Y along the cell cycle (Figure 7A). Rad11–D223Y binds to telomeres in late S-phase as the WT Rad11 except the mutant protein stays longer associated to telomeres (Figure 7A). Then, we analysed the binding of TER1 to Rad11–13myc and Rad11–D223Y–Myc (Figure 7B). The interaction of Rad11–13myc with TER1 was readily detected in the immunoprecipitates at the 90-min time point when the number of septed cells is maximum while the interaction between Rad11–D223Y–Myc and TER1 was severely reduced (four- to five-fold) under the same conditions. We next asked whether the interaction between spRPA and TER1 is affected by the absence of Rad3 that leads to a substantial telomere shortening (Naito et al, 1998; Nakamura et al, 2002; Kanoh et al, 2003). As shown in Figure 7C, inactivation of Rad3 leads to substantial reduction of the Rad11/TER1 interaction in contrast to the deletion of tel1 that has no effect on telomere length by itself and on the Rad11/TER1 interaction.
Figure 7.
The Rad11–D223Y mutation affects the interaction with TER1. (A) (Left panel) FACS analysis of cdc25-22 rad11–myc and cdc25-22 rad11–D223Y–myc strains were grown at 25°C until OD=0.4 and shifted to 36°C for 3.5 h to synchronize them. Cells were then released at 25°C from the G2 block. (Right panel) Binding of Rad11–myc and Rad11–D223Y–myc to telomeres was analysed by ChIP performed with an anti-myc antibody at the indicated time points after release. The relative precipitation of bound telomeric DNA is represented. (B) Rad11–myc and Rad11–D223Y–myc proteins have been immunoprecipitated at the indicated time points after release from the G2 block and the presence of TER1 analysed by RT–PCR. (C) rad3 deletion affects the RPA/TER1 interaction in asynchronous cells. Co-IP of Rad11 and TER1 has been performed as described above in WT, Δtel1 and Δrad3 strains.
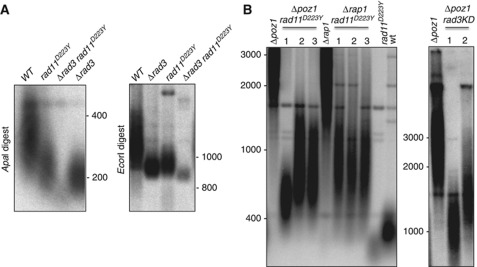
The Rad11–D223Y mutation antagonizes the Δrap1- and Δpoz1-dependent overelongation of telomeres
We next asked whether the rad3Δ and the rad11D223Y alleles act in the same pathway. As shown in Figure 8A, rad11D223Y has a synergistic reduction in telomere length with rad3Δ. Although telomeres are very short, the chromosomes are not circular (data not shown). This result suggests that the short telomere length phenotype conferred by the rad11D223Y allele was not related to Rad3 recruitment or activation.
Figure 8.
The Rad11–D223Y mutation antagonizes the Δrap1- and Δpoz1-dependent overelongation of telomeres. (A) The telomere length of WT, rad11D223Y, Δrad3 and Δrad3 rad11D223Y was analysed by Southern hybridization. Genomic DNAs were digested by either ApaI or EcoRI, fractioned by 1% agarose gel electrophoresis and hybridized with the telomeric probe as described previously (Nakamura et al, 2002). (B) (Left panel) Telomere length of WT, rad11D223Y, Δrap1, Δpoz1 and three independent clones of Δrap1 rad11D223Y and Δpoz1 rad11D223Y double mutants. Genomic DNAs were digested by ApaI and treated as above; right panel, telomere length of Δpoz1 rad3KD (kinase deleted) double mutants.
The group of M Ueno reported that the rad11–D223Y mutation reduces by itself telomere length and the telomere elongation of rap1-deleted cells (Ono et al, 2003; Kibe et al, 2007). Since the Rfa1–D228Y mutation counteracts the rif1Δ rif2Δ overelongation of telomeres in S. cerevisiae, we asked whether the rad11–D223Y mutation would reduce as well the telomerase-dependent elongation of telomeres caused by the inactivation of Poz1. Poz1 has been proposed to bridge the Taz1/Rap1 complex to the Pot1/Tpz1/Ccq1 complex and inactivation of Poz1 results in overelongated telomeres (Miyoshi et al, 2008; Tomita and Cooper, 2008). As shown in Figure 8B, the rad11–D223Y mutation suppresses the telomere overelongation of Δpoz1 cells as does the deletion of the kinase domain of Rad3 (rad3KD).
Discussion
RPA binds to resected telomeres and associates with telomerase
We have previously shown that RPA is bound at telomeres in late S-phase and proposed that RPA acts in the telomerase pathway (Schramke et al, 2004). However, the reason for which RPA is specifically found associated to telomeres in late S-phase has been questioned. Indeed, the increased time of residence of RPA at telomeres, as evidenced by ChIP, could reflect the association of RPA with the replication machinery, which is known to pause during telomere replication in late S-phase (Makovets et al, 2004). Alternatively, telomere binding of RPA could reflect the binding of RPA to telomeric ssDNA that is generated by the telomeric 5′ resection that also occurs in late S-phase. As an alternative explanation, not mutually exclusive with the two previous ones, time of residency of RPA at telomeres could be increased by its association to the telomerase complex or due to a role of RPA in synthesis of the complementary C-strand.
Using a more sensitive assay, the results presented here provide evidence for an interaction between RPA and telomerase by the time when telomerase is associated to telomeres in late S-phase. Several lines of evidence indicate that this interaction is specific. Indeed, RPA interacts with TLC1 in a narrow window in late S-phase and mutations in Rfa1 abolish the association between TLC1 and RPA. Moreover, we show that in mre11Δ cells, the decreased interaction between RPA and TLC1 is correlated to the decrease of telomere-bound telomerase (Goudsouzian et al, 2006).
Our results indicate that a significant fraction of the RPA that we are able to ChIP is localized at the lagging-strand telomere. It is likely that both the time of residence and the amount of RPA is higher at the lagging strand than at the leading strand. This may explain why we do not observe a significant reduction of the total amount of telomere-bound RPA in mre11Δ cells. Nevertheless, the fact that deleting MRE11 affects only the binding of RPA at the leading telomere suggests that RPA binds to resected telomere as does Cdc13. However, the absence of RPA/telomerase interaction in mre11Δ cells is likely to be due to the role of Mre11 in recruiting telomerase rather than to its role in resection.
The interaction between RPA and telomerase is mediated by yKu and Est1
In this study, we provide evidence for an interaction between RPA, yKu, Cdc13 and telomerase, suggesting the formation of a transient complex at chromosome ends. Further work will be required to analyse whether formation of this complex is related to replication fork progression since RPA interacts with several proteins of the replisome.
These interactions RPA/yKu and RPA/TLC1 are cell cycle regulated and still occur in the presence of DNase and RNase, indicating that these interactions are mediated by protein/protein interactions rather than by DNA or RNA. Although our experiments do not prove formally that these interactions occur at telomeres, this possibility is strongly supported by the timing of the RPA/yKu interaction that peaks when the binding of both RPA and yKu at telomeres is maximum. Consistent with our recent study (Faure et al, 2010), we found that yKu is constitutively bound at telomeres but in addition peaks in late S-phase as does Cdc13, RPA and telomerase.
The deletion of the 48-nt stem-loop of TLC1 that links TLC1 to yKu80 reduces only partially the interaction between RPA and TLC1, indicating that another protein bridges RPA to telomerase. We previously observed that a specific allele of RPA (rfa2Δ40) eliminating the RFA2 intron and amino acids 3–39 of Rfa2 resulted in a severe telomere shortening and a three-fold decrease of Est1 binding to telomeres (Schramke et al, 2004). This effect is specific since the control construct removing only the intron of RFA2 did not lead to telomere shortening (Supplementary Figure S6). However, we noticed that cells carrying the rfa2Δ40 allele gave very rapidly a very high number of revertants that can be distinguished from the original mutant by their normal growth and their WT length telomeres. These results prompted us to look at the role of Est1 in the interaction between RPA and TLC1. We found that deleting EST1 by itself reduces two-fold the interaction between RPA and TLC1 and that the double deletion tlc1Δ48 est1Δ completely abolishes the interaction between RPA and TLC1. We therefore concluded that RPA contacts telomerase via both proteins Est1 and yKu. Consistent with this assumption, Est1 interacts with RPA in vitro although with a lower affinity than with Cdc13 (Wu and Zakian, 2011).
The role of RPA in telomere length control
In this work, we show that the rfa1–D228Y allele reduces the length of both rif1Δ and rif2Δ single mutant, and overall the length of the rif1Δ rif2Δ double mutant. Rif1 and Rif2 prevent telomere elongation by telomerase by two different mechanisms. Rif2 has been shown to inhibit nucleolytic processing at telomeres and to reduce MRX and Tel1 access to telomeres (Grossi et al, 2004; Sabourin et al, 2007; Hirano et al, 2009; Bonetti et al, 2010). Consistent with these results, the absence of Rif2 abolishes the preferential binding of Tel1 to short versus WT length telomeres (McGee et al, 2010). Also, Rif1 has been recently shown to display a synthetic interaction with mutations that reduce the activity of Cdc13 or Stn1 and also of the polα–primase complex (Anbalagan et al, 2011). From this work, Longhese and collaborators propose a model in which Rif1 plays a role in assisting the CST complex in carrying out its essential telomere protection function. Since a rif1Δ mutant displays elongated telomeres, one implication of this model could be that Rif1 favours the interaction of Cdc13 with Stn1 and Ten1 to synthesize the C-strand and protect the telomere at the expense of the Cdc13–Est1 interaction. According to these results, the overelongated telomeres in rif1Δrif2Δ cells would be due to the increased access of MRX and Tel1 at long telomeres thereby favouring the action of telomerase, and also to the decreased synthesis of the C-strand. Given the fact that RPA promotes telomerase action in rif1Δ rif2Δ cells, interacts with yKu and telomerase, and is epistatic with Tel1, we favour a model in which RPA stimulates the elongation of the 5′ strand by telomerase through its interaction with yKu and Est1.
Interestingly, the rfa1–D228Y allele has a synthetic lethal interaction with conditional alleles of POL12 but not with conditional alleles of POL1, PRI1 (primase) or POL3 (Polδ) (Smith et al, 2000). The specificity of this genetic interaction led Smith et al (2000) to suggest that Rfa1 interacts physically directly or indirectly with Pol12. This genetic interaction combined to the fact that RPA stimulates polα–primase activity (Wold, 1997) and interacts with Mcm10, which in turn stabilizes polα–primase association with DNA (Ricke and Bielinsky, 2004) suggests that RPA may also regulate positively the C-strand synthesis at telomeres. In such a case, we would have expected that the rfa1–D228Y mutant would exhibit telomerase-dependent long telomeres as do conditional mutants defective in telomeric C-strand synthesis (Adams and Holm, 1996). The rfa1–D228Y allele by itself does not produce such a phenotype. However, we found that another allele of RFA1, rfa1–M2 that is synthetic lethal with conditional alleles of CDC17 (Longhese et al, 1994) causes an elongation of telomeres (Pierre Luciano, unpublished results). Therefore, rfa1–D228Y may affect at the same time the action of telomerase and the synthesis of the complementary strand explaining the telomere length phenotype of the rfa1–D228Y mutant. In support of this dual role of RPA, different alleles of RPA have opposing effects in respect to telomere length as it is the case for Cdc13 (Chandra et al, 2001).
RPA association with telomerase and function at telomeres is conserved in fission yeast
We show that fission yeast RPA also associates with telomerase in late S-phase and that the Rad11–D223Y mutation affects the association between spRPA and telomerase. This mutation leads to a clear reduction of telomere length in S. pombe. Interestingly, in S. pombe, spKu does not interact with TER1, the RNA component of telomerase. This may suggest that, in fission yeast, the absence of interaction between telomerase and spKu (Webb and Zakian, 2008) renders the activity of telomerase more dependent on the association between RPA and its RNA component. Interestingly, in S. pombe, the recruitment of RPA to telomeres coincides with one of the checkpoint protein Rad26 (orthologue of the mammalian ATRIP protein) (Moser et al, 2009) and both RPA and Rad3–Rad26 (ATR-ATRIP) have been shown to be important for telomere maintenance in fission yeast (Nakamura et al, 2002; Ono et al, 2003). However, our data suggest that Rad3 and RPA act in different pathways to stimulate telomerase activity. Nevertheless, we show that the association of the telomerase RNA to RPA and Est1 coincides in fission yeast. These observations raise the possibility that RPA recruitment to telomeres might be shortly followed by association of RPA to the telomerase complex and that this association plays an important role in telomere maintenance.
We found in addition that the Rad11–D223Y mutation strongly reduces the telomerase-dependent overelongation of telomeres of Δrap1 or Δpoz1 cells. It is not known how the absence of SpRap1 (or Poz1) leads to extended G-tails and elongated telomeres (Miller et al, 2005), but the absence of either SpRap1 or Poz1 somehow mimics the situation of budding yeast deleted of RIF1 and RIF2. The fact that the equivalent mutation of RPA produces the same effect in both yeast points out the conservation of the RPA function at telomeres. Our results suggest that in both yeast, RPA stimulates the elongation of the 5′ strand by telomerase. This assumption is further reinforced in S. pombe by the fact that the Rad11–D223Y mutation leads by itself to a clear reduction of telomere length (Ono et al, 2003). The mechanism by which the C-strand is synthesized is unknown in S. pombe. It will be interesting to determine whether RPA plays a role in this process.
Overall, our work indicates that RPA directly contributes to telomerase action at yeast telomeres. In mammalian cells, the recognition of telomeres by telomerase depends on TPP1, a protein containing the predicted structural protein domains OB-folds present in Cdc13 and Est1 (Abreu et al, 2010; Tejera et al, 2010). An additional OB-fold containing protein, related to an RPA large subunit was found to be associated with a high processive form of telomerase in the ciliate Tetrahymena thermophila (Min and Collins, 2009). Collectively, these findings converge towards the idea that various types of RPA and RPA-like proteins have been selected during evolution to bridge telomerase to telomeres to increase telomerase processivity, and to participate to C-strand synthesis (Giraud-Panis et al, 2010). Whether RPA increases processivity of yeast telomerases remains to be established. Future experiments will certainly shed new lights in this aspect of the increasingly complex world of telomerase regulation.
Materials and methods
Oligonucleotides and strains are shown in Supplementary Tables S1 and S2, respectively.
Co-IP with telomerase RNA
The interaction between RPA and TLC1 and between spEst1 and Rad11 with TER1 was analysed as described by Fisher et al (2004) and Webb and Zakian (2008), respectively.
Co-IP of RPA
For asynchronous cell cultures, yeast were grown at 30°C in YPD to OD600=0.6–0.8. Extracts were lysed with glass-beads in TMG-50 (10 mM Tris–HCl, pH 8.0, 0.1 mM MgCl2, 10% (V/V) glycerol, 50 mM NaCl, 0.1 mM EDTA, 0.1 mM DTT) containing protease inhibitors, 10 μM MG-132 and 10 μg/ml RNase inhibitor (Sigma). Extracts were adjusted to 0.5% (V/V) Tween-20 and incubated overnight at 4°C with monoclonal anti-Myc (9E10) or polyclonal anti-Rfa1. Dynabeads protein G (Invitrogen) equilibrated with TMG-50 plus 0.5% Tween-20 were then added and incubated for 4 h at 4°C. Beads were washed three times with TMG-50 plus 0.5% Tween-20, once with TMG-50 and resuspended in 1 × loading buffer. For DNase and RNase treatments, cell extracts were pre-incubated in TMG-50 without RNase inhibitor before IP either with 20 U of DNase I (Roche) or with 1 μg/ml of RNase A (Roche) for 15 min at RT.
For synchronous cell cultures, yeast cells grown at 30°C in YPD to OD600=0.6 cells were arrested in G1 by the addition of 10 μg/ml α-factor for 2 h. α-Factor was removed and cells were allowed to progress synchronously through the cell cycle. Samples were taken every 15 min for FACS and co-IP analysis.
Telomere Southern blot analysis
Genomic DNA was prepared from 15 ml of cells at OD600 nm=1 and digested with XhoI for S. cerevisiae and ApaI or EcoRI for S. pombe samples. The digested DNA was resolved in a 1.2% agarose gel and blotted onto a Hybond-N+ membrane. The DNA was hybridized with a radiolabelled telomeric repeats DNA fragment.
Chromatin immunoprecipitation
Cell-cycle progression and BrdU labelling in S. cerevisiae are described in the Supplementary data. In all experiments, errors bars represent the standard deviations from three independent experiments.
In S. pombe, cells were processed for ChIP analysis as previously described (Nakamura et al, 2002) with minor modifications. 9E10 monoclonal anti-myc antibody (Santa Cruz) was added to whole cell extracts and incubated over night at 4°C on rotator wheel, then magnetic Dynabeads (Invitrogen) were added for 4 h at 4°C. Recovered DNA was analysed by triplicate SYBR Green-based real-time PCR (Takara) using Telomeric and non-specific primers that are listed in Supplementary Table S1. Septation index or FACS analysis were monitored to ensure the correct cell synchronization.
Supplementary Material
Acknowledgments
We are very grateful to M Ueno, F Ishikawa, T Nakamura R, J Cooper, PM Dehé, R Rothstein, P Pasero, S Marcand and V Zakian for sharing yeast strains and plasmids. We thank Frederic Jourquin and Julien Audry for technical help, Raphaël Guerois for the structural analysis of the Rfa1–D228Y mutation and D Churikov for correcting the MS. VG and EG laboratories were supported by the ‘Agence Nationale de la Recherche’ (ANR programme blanc) and by the ‘Ligue Nationale contre le Cancer (LNCC) (équipe labellisée)’. SC has been supported by the ‘Association pour la Recherche sur le Cancer’ (ARC).
Author contributions: PL generated the data shown in Figures 1, 2, 3, 4 and 5 and Supplementary Figure S2; VF generated the data shown in Supplementary Figure S1; SC generated the data shown in Figures 6, 7 and 8; YC, MNS and YC generated the data shown in Figures 1 and 5; SB did the anti-Rfa1 and Rfa2 antisera; JB did in vitro work, all contributors participate in the design and interpretations of the results; PL, EG, MNS and VG wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP (2010) TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30: 2971–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AK, Holm C (1996) Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol Cell Biol 16: 4614–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbalagan S, Bonetti D, Lucchini G, Longhese MP (2011) Rif1 supports the function of the CST complex in yeast telomere capping. PLoS Genet 7: e1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arneric M, Lingner J (2007) Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep 8: 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Negrini S, Shore D (2004) Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell 16: 139–146 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D (2007) Increased association of telomerase with short telomeres in yeast. Genes Dev 2: 1726–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Shore D (2008) How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol Cell 31: 153–165 [DOI] [PubMed] [Google Scholar]
- Binz SK, Sheehan AM, Wold MS (2004) Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair 3: 1015–1024 [DOI] [PubMed] [Google Scholar]
- Bonetti D, Clerici M, Anbalagan S, Martina M, Lucchini G, Longhese MP (2010) Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet 6: e1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP (2009) Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell 35: 70–81 [DOI] [PubMed] [Google Scholar]
- Bourns BD, Alexander MK, Smith AM, Zakian VA (1998) Sir proteins, Rif proteins, and Cdc13 bind Saccharomyces telomeres in vivo. Mol Cell Biol 9: 5600–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A, Boule JB, Zakian VA (2008) Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet 4: e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev 15: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandjinou AT, Levesque N, Larose S, Lucier JF, Abou Elela S, Wellinger RJ (2004) A phylogenetically based secondary structure for the yeast telomerase RNA. Curr Biol 14: 1148–1158 [DOI] [PubMed] [Google Scholar]
- Dezwaan DC, Freeman BC (2009) The conserved Est1 protein stimulates telomerase DNA extension activity. Proc Natl Acad Sci (USA) 106: 17337–17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezwaan DC, Freeman BC (2010) Is there a telomere-bound ‘EST’ telomerase holoenzyme? Cell Cycle 9: 1913–1917 [DOI] [PubMed] [Google Scholar]
- Dionne I, Wellinger RJ (1998) Processing of telomeric DNA ends requires the passage of a replication fork. Nucleic Acids Res 26: 5365–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (1999) Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (2002) The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure V, Coulon S, Hardy J, Géli V (2010) The telomeric single-stranded DNA-binding protein Cdc13 and telomerase bind through different mechanisms at the lagging- and leading-strand telomeres. Mol Cell 38: 842–853 [DOI] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA (2004) Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Frank CJ, Hyde M, Greider CW (2006) Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell 24: 423–432 [DOI] [PubMed] [Google Scholar]
- Gao H, Toro TB, Paschini M, Braunstein-Ballew B, Cervantes RB, Lundblad V (2010) Telomerase recruitment in Saccharomyces cerevisiae is not dependent on Tel1-mediated phosphorylation of Cdc13. Genetics 186: 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V (2007) How telomeres are replicated. Nat Rev 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Teixeira MT, Géli V, Gilson E (2010) CST meets shelterin to keep telomeres in check. 2010. Mol Cell 39: 665–676 [DOI] [PubMed] [Google Scholar]
- Goudsouzian LK, Tuzon CT, Zakian VA (2006) S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol Cell 24: 603–610 [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M (2000) Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol Cell Biol 20: 8397–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D (2004) Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW (2007) Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell 27: 851–858 [DOI] [PubMed] [Google Scholar]
- Hirano Y, Fukunaga K, Sugimoto K (2009) Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell 33: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Evans SK, Weilbaecher RG, Lundblad V (2000) The Est3 protein is a subunit of yeast telomerase. Curr Biol 10: 809–812 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Francesconi S, Collura A, Schramke V, Ishikawa F, Baldacci G, Géli V (2003) The fission yeast spSet1p is a histone H3-K4 methyltransferase that functions in telomere maintenance and DNA repair in an ATM kinase Rad3-dependent pathway. J Mol Biol 326: 1081–1094 [DOI] [PubMed] [Google Scholar]
- Kibe T, Ono Y, Sato K, Ueno M (2007) Fission yeast Taz1 and RPA are synergistically required to prevent rapid telomere loss. Mol Biol Cell 18: 2378–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ (2004) The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V (1996) Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi J, Box JA, Bunch JT, Baumann P (2008) TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol 15: 26–33 [DOI] [PubMed] [Google Scholar]
- Levy DL, Blackburn EH (2004) Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol 24: 10857–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH (2009) Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136: 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA (1996) The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA 93: 13760–13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567 [DOI] [PubMed] [Google Scholar]
- Longhese MP, Plevani P, Lucchini G (1994) Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol Cell Biol 14: 7884–7890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee JS, Phillips JA, Chan A, Sabourin M, Paeschke K, Zakian VA (2010) Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat Struct Mol Biol 17: 1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S, Herskowitz I, Blackburn EH (2004) Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol 24: 4019–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory JC, Bashkirov VI, Trujillo KM, Solinger JA, Dominska M, Sung P, Heyer WD, Petes TD (2003) Amino acid changes in Xrs2p, Dun1p, and Rfa2p that remove the preferred targets of the ATM family of protein kinases do not affect DNA repair or telomere length in Saccharomyces cerevisiae. DNA Repair 2: 1041–1064 [DOI] [PubMed] [Google Scholar]
- Miller KM, Ferreira MG, Cooper JP (2005) Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J 24: 3128–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Collins K (2009) An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell 36: 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F (2008) Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344 [DOI] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM (2009) Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J 28: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, Podell ER, Cech TR (2008) Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol Cell Biol 28: 4152–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Matsuura A, Ishikawa F (1998) Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet 20: 203–206 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Moser BA, Russell P (2002) Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161: 1437–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Ribaud V, Bianchi A, Shore D (2007) DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev 21: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Ono Y, Tomita K, Matsuura A, Nakagawa T, Masukata H, Uritani M, Ushimaru T, Ueno M (2003) A novel allele of fission yeast rad11 that causes defects in DNA repair and telomere length regulation. Nucleic Acids Res 31: 7141–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE (2001) The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet 27: 64–67 [DOI] [PubMed] [Google Scholar]
- Petreaca RC, Chiu HC, Eckelhoefer HA, Chuang C, Xu L, Nugent CI (2006) Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol 8: 748–755 [DOI] [PubMed] [Google Scholar]
- Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D (2008) Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J 27: 2328–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, Bielinsky AK (2004) Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol Cell 16: 173–185 [DOI] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Zakian VA (2007) Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell 27: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V, Luciano P, Brevet V, Guillot S, Corda Y, Longhese MP, Gilson E, Geli V (2004) RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat Genet 36: 46–54 [DOI] [PubMed] [Google Scholar]
- Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR (2002) A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev 16: 2800–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Smith FW, Schultze P, Feigon J (1995) Solution structures of unimolecular quadruplexes formed by oligonucleotides containing Oxytricha telomere repeats. Structure 3: 997–1008 [DOI] [PubMed] [Google Scholar]
- Smith J, Rothstein R (1995) A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol Cell Biol 15: 1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Rothstein R (1999) An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics 151: 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Zou H, Rothstein R (2000) Characterization of genetic interactions with RFA1: the role of RPA in DNA replication and telomere maintenance. Biochimie 82: 71–78 [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE (2003) Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 1: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA (2002) Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Tejera AM, Stagno d’Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA (2010) TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell 18: 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Cooper JP (2008) Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SF, Lin JJ, Teng SC (2006) The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res 34: 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodenicharov MD, Wellinger RJ (2006) DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell 24: 127–137 [DOI] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA (2008) Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol 15: 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA (1996) Evidence for a new step in telomere maintenance. Cell 85: 423–433 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA (1993) Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72: 51–60 [DOI] [PubMed] [Google Scholar]
- Wold MS (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66: 61–92 [DOI] [PubMed] [Google Scholar]
- Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11: 748–760 [DOI] [PubMed] [Google Scholar]
- Wu Y, Zakian VA (2011) The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc Natl Acad Sci USA 108: 20362–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR (2004) Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci USA 101: 10024–10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich K, Cech TR (2005) A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol 12: 1072–1077 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.