Summary
Mathematical modeling has proved to be a critically important approach in the study of many complex networks and dynamic systems in physics, engineering, chemistry, and biology. The nuclear factor κB (NF-κB) system consists of more than 50 proteins and protein complexes and is both a highly networked and dynamic system. To date, mathematical modeling has only addressed a small fraction of the molecular species and their regulation, but when employed in conjunction with experimental analysis has already led to important insights. Here, we provide a personal account of studying how the NF-κB signaling system functions using mathematical descriptions of the molecular mechanisms. We focus on the insights gained about some of the key regulatory components: the control of the steady state, the signaling dynamics, and signaling crosstalk. We also discuss the biological relevance of these regulatory systems properties.
Keywords: computational models, IκB control, temporal control, signaling dynamics, dose–response control, negative feedback
Introduction
Efforts to apply the language of mathematics to biology can be traced back to the 13th century (1). The promise of this approach is in the power of mathematical tools for the analysis of complex, often dynamic relationships. One important such tool is a mathematical model in which these relationships are represented in terms of interacting equations, that break the limitations of measured data (which represent the past and are limited to those events that are measurable), but may be used for simulating and predicting the future. In modern biology, different types of mathematical models have been successfully utilized to address questions pertaining to fisheries, organ function, tumor growth, molecular networks within cells, or the interaction between atoms within biological macromolecules. Indeed, broad recognition of the importance of mathematical modeling in Molecular Biology contributed to the articulation of the systems biology approach (2), in which mathematical modeling is used to formulate testable hypotheses and guide experimental design to maximize the information obtained.
Biological regulatory systems are highly networked (each gene or protein engages in numerous physical and functional interactions), and they are kinetic, often showing complex dynamic regulation in response to perturbations. Studies have identified synergistic, competitive, or redundant gene functions, feedback and feed-forward loops, combinatorial gene controls, intricate dose–response relationships and dynamic behavior. Such emergent systems properties are best explored with kinetic mathematical models using ordinary differential equations (ODEs), which may involve stochastic or thermodynamic formulations for molecular interactions. Using parameter sensitivity analyses, modeling may then identify sensitive/robust parameters or signaling behavior. Modeling may explore how distinct signals are integrated or how the steady state may affect the dynamic response to perturbation. Though many models have been constructed to describe biological regulatory systems, mathematical models of the NF-κB signaling system are unusually well-grounded in biochemical and cell biological measurements. Iterative refinements of the NF-κB mathematical model over the past decade have established it as a virtual experimental model system that yields regulatory and biological insights through repeated model simulations and analysis of the simulation data (3,4).
Modeling the NF-κB signaling system: a personal narrative
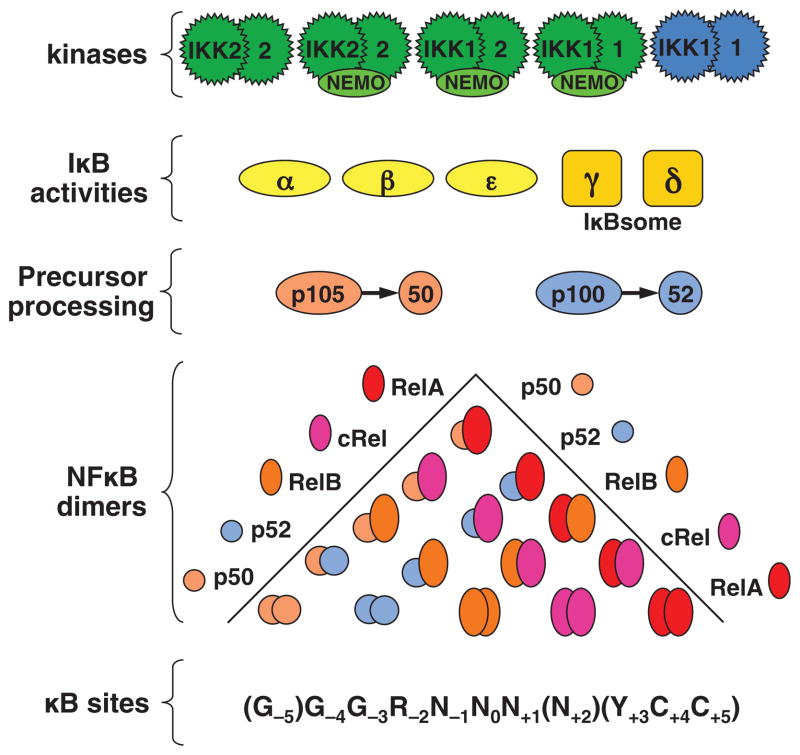
The NF-κB signaling system plays myriad roles in mammalian physiology and disease, as suggested by a rapidly expanding clinical and biomedical literature. It is also a complex dynamic protein interaction network of several interacting components that regulate each other (Fig. 1): five transcriptional monomers that form obligate dimers (in fact 15 NF-κB dimers are possible; (5); two precursor protein that mediate self-inhibition and within a multimeric complex (IκBsome) also trans-inhibition of other dimers (6, 7); three stimulus-responsive inhibitor proteins (classical IκBs); and several other highly homologous ankyrin-repeat containing proteins (e.g. Bcl3, IκBζ, IκBNS) that may interact with NF-κB dimers to regulate their activities (8). Most of these components are regulated by each other or by upstream, downstream, or neighboring signaling modules, at the level of protein synthesis, protein degradation, or post-translational modifications that alter their biochemical activities.
Fig. 1. The NF-κB signaling system.
Schematic of the molecular components that make of the NF-κB signaling system: a family of kinases controls the half-life of a family of inhibitors and protein precursors, thereby regulating the activity of 15 dimers (which consist of 5 monomer combinations) which control gene expression through a degenerate regulatory DNA element.
Thorough biochemical and molecular biological studies have identified two distinct signaling pathways that are mediated by this complex interaction network, the canonical and noncanonical pathways and regulate the activity of distinct NFkB dimers in a stimulus responsive manner. In the canonical NF-κB-IκB Signaling Module, NF-κB activation is achieved through NEMO-dependent phosphorylation and proteasomal degradation of three NF-κB inhibitors, IκBα, IκB-β, and IκB-ε, that promotes nuclear translocation of the RelA:p50 dimer in response to a variety of inflammatory cytokines and pathogen-derived substances. Activation is typically transient as several NF-κB target genes mediate postinduction termination of NF-κB response thus forming negative feedback loops. However, transient RelA:p50 activity is sufficient to induce the expression of inflammatory cytokines, chemokines, proteinases, and other inflammatory effectors, whose hyper-expression is often associated with disease pathology (9).
In contrast, the noncanonical Signaling Module regulates the production of the RelB:p52 dimer. Here, NF-κB-inducing kinase (NIK) promotes phosphorylation of the NF-κB precursor protein p100/nfkb2 in an IKK1-dependent manner. Subsequently, p100 undergoes partial proteolysis to generate the mature p52 subunit, which then dimerizes with RelB to appear as RelB:p52 dimer in the nucleus. Noncanonical signaling is not as rapid as canonical signaling, as it involves the build-up of newly synthesized RelB and p52, but it may last for days, consistent with the long-term nature of survival control, maturation, or developmental changes that it controls (10).
Construction of Model 1.0
Our first mathematical model (11) focused on canonical NF-κB activation grew out of an interest to delineate the differential functions of IκB proteins. Timecourse studies of NF-κB activation and IκB abundance using biochemical tools supported the hypothesis that the three IκB proteins have differential functions in controlling the temporal activity profile of NF-κB in response to inflammatory stimuli; only the coordinated function of all three would produce the NF-κB activation profile observed in response to tumor necrosis factor (TNF).
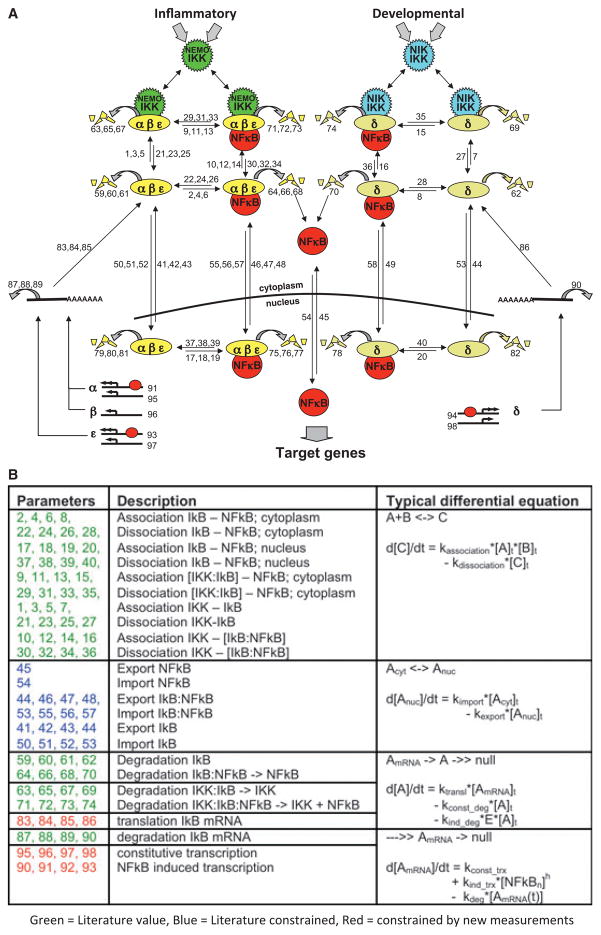
To study the specific roles of IκBα, IκBβ, and IκBε in the temporal control of NF-κB, a kinetic mathematical model was needed (Fig. 2A, adopted from a later model version 3.0), prompting a fruitful partnership between a computational biologist (Andre Levchenko) and a biochemist (Alexander Hoffmann). The questions to be addressed in the project dictated the model scope: three IκB proteins interacting with the NF-κB dimer RelA:p50 in the cytoplasm and nucleus; whereas the metabolism (synthesis, interactions, degradation) and localization of IκB proteins needed to be modeled in mechanistic detail, NF-κB’s cellular abundance could be assumed to remain unchanged, its activity being defined as the form in the nucleus that was not bound to IκBα or IκBε (NF-κB–IκBβ complexes had been reported to bind DNA and contribute to target gene activation) (11). This led to 70 parameters in the model representing about 25 independent kinetic rate constants (Fig. 2B, adopted from a later model version 3.0). Based on an abundant literature, about one-third of the rate constants came from biochemical measurements; another one-third could be constrained using published cell biological data (e.g. nucleo-cytoplasmic shuttling) (13).
Fig. 2. The NF-κB signaling system.
(A) Schematic of the molecular interactions contained in a sample kinetic model [version 3.0 (37) and version 3.1 (38)]. Each number indicates a kinetic parameter within an ordinary differential equation. (B) Table of the kinetic parameter classes [contained in (A)] and the type of ordinary differential equation used to describe the molecular interaction. Parameter values were obtained from literature (green), fitted predominantly based on literature data (blue), or fitted predominantly based on new experimental data (red).
To determine the remaining parameters and test the assumptions made determining the model topology, we took a genetic approach to generate three cell lines containing simplified signaling modules, each being controlled by only one IκB. The resulting double knockout cells indeed proved instructive as they revealed control of NF-κB by a single IκB and enabled parameterization of all reactions involving one IκB in a reduced single-IκB-containing model. Interestingly, combining the three reduced models did not immediately account for NF-κB activation profiles in wildtype cells, but required a scaling factor; this prompted subsequent studies into compensation between IκB isoforms (14).
What began as a research tool for a single study to address a specific question turned out to be a research tool with numerous applications. There are several reasons for that. First, the model functions as a repository of current knowledge about the signaling pathway, which may be used to determine (i) whether current knowledge is sufficient to account for observed signaling behavior, and (ii) which reported mechanisms seem to contribute to signaling in particular conditions/cell types. It is thus the foundation for numerous biochemical and cell biological studies that allow for iterative refinement of the model as described in the next section. Second, articulation of the mechanisms known to the experimental biochemists/cell biologists in the language of math, brought in researchers from physics, mathematics, computer science, or bioengineering interested in dynamic or metabolic control. These researchers strove to understand the dynamical control of the system at a more fundamental level through large-scale computation or model reduction and analytical methods (15–30).
The systems biology approach: iteratively refined model versions of NF-κB signaling
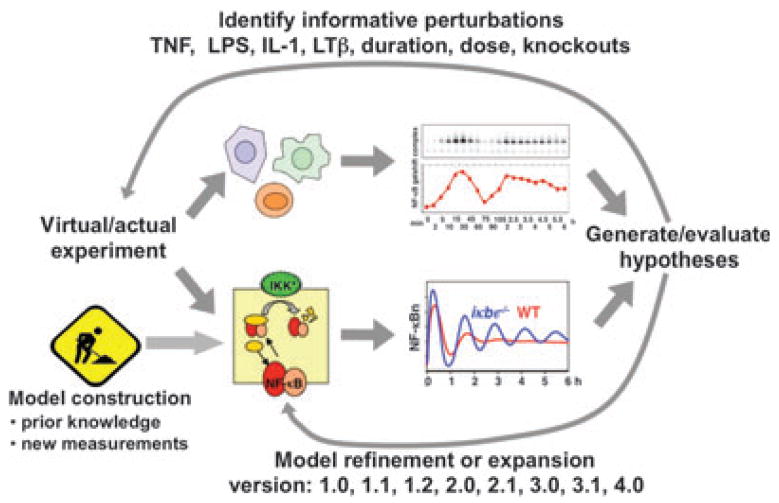
In 2001, shortly before the publication of NF-κB model version 1.0, Hood and co-workers (2) coined the phrase ‘Systems Biology’ and defined it as an approach in which parallel and iterative mathematical modeling and experimentation would reveal the functions of biological control systems. The nature of model version 1.0 proved amenable to subsequent Systems Biology studies, in which the model was subjected to increasingly demanding experimental tests (Fig. 3). Some of these resulted in revisions of kinetic rate constants that were previously obtained from published studies or in expansion of the model to include novel molecular mechanisms or regulators (Table 1).
Fig. 3. The Systems Biology approach to elucidate NF-κB signaling.
The Systems Biology approach was defined as involving iterative mathematical modeling and experimentation (2). To elucidate NF-κB signaling, a kinetic model was constructed based on literature. Simulations were used to develop hypotheses, and then identify perturbations (such as TNF duration dose–responses) that may test these informatively. Subsequent experimental studies were carried out accordingly, allowing evaluation of the hypothesis, and may demand a refinement or expansion of the kinetic model.
Table 1.
Iterative development of successive NF-κB model versions: their characteristics, capabilities, and shortcomings
| Version (REF)
|
Characteristics and capabilities (all models consider only the RelA:p50 dimer)
|
Shortcomings addressed in subsequent versions
|
|---|---|---|
| 1.0 (11) |
|
|
| 1.1 (14) |
|
|
| 1.2 (34) |
|
|
| 2.0 (37) |
|
|
| 2.1 (D. Barken, unpublished data) |
|
|
| 2.2 (51) |
|
|
| 2.3 (32) |
|
|
| 3.0 (39) |
|
|
| 3.1 (40) |
|
|
| 4.0 (38) |
|
|
In the first study (11), the response to the system to different durations of TNF exposure was investigated, given that TNF has a short half-life and is secreted transiently by macrophages within tissues. Computational simulations of stimuli with varying durations (IKK curves of different durations) resulted in distinct temporal profiles that nevertheless shared an invariant first phase of 1 h of activity. To quantitate this response to input signals of varying duration we developed the duration dose–response curve by thresholding by plotting on the y-axis the duration of activity above a certain threshold. This duration dose–response curve was quite different in IκBα-knockout models, indicating that IκBα was responsible for limiting the response to transient stimuli to 1 h. Gene expression studies indicated that durations longer than 1 h can result in different sets of genes being activated.
Next, we addressed the control of the steady state of the NF-κB system. Model simulations were used to identify important (‘sensitive’) kinetic rate constants. Aside from IKK-mediated degradation of IκBs, a stimulus and IKK independent pathway targeting free IκB for degradation was found to be critically important, prompting new experimental measurements of the free IκB half-life. Using NF-κB-deficient cells, the new measured half-lives of endogenous IκB proteins were found to be much shorter than previous estimates that were based on transient transfection experiments (31), leading to version 1.1 of the model (14). Subsequent studies revealed that this degradation pathway to be proteasome-dependent yet ubiquitin-independent (32) and involving partially unfolded regions in the C-terminal ankyrin repeats of IκBα (33). A second mechanism providing for homeostatic control of NF-κB activity turned out to be NF-κB-dependent synthesis of IκBε, which also provided for the compensation mechanism lacking in model 1.0. NF-κB-inducible IκBε expression was quantified leading to model 1.2 (34). Interestingly, an apparent 30–45 min delay in the IκBε negative feedback loop with respect to IκBα, led to the computational prediction that IκBε provides negative feedback in anti-phase thus dampening IκBα-mediated oscillations. This prediction was confirmed through the biochemical experiments. An alternative hypothesis that would account for the biochemical observations is that IκBε feedback introduces cell-to-cell variability into the secondary peaks of NF-κB oscillatory activity (35) and is awaiting experimental confirmation at the single cell level.
A parallel line of research addressed the concentration dose–response of NF-κB to TNF. Whereas experimental studies showed that the first phase of NF-κB activity was largely independent of TNF doses, the model was unable to reproduce this observation, even following an exhaustive exploration of the parameter space (36). These computational studies led to the prediction that the assumption of a linear relationship between TNF dose and IKK activity must be incorrect and predicted a much more dynamic IKK activity profile. Fine-grained IKK time course measurements indeed revealed these. The realization that IKK activity may be dynamically regulated itself led to a new formulation of the model (version 2.0) (37) that used dynamically regulated input curves that can be numerically (rather than algebraically) defined providing for maximum flexibility in simulations and the direct use of experimentally measured time courses. Model 2.0 allowed for exploration of the signal processing properties of the IKK–IκB–NF-κB signaling module, indicating that while differences in the early phase IKK activity were not perceived, the signaling module is very sensitive to small changes in the late activity. Indeed, the late activity turned out to be much more stimulus-specific (comparing TNF and LPS) and subject to the control of both positive and negative feedback regulators (37). Subsequent studies began to delineate the mechanisms that produce IKK activity in response to diverse stimuli; a first version of the TNF pathway model (model version 4.0) (38) enabled comparisons between the immediately induced negative feedback regulators IκBα and A20 and revealed them to have entirely different functions, with negative feedback of A20 not providing for dynamic control, but rheostat-like dose–response function.
However, the three IκB–NF-κB models proved insufficient in recapitulating NF-κB/RelA:p50 activation by LTβR agonist: at low stimulus concentrations, experiments showed much more RelA:p50 activity than calculated by the model given the measured miniscule amount of NEMO-kinase activity during LTβR stimulation. This discrepancy led to a search for additional RelA:p50 regulators and the identification of an oligomer of p100 (39). This protein complex has IκB-like functions, termed IκBδ, as it traps RelA:p50 dimers in a latent, readily activatable state, and it undergoes stimulus-responsive degradation akin to IκBα, IκBβ, and IκBε. However, its degradation is triggered by NIK-inducing (i.e. noncanonical) stimuli. Inclusion of IκBδ into model versions 3.0 (39) and 3.1 (40) led to model-directed experimental studies that characterized IκBδ as a negative feedback regulator particularly relevant in response to stimuli such as LPS that lead to long lasting IKK activity (40). Because of IκBδ’s long half-life, transient inflammatory exposure may result in increased IκBδ levels that alter the cellular steady state in such a way as to strengthen the capacity for noncanonical activation of RelA:p50, where it may produce expression of inflammatory genes (39).
While the present models of the NF-κB signaling module appear sufficient in recapitulating a number of physiological conditions, the Systems Biology approach of iterative refinement may well continue, certainly when considering an expansion of the scope of the model to include the signaling modules associated with other inflammatory receptors, include the downstream DNA interactions on target genes, or considering the important topic of the mechanisms that actually determine the cellular repertoire of dimers, including those inducible mechanisms controlled by the noncanonical pathway (see Perspectives). Furthermore, the present models may well function as seeds for the construction of a predictive model of the inflammatory network that includes other transcription factors, notably AP-1 and IRF/ISGF3, and coordinated regulation between them.
Mathematical models and single cell studies
The dynamic control of NF-κB described in the 2002 study (9) attracted not only theoretical and computational biologists but also microscopists interested in examining the dynamic responses in single cells. The traditional approach we followed involved immunocytochemistry to stain for activated nuclear NF-κB/RelA in single cells at various timepoints. When many cells are examined in this way, the average indeed conformed to the results of biochemical studies of populations of cells in terms of the TNF-induced time course (41) and TNF dose–responses (42). The alternative approach of live cell microscopy avoids the limitation of timepoint studies in which no trajectory of a particular cell can be obtained no matter how close the timepoints are spaced. It, therefore, has the potential to produce data of unprecedented resolution resulting in new insights. Indeed, live cell microscopy studies published by all groups have revealed NF-κB activity trajectories with very detailed dynamic features. These continue to pose a challenge to the available mathematical models, which are based largely on biochemical/population data and do not account for them.
However, technical concerns remain with this approach. One set of concerns relates to the need for special culture conditions (cell density, medium etc.) and photo-toxicity caused by exposure to lasers or the production of reactive oxygen species by fluorescent protein moieties. A second set relates to the live cell reporter design. Live cell imaging studies rely on a live cell reporter protein that ideally has no effector function within the signaling system. When it is fused to an effector (such as RelA) it may provide more direct information about signaling dynamics, but at the cost of two caveats: altered cellular level of the signaling effector (due to altered expression and/or degradation) and potentially altered biochemical behavior of that effector.
Single cell studies therefore provide data that is quite different and remains to be connected to the biochemical knowledge and the genetic knowledge available about NF-κB. Thus, it remains important to address the two caveats and their potential for artefact. A knockin RelA-GFP mouse has addressed the worry about overexpression, and tour de force microscopy work (43) overcame the challenges of significantly weaker signals that in the ectopically transfected (44) or transduced cell studies (45). These cells also produce spiky NF-κB translocation at the detectable limit where technical noise makes observations more challenging.
To address the second caveat of the effect of the fusion protein (e.g. GFP variants used in published studies have propensity to dimerize), biochemical data (cytoplasmic/nuclear immunoblots and EMSAs) from cells harboring the reporter-effector fusion is needed to be compared with genetically non-modified wildtype cells. Such biochemical data, when compared to single cell imaging data, may also address the disconnect that results from focusing on nuclear localization in live cell imaging studies, in contrast to DNA binding activity of nuclear NF-κB in biochemical EMSA studies. This disconnect is amplified in experiments that use leptomycin D (44), which is an inhibitor of nuclear export not only of NF-κB but also of IκBα. In principle, and as has been stated, biochemical studies represent the average of the behavior of many cells, but surprisingly no study has as yet examined whether this is in fact the case (i.e. whether the average of many single cell tracks account for parallel biochemical data) or whether such a comparison is confounded by distortions introduced by the technicalities of the two experimental approaches.
When the above-described analysis pipeline has been established, important controls and genetic perturbation studies must be pushed in single cell studies to catch up with the biochemical characterization found in the literature. At this time, it remains unclear which feature in the jittery tracks revealed by live cell imaging is present in medium alone (no cells), in cells without stimulus, in NEMO-deficient cells with stimulus, or in IκBαβε-deficient cells that show no biochemical response to TNF (Alexander Hoffmann, unpublished data). Then studies with single IκB knockouts and mutants thereof may establish how each IκB and the described feedback and control mechanisms contribute and to which features of the NF-κB temporal profile. Such data may then allow revision and further refinement of the mathematical model of NF-κB activation, potentially including stochastic components in a few or many reactions to account for the observables.
A word of caution that applies to both biochemical and single cell signaling studies: while highly dynamic features in the activity profiles of signaling mediators are information rich and thus useful for model parameterization, some of these may depend on how the external stimulus is applied. Given that we do not know precisely what a physiological stimulus looks like (e.g. TNF dose, duration, or temporal profile), to address the concern of physiological relevance, in vitro signaling studies ought to either focus on dynamic features that are conserved for a range of stimulus-applications or image cells in their natural environment in situ. These considerations ought to impact the design and appreciation of NF-κB signaling studies in the near future.
Can the NF-κB mathematical model help us understand disease?
The goal in pursuing a mathematical modeling approach is of course to develop a tool for understanding disease processes and directing therapeutic intervention. At the cellular scale, combined computational and experimental studies have already guided pharmacological studies. However, to have impact in understanding various types of diseased cells, there is an often-asked question and another that is less well appreciated. The answer to the former turns out to be straightforward, but the latter poses greater challenges for applying models to disease.
The often-asked question is whether the model can be applied to other cell types, given that the bulk of the experiments that validate the model have been in MEFs (MEFs were chosen to take advantage of their responsiveness to diverse NF-κB-inducing stimuli, a protocol to develop immortal cell lines without the use of oncogenes that may affect the pathways we are studying, and the ability to produce such cells from knockouts that are embryonic lethal). The answer is straightforward in that the model in fact implies a powerful prediction: the kinetic parameters that are based on protein interaction or enzymatic function are rooted in the biophysical characteristics of molecules that – in turn – are a function of their genetic sequence not the cell type. It is true that E3 ligase, proteasome, or shuttling machineries may be different between different cells, but the current understanding is that they are not rate limiting. Indeed, discovering that cellular ‘house-keeping’ functions (e.g. proteasome activity, shuttling machineries) vary between cell types in a manner that affects NF-κB regulation would constitute an exciting finding. However, cells do differ in their chromatin regulation, which controls the transcriptional parameters of the model, and the way they perceive signals (receptor availability and adapter proteins), which determines the IKK input of the model. Thus, the implicit prediction that the model of the IKK–IκB–NF-κB signaling module can be easily adapted to different cell types by measuring IκB mRNA and IKK activity time courses.
What limits the application of the model to understanding disease more seriously is its limited scope: IKK to NF-κB signaling is only a small part of the relevant regulatory network and the causes for misregulation as well as promising therapeutic targets are often outside this ubiquitous signaling module. The work on B-cell lymphomas has been instructive: though a fraction of Hodgkin lymphomas show deletions in IκBα (resulting not only in deficiency in postinduction repression, but elevated basal levels) (46), elevated NF-κB signaling in several non-Hodgkin lymphoma sub-types have led to the identification of mutations in Malt1, A20, and Myd88, which function upstream of the pleiotropic signaling module (47–49). Similarly, whereas ubiquitin-dependent and -independent proteasome pathways are represented in the model (and efficacy of drugs targeting them can be explored), other drug targets being explored in the clinic are not. Recent results from studies into the causes of NF-κB misregulation in disease only emphasize the urgency and importance of expanding the scope of the mathematical models of NF-κB regulation.
Insights into the regulation of NF-κB
The steady state
Steady state control of the NF-κB signaling system determines not only the steady state NF-κB activity (also known as ‘basal’ NF-κB), it also determines its responsiveness to signals. Mathematical modeling has confirmed and quantified known mechanisms of regulation and has helped discover unexpected mechanisms as well.
Fail-safe control among IκBs
Steady-state NF-κB activity in unstimulated cells (basal activity) is not zero, but must be kept at a low level, as NF-κB is a powerful transcriptional activator of inflammatory effectors including secreted cytokines and chemokines that initiate and amplify an adaptive immune response, metalloproteases and other proteins that can effect tissue remodeling and damage, as well as survival and proliferation regulators. Indeed, many inflammatory diseases and cancer types show elevated basal NF-κB activity. The first NF-κB model suggested that the system contains an as yet unknown compensation mechanism to ensure low NF-κB activity when cells are deficient in IκBα (9). Some years later this compensation mechanism was found to be NF-κB-inducible expression of IκBε, which lags behind the inducible expression of IκBα, but shows an equivalent dynamic range (34) and thus efficiently compensates at least in the cell type tested. Indeed, cells lacking both IκBα and IκBε show much higher basal NF-κB, than either of the single knockouts (14). However, the model predicted an even greater elevation in the double knockout, which was one motivation for the search of additional NF-κB-inducible IκB activities and led to the discovery of IκBδ, an oligomer of the NF-κB-inducible nfkb2/p100 protein (39). Thus while IκBα is the primary inhibitor of NF-κB in the steady state, there appear to be two fail-safe inhibitors, IκBε and IκBδ.
Kinetic buffering
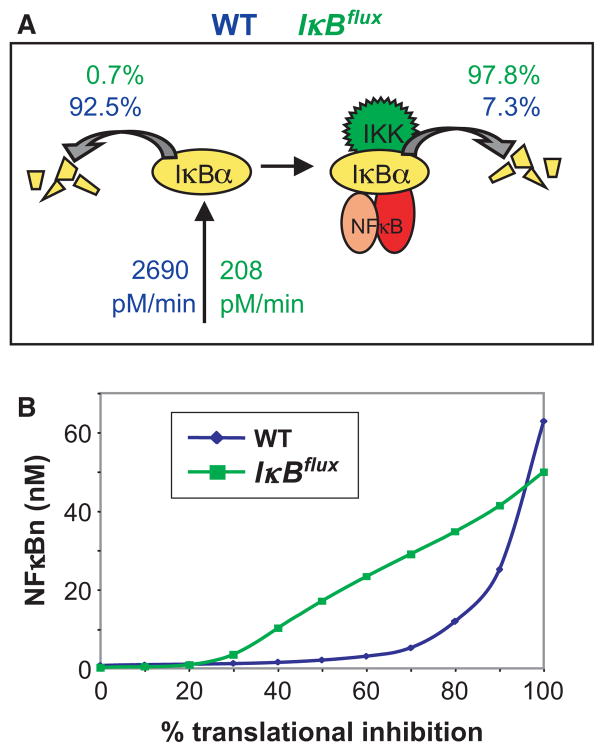
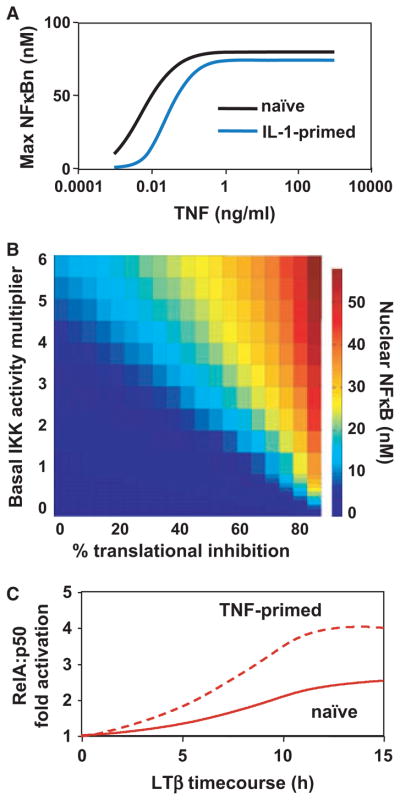
A different and unexpected mechanism to ensure low NF-κB activity in the steady state (even in the face of transient or minor perturbations) was revealed by a sensitivity analysis of the model’s parameters. The abundance of any protein is determined by its synthesis and degradation rate. For IκBs there are two degradation pathways, one that is IKK-triggered and one that is IKK-independent. Simulations revealed that the strength of the IKK-independent pathway to be critical for IKK-dependent signaling (14), which prompted a research program to understand the molecular mechanisms controlling this pathway. While IKK-does not prefer NF-κB-bound over free IκB (revealed by combined mathematical modeling experimental studies; 32), the IKK-independent pathway only applies to free IκB. Free IκB is degraded by a proteasome-dependent, but ubiquitin-independent pathway (32) that appears to require the incompletely folded C-terminal ankyrin repeat domains of IκBα (33, 50) rather than the previously described PEST sequence in the C-terminal extension (31). In fact the folded state of those non-consensus ankyrin repeat domains appears to be critical for determining the half-life of free IκBα, suggesting that IκBs have evolved away from the ankyrin repeat consensus to ensure a short half-life of free IκB. IκB is stabilized by folding upon binding NF-κB, thereby kinetically sensitizing it to the IKK-responsive, ubiquitin-dependent degradation pathway. In the unstimulated state, the high difference in half-life between free and NF-κB-bound IκBs (Fig. 4A) proved to be highly homeostatic. This insight was further tested with experimental studies that demonstrated cellular insensitivity to ribotoxic stress inducers such as ultraviolet (UV) or the UPR-inducing drug thapsigargin (51). Computational studies using a virtual flux mutant in which free IκB half-life was prolonged, but synthesis was decreased to achieve the same level of excess free IκBα, demonstrated that high flux necessitated by the short half-life of free IκB can produce kinetic buffering that renders the NF-κB signaling module resistant to small perturbations, whereas maintaining its responsiveness to stimuli above a threshold of about three times over basal (51) (Fig. 4B). Only when the steady-state abundance of free IκB is reduced (for example by chronic inflammatory signaling), the NF-κB system regains sensitivity to UPR-inducers (Fig. 6B, discussed later).
Fig. 4. Kinetic buffering desensitizes the NF-κB system to metabolic perturbations.
(A) Schematic of the key reactions controlling IκB abundance. In resting cell, the short half-life of free IκB necessitates a high synthesis rate (2690 pM/min) to maintain the observed pool of free IκB proteins (51). When the free IκB is stabilized to the same half-life as bound IkB the required synthesis rate is reduced >10-fold (208 pM/min). This alteration defined the virtual IκB-flux mutant. (B) Where as normal cells show a high degree of insensitivity to ribotoxic stress (translational inhibition must be >70% to elicit a response), the IκB flux mutant shows NF-κB activation with >30%, a level achieved by UV and during UPR.
Fig. 6. Signaling crosstalk within the NF-κB signaling system: some examples.
(A) Dose–response graph of NF-κB to TNF. A prior exposure to IL-1 renders cells less sensitive to subsaturating doses of TNF. This crosstalk effect is mediated in part by A20 (38). (B) Heat map of peak NF-κB activity (color scale) in response to ribotoxic stress signals effecting translational inhibition (x-axis) in cells containing constitutive IKK activity at various values (y-axis; 51). (C) Timecourse of RelA:p50 activation to LTb. Whereas naıve cells only show modest activation of RelA:p50, prior TNF exposure enhances the activation (compare solid and dashed lines) to such an extent that inflammatory target genes are activated as well (not shown here). This priming effect is mediated by IkBd (39).
Alternate cellular states
Many biological regulatory systems are capable of assuming alternate quasi-steady states that last for several hours or days often in response to prior stimulation or priming. The short half-life of free IκBα, IκBβ, and IκBε proteins ensures that they do not mediate transitioning of the NF-κB signaling network to a different state of stronger or weaker signaling. However, with the identification of the IκBsome and its associated IκBδ activity, a mechanism was revealed for regulating the abundance of the latent pool of NF-κB that is capable of being activated in a classical IκB-independent manner. Indeed, both components of the IκBsome, p105 and p100, are expressed from NF-κB-responsive promoters. Mathematical modeling focused on the role of the p100-derived IκB activity, termed IκBδ (39, 40). When experimental evidence indicated that IκBδ has a long half-life (greater than 8 h), mathematical modeling could be employed to quantitatively predict the effect of prior inflammatory exposure (that may increase IκBδ expression) on the responsiveness of the signaling system. Examining the canonical signaling pathway, the attenuation was unexpectedly minor, reflecting that the abundance of IκBδ activity constitutes only a minor fraction of the total cellular IκB pool (39), except in cases where prior signaling was prolonged (40), where IκBδ induction could have tolerizing functions for inflammatory signaling. However, on the flip-side in examining the responsiveness to noncanonical signals, we found that even transient inflammatory exposure could lead to substantial increases in NF-κB/Rel:p50 activation (39), a reflection of the generally lower amplitude of non-canonical versus canonical pathway signaling. Indeed, experimental studies using a transient 15 min pulse of TNF followed by a rest period confirmed a higher degree of noncanonical activation of RelA:p50 by LTβR agonist that was sufficient to drive expression of a variety of inflammatory genes. As cells in tissues are not naive as in cell culture, IκBδ may thus integrate the history of exposure to inflammatory agents and thereby tune the strength of the noncanonical pathway and its ability to induce inflammatory RelA:p50 target genes (Figs 5 and 6C).
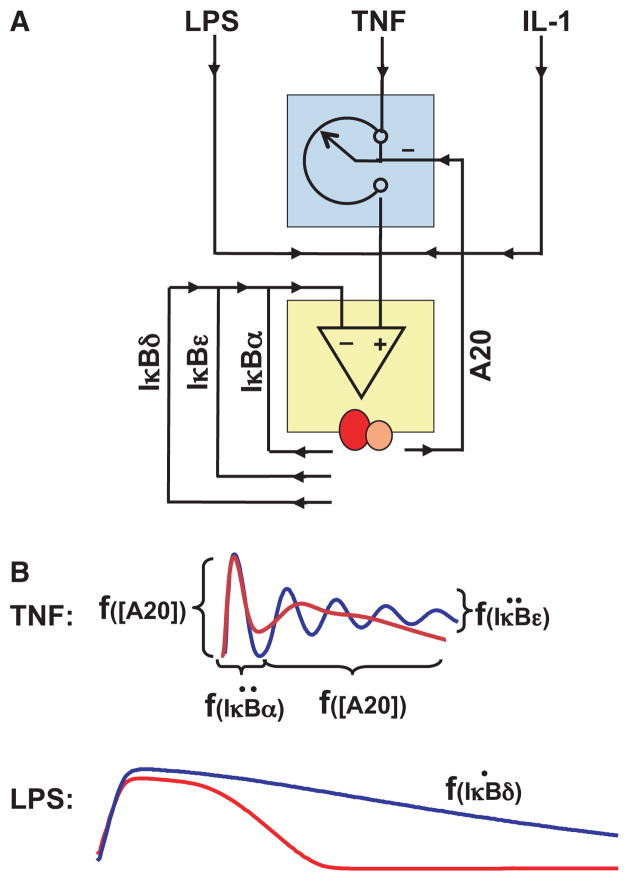
Fig. 5. Encoding dynamic control of NF-κB in response to TNF and LPS.
(A) Schematic of negative feedback regulators involved in shaping NF-κB responses to inflammatory stimuli. The depicted IκBs function is dynamic feedback regulators, whereas A20 provides integral feedback and a rheostat function to limit NF-κB responses. (B) Schematics of the TNF and LPS-responsive NF-κB profiles observed in biochemical experiments in wild-type (red) or specific knockout (blue) cells. In response to TNF, the duration of the first phase of NF-κB activation is determined by the inducibility of IκBα synthesis, whereas dampening of the responses is a function of the inducibility of IκBε. The amplitude of the first phase and duration of the second phase is a function of A20’s abundance. In response to LPS, the duration of NF-κB signaling is a function of the rate at which IκBd is produced. This graphic summary is based on work described and cited in the text.
Following the extension of the mathematical model to include the TNFR-associated signaling module that produces IKK activity, computational simulations predicted a role for the NF-κB-inducible inhibitor A20 in regulating the dose–response characteristics of the TNF signaling pathway in contrast to the prior understanding of A20 as a terminator of NF-κB activity. The new prediction suggested that A20 with its own long protein half-life may integrate the inflammatory exposure history to establish a quasi-steady state to determine the strength of the canonical signaling pathway (38) (Figs. 5 and 6A). Indeed, experimental studies confirmed this prediction of A20’s rheostat function; after inducing A20 expression with the cytokine IL-1, cells displayed a less sensitive dose–response curve to TNF in activating RelA:p50. This characterization should inform further studies on a range of disease pathologies including atherosclerosis (52), rheumatoid arthritis (53), and a variety of B-cell cancers (47), where A20 has been implicated.
We conclude that mathematical modeling helped reveal that inflammatory exposure of cells results in a quasi-steady state that in which the canonical pathway is weaker, but the non-canonical pathway is stronger in activating RelA:p50. The stability of this quasi-steady state is determined by the half-life of the relevant regulators IκBδ and A20 (Fig. 5).
Dynamic control
The initial construction of the mathematical model for the NF-κB signaling module was motivated by the intricate dynamic control apparent in time course experiments. Since then, the mathematical model has proved to be a useful tool for dissecting the mechanisms that control NF-κB dynamics and for guiding experimental design.
Dynamic control of NF-κB is stimulus specific. The stimulus may be described in terms of its chemical identity as well as the concentration dose and duration dose. The latter two may be related when the half-life of the stimulus is short, as low doses decay quickly below the level of effective concentration. In short, the dynamic control of NF-κB is a function of the concentration dose, and exposure duration, and importantly the identity of the stimulus.
The dynamic control of NF-κB activity determines the NF-κB-mediated gene expression program. Two lines of evidence support this statement. First, transient or pulse stimulation with TNF results in the expression of some genes, but long-term stimulation or repeated pulse stimulation results in the expression of others, as exemplified by the chemokine CCL5/RANTES (11, 54). Further, inactivation of the negative feedback by cycloheximide treatment or in IκBα-deficient cells allows expression of RANTES in response to pulse stimulation (11). Second, LPS and TNF activate distinct though overlapping sets of genes. Inactivating cytokine-mediated autocrine loops triggered by LPS alters the LPS-induced NF-κB dynamic control and also affects the expression of a subset of NF-κB-response genes (37). These observations led to the hypothesis of a Temporal Signaling Code (5), which states that information about the stimulus is encoded in the temporal profile or dynamics of NF-κB activity to specify which genes are to be expressed. How gene promoters or gene regulatory networks decode the temporal profile of NF-κB activity remains unclear, but iterative Systems Biology studies have begun to delineate how cells encode exposure to TNF and LPS in the dynamic of NF-κB activity. Indeed, mathematical modeling has proved critical in understanding the specific functions of feedback regulators in shaping stimulus-induced NF-κB dynamics (Fig. 5A). These are summarized below.
IκBα
Mathematical modeling was instrumental in revealing specific functions of the negative feedback regulator IκBα. IκBα plays key roles in shaping NF-κB dynamics in response to TNF. Its rapid IKK-responsive degradation allows for rapid activation of NF-κB, within 5–10 min following TNF exposure, as observed in biochemical and single cell microscopy studies (11, 44, 55). Despite a readily responsive promoter capable of wide dynamic range (56) that ensures that IκBα promoter activity generally mirrors nuclear NF-κB activity, the delay intrinsic in mRNA production, processing, export, and translation (as well as protein location and interaction with NF-κB) allows for NF-κB activity to last for about 40 min, even if IKK activity is short lived, as it is in the case of transient TNF stimulation (11). Indeed, even a 1 min exposure to TNF provides for this invariant ‘quantum’ of NF-κB activity as control of IKK is similarly ‘excitable’ and lasts a minimum of 10–15 min (38). Turning-off NF-κB activity after 40–60 min requires NF-κB-mediated induction of IκBα expression and cannot be achieved by any level of constitutive expression (38), emphasizing IκBα’s role as a dynamic feedback regulator.
TNF stimulation that extends beyond 60 min in duration results in a second phase of NF-κB activity that is separated by a transient trough caused by an excess negative feedback production of IκBα. Long lasting stimulation with high concentration doses of TNF can result in ringing (damped oscillations) of NF-κB activity observed biochemically in special conditions (34, 37, 41, 57) and in a broader set of conditions in single cell studies (43, 44). However, mathematical modeling clarified that IκBα would have remarkably little role in regulating the overall duration of late NF-κB activity or in shaping LPS-induced NF-κB responses (37, 40). We may rationalize this prediction by considering that long-lasting IKK activity effectively neutralizes the IκBα feedback, as IκBα is of course readily phosphorylated by the NEMO IKK complex causing IκBα degradation through the ubiquitin-proteasome system. The stimulus-specificity of IκBα for cytokine, and in particular TNF, was confirmed not only experimentally in cell culture but also in animals: removing the inducing stimulus TNF by genetic ablation, rescued the perinatal lethality of IκBα-deficient mice (40).
IκBε
Mathematical modeling was also instrumental in revealing IκBε’s role as a negative feedback regulator functioning in coordination with IκBα. Whereas IκBε expression is highly inducible, its inductions appears to be delayed in part because of a longer mRNA half-life, but possibly also because the NF-κB-response elements are probably not promoter proximal (they have not yet been conclusively identified). Model simulations suggested that delayed IκBε feedback may function in antiphase with the strong negative feedback control mediated by IκBα (34) to dampen the propensity of the system to oscillate (Fig. 5B). Indeed, further studies found that the measured delay and abundance of IκBε induction match those parameters that maximize the damping effi-ciency in the mathematical model (Longo and Kearns, unpublished results). An alternative hypothesis for the apparent damping observed in biochemical assays is that the presence IκBε renders oscillatory NF-κB activity in different cells more asynchronous, but genetic or mechanistic studies have yet to address this hypothesis.
A20
Mathematical modeling resulted in new insights about the function of the negative regulator A20, which along with IκBα is one of the most rapidly and highly inducible NF-κB target genes. Previous experimental studies had established that A20 is required for effective repression of NF-κB following initial signaling. Mathematical modeling confirmed that A20 is required for inhibiting NF-κB, specifying that it is not required for postinduction repression of NF-κB following transient TNF stimulation, that is the role of IκBα, but instead is required for attenuating the second phase of NF-κB activity in response to longer term TNF stimulation (Fig. 5B). A further surprising insight emerged: the inducible expression of A20 was not required for this function, but merely the cellular steady-state abundance of A20 protein (38). This model-derived prediction was confirmed experimentally by reconstituting A20-deficient cells with either inducibly or constitutively expressing A20 transgenes: both provided for functional reconstitution of TNF-induced NF-κB signaling. Computational simulations provided a rationale for this surprising conclusion: although A20 mRNA is rapidly induced, the protein being large takes longer to be synthesized and has a long half-life; its function is further upstream in the pathway thus impacting events that precede NF-κB activity by several minutes, and its function is an enzymatic rather than stoichiometric inhibitor (such as IκBα), thus its functional effect builds up over time.
Given that constitutive expression of A20 was sufficient for its role in TNF signaling, subsequent work addressed why its expression is inducible. The answer turns out to be rather simple: any NF-κB inducing stimulus will also induce the expression of A20, thus altering the cellular steady state at least for a period of time. IL-1 for example, whose signaling to NF-κB is unaffected by A20 (at least in MEFs), induces A20 expression and may thus impact subsequent signaling of TNF to NF-κB. Mathematical modeling identified A20 as a rheostat negative regulator that can be set by diverse inflammatory signals to mediate a degree of tolerance to TNF exposure. A20 mediates signaling crosstalk to limit the late phase of NF-κB activation and dose responsiveness to TNF.
IκBδ
Mathematical modeling revealed functional roles of the newly identified IκBδ activity, a component of the multimeric IκB-some complex, which is able to associate with RelA:p50 dimers and sequester them in an inactive, but latent state (7, 39, 40). IκBδ activity is mediated by the oligomerized p100 polypeptide which is encoded by the nfkb2 gene whose expression itself is inducible by the RelA:p50 dimer (58). IκBδ forms a potential negative feedback loop, but its importance or physiological relevance in the context of the much more prominent IκBα (and IκBε) negative feedback regulator was unclear. After measuring relevant kinetic parameters, the IκBδ feedback was incorporated into the mathematical model (version 3.0 and version 3.1), enabling computational simulations to explore its functionality (39, 40). As different cellular stimuli elicit different temporal profiles of IKK activity, we asked whether IκBδ feedback may be particularly relevant in the attenuation of signaling in response to a subset of potential IKK temporal profiles. Using a library of hypothetical IKK curves, the mathematical model was used as a computational phenotyping tool to distinguish the functions of IκBα and IκBδ negative feedback. By comparing wildtype and knockout models, the analysis confirmed that IκBα feedback acts rapidly to terminate transient IKK-induced NF-κB responses and revealed that IκBδ feedback was required for attenuating longer term IKK-induced NF-κB activities (Fig. 5B). Further parameter sensitivity studies suggested that the irreversibility of the IκBδ negative feedback loop (IκBδ is not a substrate for canonical IKK) combined with its slow induction renders it dominant over the faster acting IκBα and IκBε at late times of sustained signaling. Subsequent biochemical analyses con-firmed a role of stimulus induced IκBδ in terminating PAMP-triggered NF-κB responses that are generated through sustained IKK signaling (40). An impaired IκBδ feedback resulted in heightened inflammatory gene expressions in response to bacterial LPS. In contrast, IκBα-mediated negative feedback primarily controls short-lived cytokine signaling. Therefore, IκBδ appears to execute a non-redundant task within the NF-κB system to encode the NF-κB temporal pro-file in response to specific stimuli, such as LPS in fibroblasts. This conclusion is also supported by observations in T cells, in which TCR stimulation induces the expression of p100 in CD4+ T cells in a NEMO-IKK2 dependent manner. Late acting negative feedback mediated by p100 was shown to similarly dampen TCR-induced NF-κB responses and IL-2 production (59). Further multidisciplinary studies are required to illuminate IκBδ function in integrating chronic signals derived from tissue microenvironment with acute immune response signals.
Signaling crosstalk
Mathematical modeling allows for systematic studies to identify the regulators that may tune the responsiveness of the network to specific stimuli. When these dose–response regulators are inducible or otherwise subject to control by another stimulus or signaling pathway, they mediate signaling crosstalk, i.e. a case in which exposure of cells to one stimulus will affect signal transduction in response to exposure to a second stimulus. Given the highly networked nature of the NF-κB signaling system and its responsiveness to numerous stimuli, there is the capacity for integrating several signals to produce an appropriate response. Just a few examples of signaling crosstalk are described below (Fig. 6).
Tuning TNF-NF-κB dose–responses
Mathematical modeling has contributed to studies of the dose-responsiveness to TNF. Two inducible long-lived inhibitors have been identified that may play important roles. IκBδ, a component of the multimeric IκBsome, controls the pool of latent NF-κB that may be activatable by inflammatory stimuli. Mathematical modeling has shown that because the inducibility of the nfkb2 gene and hence IκBδ activity is rather sluggish, only long term inflammatory exposure will have a substantial impact on the amount of NF-κB that can be activated (39, 40). However, it is interesting that IκBγ and IκBδ activities may also be subject to half-life control by some stimuli in some cell types, such as LPS in B cells or LTβ in MEFs, and so their impact on TNF-dose–responses or the LPS-refractory period may be further explored.
Mathematical modeling showed that A20 controls the efficiency of receptor proximal signaling and thus determines the sensitivity of the pathway to low doses of TNF (38). Because A20 is rapidly and highly inducible, even a transient inflammatory exposure, such as a pulse of IL-1, has been shown to alter the sensitivity to a subsequent sub-saturating dose of TNF (Fig. 6A). Although IκBδ’s and A20’s mode of action is distinct and would therefore be predicted to have distinct effects on the dose–response curve with A20 controlling the threshold for activation and steepness of the dose–response curve, and IκBδ controlling the saturation plateau. Future studies may focus on single cells to address these predictions.
Tuning NF-κB responsiveness to ribotoxic stress
Mathematical modeling was instrumental in elucidating the mechanisms by which cells may respond to ribotoxic stress agents such as UV or UPR (51). Previous literature had established that IKK activity is required, but was not able to establish that IKK activity is induced. Mathematical modeling allowed us to delineate how basal or steady-state activity controls the cells responsiveness to ribotoxic stress. Remarkably, at low basal IKK activity typically found in cells that are not exposed to inflammatory agents or that do not harbor IKK-activating mutations, the dose–response curve for ribotoxic stress agents is so low that barely any NF-κB activity can be elicited even with lethal doses of UV (due to kinetic buffering described above). However, this dose–response curve is shifted significantly when the basal IKK activity is increased (Fig. 6B). These predictions were examined experimentally; cells grown in the presence of very low LPS concentrations that on their own did not substantially raise the basal NF-κB activity were now highly responsive to UV or thapsigargin producing substantial NF-κB activation. These results were also confirmed by using the translation inhibitor cycloheximide.
Tuning RelA:p50 activation by the non-canonical pathway
Mathematical modeling predicted that the degree of RelA:p50 activation by the noncanonical pathway is a function of prior exposure to inflammatory stimuli (39). The noncanonical (NEMO independent) pathway is mediated by IKK1 and p100 and triggers not only an accelerated processing of newly synthesized p100 to p52 to produce a RelB:p52 dimer, but also degradation or processing of the ankyrin-repeat domain within p100-containing IκBsomes, to release pre-formed latent RelA:p50 dimer. Modeling showed that the second signaling mechanism is tunable by inflammatory stimuli, i.e. that the inducibility of nfkb2 and half-life of the resulting IκBδ activity within the IκBsome allows the signaling system to integrate the prior inflammatory exposure to tune the degree of RelA:p50 activation by stimuli activating the noncanonical pathway. The model predicted that cells that are primed by inflammatory stimuli would produce an inflammatory response to developmental stimuli (Fig. 6C). Experimentally, this prediction was confirmed by pre-treating cells with a small dose of TNF for just 15 min before stimulating them with the LTβR agonist antibody: the activation of the RelA:p50 dimer was significantly enhanced over unprimed cells. Similarly, cells that lacked IκBα (and other classical IκBs) were predicted to contain a higher proportion of IκBδ within the total IκB pool and thus to allow for greater RelA:p50 activation in response to LTβ - this prediction was also confirmed with a panel of IκB knockout cell lines.
Interestingly, mathematical modeling has not yet explored whether the first signaling mechanism leading to RelB:p52 activation may also be tuned by inflammatory exposure. Some studies have suggested that RelA:p50 responsive nfkb2 expression be a way of tuning RelB:p52 activation (60), but RelA:p50-dependent expression of RelB may be at least as important (6, 66). However, the degree and limitations of this crosstalk have yet to be examined quantitatively.
Insights into the biology of NF-κB
As mathematical modeling addresses the dynamic network of interactions within a functional module, it reveals network emergent properties such as dose–response control, temporal control, the ability to integrate or insulate distinct input signals, or network memory. These emergent network properties may directly relate to biological functions that have consequences in physiology and/or disease. Below, four examples of such properties are described.
NF-κB is primarily a transducer of extracellular signals
Mathematical modeling provided a means to examine the homeostatic control of the steady state. Aside from quantifying the compensation among IκB proteins, the model helped reveal an extraordinarily high flux of IκB synthesis and degradation in the steady state (14). Given its high metabolic cost, mathematical modeling was employed to examine why it may have evolved. These studies revealed that high IκB flux renders the cells less sensitive to perturbations such as ribotoxic stresses that may reduce cellular translation capacity by 50%. However, activation of NF-κB in response to signals that induce IKK remains substantial and is only marginally affected by the high flux. We therefore conclude that the instability of free IκB and the resulting high flux system has evolved to provide kinetic buffering against metabolic perturbations, but focusing this signaling system primarily on to extra cellular inflammatory signals that activate IKK. Unlike several other signaling pathways, the biological function of the NF-κB system is not primarily about sensing intracellular stress, but about transducing information about the extracellular environment to the nuclear gene expression machineries. However, as described below, information about the metabolic conditions is taken into account in the processing/transduction of signals from the extracellular environment.
NF-κB may mediate stimulus-specific biological functions
Mathematical modeling has been instrumental in studying NF-κB dynamic control. Aside from understanding which factors and mechanisms are responsible for specific features of the dynamic activation profile (which is discussed in the previous section), mathematical modeling has been used to examine the capacity of the NF-κB signaling module to generate distinct temporal profiles (37). Using a library of hypothetical IKK input profiles, modeling revealed that the signaling module can discriminate between many different amplitude temporally modulated signals. This capacity to distinguish between signals allows the NF-κB signaling pathway to faithfully transduce stimulus-specific IKK activity profiles to the nucleus. In other words, as a signaling hub, the IKK-NF-κB signaling module can transduce distinct signals through temporal insulation (61). These insights have led to the Temporal Code hypothesis (5) (described above), which states that information about the stimulus is encoded in the temporal profile or dynamics of NF-κB activity to specify which genes are to be expressed. This is an important concept, as it may underlie the remarkably diverse physiological function of NF-κB in immune, inflammatory, and developmental signaling. Further investigations are required to understand the mechanisms by which gene regulatory networks may decode temporally distinct NF-κB activities.
NF-κB integrates diverse signals to control inflammation
Mathematical modeling has revealed the extent and the mechanisms by which diverse signals may combine to produce additive, non-additive/synergistic, or qualitatively distinct NF-κB activities. Such findings are of biological interest as cells are conditioned within different micro-environments or histories of prior exposure prior to responding to noxious agents. Specifically, these micro-environments may fine-tune NF-κB responses to specific agents. In one example, we examined what factors may determine the responsiveness of the NF-κB signaling system to ribotoxic stress agents. Whereas healthy primary fibroblasts in cell culture showed little NF-κB activation to UV and the UPR-inducing agent thapsigargin (due to the kinetic buffering mechanism discussed in a previous section), modeling predicted that cells grown in the presence of a low inflammatory environment would show substantial NF-κB activation. This prediction was confirmed experimentally. The biological dimension of this work is that ribotoxic stress agents elicit a much stronger effect in cells that show already some (though may be small) NF-κB upregulation; thus UV may amplify a pre-existing inflammatory conditions, making matters worse; or ribotoxic chemotherapeutic compounds may amplify elevated NF-κB activity in tumor cells, rendering them more resistant to apoptotic-inducing signals. Thus the mathematical model may prove useful in fine-diagnosis of disease-subtypes to predict and evaluate the efficacy of therapeutic agents.
Inflammatory history tunes developmental NF-κB functions
Mathematical modeling has contributed to our understanding of how inflammatory signaling contributes to developmental of secondary lymphoid organs. Signaling through LTβR on stromal cells plays a critical role for the development of lymph nodes during early embryogenesis (62, 63). However, TNFR was also shown to be important for the development of lymphoid tissues such as payer patches and marginal zones (64). By abrogating TNFR or LTβR signaling with decoy receptors in utero, a cooperation between these two receptor pathways in lymph node formation was further suggested (63). Given LTα3 binds to TNFR, it was postulated that such cross-connectivity at the level of receptor–ligand pair integrate TNFR pathway with developmental processes. Biochemical studies guided by studies with the mathematical model (version 3.0), instead revealed a signaling network mechanism by which the inflammatory history determines the strength of the NF-κB response to lymph node-inducing stimuli (39). Following the discovery of IκBδ as the IκB dedicated to RelA:p50 activation by the noncanonical pathway, modeling studies focused on identifying the mechanisms that determine its cellular abundance. Given its long half-life, IκBδ has the capacity to integrate the cellular exposure history of a wide variety of inflammatory cues. While TNF has been implicated in lymph node development, further studies may investigate the role of other inflammatory agents, the cell-to-cell network, and the broader physiological implications of crosstalk between inflammatory and developmental signals.
Modeling studies also helped reveal that ikba−/− cells may be hyper-responsive to noncanonical signaling (39). Malignant Reed Sternberg cells in Hodgkin’s lymphoma with an inactivating mutation in the ikba gene were shown to rely upon noncanonical BAFF signaling for proliferation and survival (65). Indeed, a variety of pathological conditions, especially neoplastic disorders, may show hyper-responsiveness to noncanonical signals, potentially rendering them addicted to tonic developmental signals (6). The implication that tonic developmental signals may constitute promising drug targets to reduce disease-associated NF-κB activity remains to be examined.
Perspectives: conclusions and outstanding questions
While mathematical modeling has advanced our understanding of the regulation and biology of NF-κB, there remains enormous potential for mathematical modeling to generate further insights about the NF-κB signaling system in physiology and disease. Further insights will likely involve single cell studies and the expansion of the model, to include the interdependent control of different NF-κB dimers and/or the control of upstream, downstream, and parallel signaling modules. With an expansion of the mathematical model, an increasing number of biological questions may be addressed with increasing degree of sophistication and predictive power.
The control and biological significance of single cell dynamics
Live cell imaging studies (43–45) have revealed that fluorescent RelA-fusion proteins show a much greater of degree of dynamic control than biochemical studies of endogenous RelA in populations of cells (11–37) or immunofluorescence studies of endogenous RelA at timepoints in single cells (41, 42). Future studies ought to aim to bridge this disconnect (for example by testing whether the sum of many fluorescence traces in fact accounts for the biochemical data) and establish which dynamic features observed in single cells are determined by the known kinetic regulatory network (prompting further refinement of the mathematical model) or are a function of unknown mechanisms (e.g. the phenomenon of transcriptional bursting). In other words, mathematical modeling ought to be brought to bear to gain insights into the dynamic control of NF-κB observed in single cells, as well as the apparent cell-to-cell variability. With an understanding of the underlying regulatory mechanisms, we may also gain insights about the biological roles and significance of single cell dynamics.
The generation and control of multiple NF-κB dimers
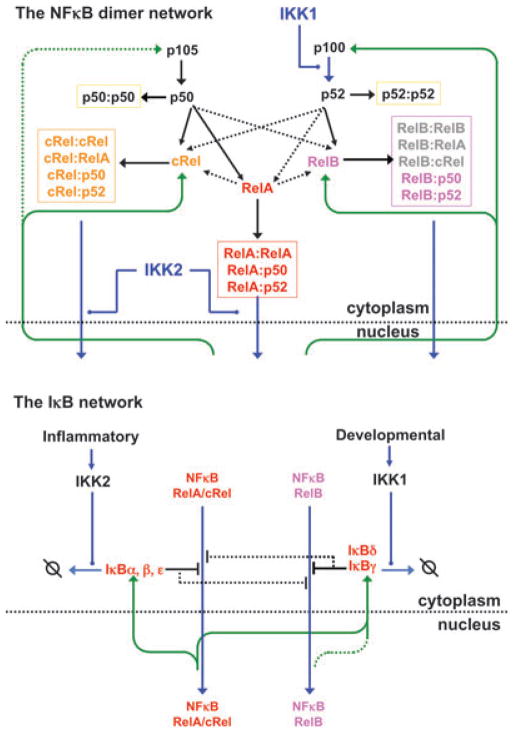
All modeling studies thus far have been focused on the regulation of the NF-κB RelA:p50 dimer, the most prominent and ubiquitous among the NF-κB family members. However, the NF-κB family is composed of 15 possible homo- and heterodimeric transcription factors (5). These differ in the manner in which they are regulated and which signals they are responsive to, as well as in which NF-κB target genes they effectively activate. Therefore understanding the biology of NF-κB more fully in terms of underlying molecular mechanism requires studies of the mechanisms that control dimer generation as well as the regulation of their DNA binding activities. Dimer generation is a function of monomer abundance, dimerization affinities, and the presence of possible dimerization chaperones. This network is complicated, because the expression of some monomers is inducible by the activity of particular dimers, because the processing of precursors is responsive to cell-intrinsic and stimulus-inducible control mechanisms, because dimers may be targeted for degradation (66) or may be stabilized (67), and because monomers compete for binding of shared binding partners (68). Thus, studying the network that controls the NF-κB dimer repertoire in a given cell will likely benefit from an experimentally grounded kinetic mathematical model. Current ‘IκB-focused’ mathematical models, therefore, must be expanded to depict generation and regulation of multiple NF-κB dimers (6) (Fig. 7).
Fig. 7. Perspective: accounting for the generation and activation/inactivation of multiple NF-κB dimers.
The NF-κB Signaling system may be described in terms of two interconnected networks: the NF-κB network (left) that depicts monomer expression (inducible in green arrows), pre-cursor processing and other regulated steps (blue), and dimer formation (black arrows); and the IκB network which depicts IκB synthesis (green arrows), IκB interactions with NF-κB dimers (black inhibitory connectors), and IκB degradation (blue arrows). Understanding how the kinetic mechanisms in each network combine to control the activity of multiple NF-κB dimers in different cell types remains an important future goal.
Integrating diverse signals and cellular steady states
With the availability multidimer NF-κB models, mathematical modeling will enable a quantitative understanding of how cells integrate many more diverse signals and exposure histories, whether they are inflammatory, growth factor, or developmental, to produce specific functional responses. Although experimental designs usually involve highly purified and specific stimulation reagents, cells are exposed to a variety of cues, concurrently or sequentially, during the process of an actual immune response. Moreover, the tissue microenvironment mediates tonic signaling that affects signaling of resident cells. How such tonic signaling interfaces with the inflammatory pathways induced upon pathogen recognition during immune response remains to be studied. Mathematical models may guide experimental research to identify signaling crosstalk. Parameter sensitivity analysis combined with biochemical and genetic experiments may thus establish key mechanisms for crosstalk in specific biological settings, such as lymph node formation, B-cell differentiation, DC maturation, as well as inflammatory and immune responses. Finally, disease-associated changes in the NF-κB signaling network alter its steady state control and its responsiveness to external or tonic signals. Mathematical modeling will enable a predictive understanding of how multiple perturbations (whether they are genetic, involving the tissue micro-environment, pharmaceuticals or noxious stimuli) are integrated to affect cellular function.
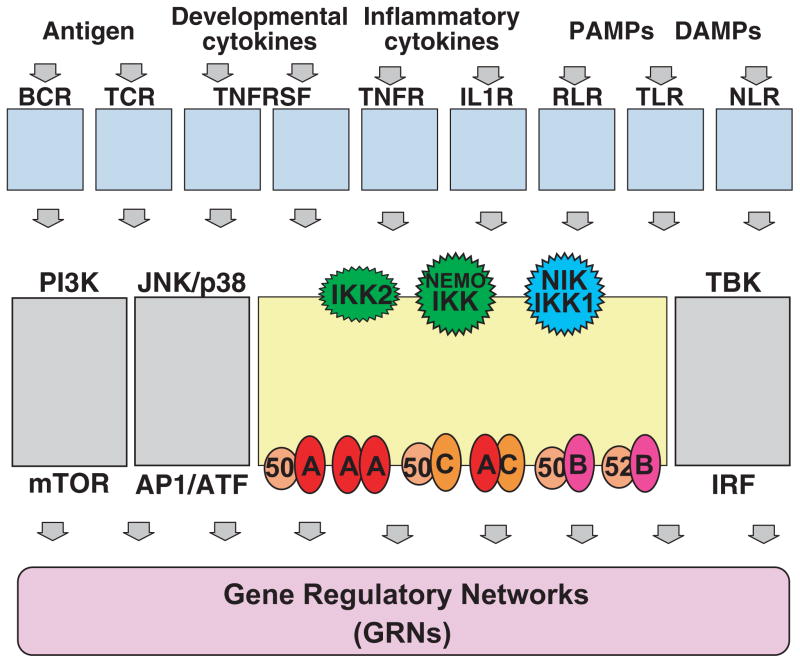
The NF-κB signaling system within proteomic cellular network
The NF-κB signaling system is embedded within a larger network that determines inflammatory, proliferative, and developmental responses to cell-intrinsic and external cues (Fig. 8). As a signaling module, NF-κB receives and transduces signals from upstream signaling modules often associated with receptors that sense the presence of pathogen or cytokine. In the case of TNF, a mathematical model that recapitulates TNF-induced IKK activity could be linked to the model of the NF-κB module to allow studies of TNF-NF-κB dose–responses and dynamic control mechanisms (36). Similar studies will be insightful for other cytokine and pathogen receptor-associated signaling modules. Further, NF-κB signaling is coordinated with signaling in other pathways (e.g. MAPK, interferon, PI3kinase) that are also triggered by receptor-associated signaling modules and may in some cases interact or crosstalk with NF-κB signaling. However, when pathways appear insulated, their effectors may cooperate, synergize, and antagonize in producing a specific cellular response. Finally, NF-κB, as a transcription factor, achieves its biological effects through downstream gene regulatory networks (GRNs) that determine which genes are expressed and to which degree. As promoters contain numerous transcription factor binding sites, transcription initiation and elongation, as well as mRNA processing and degradation are kinetic processes, mathematical modeling will be required to develop a predictive understanding of when and to what degree which genes are expressed. Achieving a predictive understanding of cellular immune responses is thus possible, but it will require a long term commitment from a broad and diverse community of experimentalists to employ the language of mathematics to codify their understanding of the molecular regulatory mechanisms.
Fig. 8. Perspective: the NF-κB signaling system within the context of upstream, downstream, and coordinated regulatory networks.
The NF-κB signaling system receives inputs from signaling modules associated with a variety of receptors that respond to the presence of distinct classes of molecules. These same signaling modules also activate a subset of other signaling systems. The coordinated activities of the effectors of theses signaling systems together determine the cellular response via gene regulatory networks. Understanding how the coordinated activation of multiple signaling systems produce stimulus-appropriate cellular responses remains an important future goal.
Acknowledgments
We thank Andre Levchenko and all past and present laboratory members and collaborators who contributed to the approaches and insights described in this review, as well as the NF-κB community for the wealth of biochemical data that made mathematical modeling possible. We acknowledge funding from the NIH GM071573, CA141722, AI08345 and also intermediate fellowship from Wellcome Trust DBT India Alliance to SB. The authors have no conflicts of interest to declare.
References
- 1.Fibonacci L. Liber Abaci. p. 1202. [Google Scholar]
- 2.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 3.Cheong R, Hoffmann A, Levchenko A. Understanding NF-kappaB signaling via mathematical modeling. Mol Syst Biol. 2008;4:192. doi: 10.1038/msb.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kearns JD, Hoffmann A. Integrating computational, biochemical studies to explore mechanisms in NF-{kappa}B signaling. J Biol Chem. 2009;284:5439–5443. doi: 10.1074/jbc.R800008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 6.Basak S, Hoffmann A. Crosstalk via the NF-kappaB signaling system. Cytokine Growth Factor Rev. 2008;19:187–197. doi: 10.1016/j.cytogfr.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savinova OV, Hoffmann A, Ghosh G. The Nfkb1, Nfkb2 proteins p105, p100 function as the core of high-molecular-weight heterogeneous complexes. Mol Cell. 2009;34:591–602. doi: 10.1016/j.molcel.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin AS. Regulation of cell death and autophagy by IKK and NF-jB: critical mechanisms in immune function and cancer. Immunol Rev. 2012;246:327–345. doi: 10.1111/j.1600-065X.2012.01095.x. [DOI] [PubMed] [Google Scholar]
- 10.Sun S-C. The noncanonical NF-jB pathway. Immunol Rev. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann A, et al. IkappaB-NF-kappaB the signaling module: temporal control, selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 12.Suyang H, et al. Role of unphosphorylated, newly synthesized I kappa B beta in persistent activation of NF-kappa Molecular B. Mol Cell Biol. 1996;16:5444–5449. doi: 10.1128/mcb.16.10.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlotti F, Dower SK, Qwarnstrom EE. Dynamic shuttling of nuclear factor kappa B between the nucleus, cytoplasm as a consequence of inhibitor dissociation. J Biol Chem. 2000;275:41028–41034. doi: 10.1074/jbc.M006179200. [DOI] [PubMed] [Google Scholar]
- 14.O’Dea EL, et al. A homeostatic model of I-kappaB metabolism to control constitutive NF-kappaB activity. Mol Syst Biol. 2007;3:111. doi: 10.1038/msb4100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipniacki T, et al. Stochastic regulation in early immune response. Biophys J. 2006;90:725–742. doi: 10.1529/biophysj.104.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho KH, et al. Investigations into the analysis and modeling of the TNF alpha-mediated NF-kappa B-signaling pathway. Genome Res. 2003;13:2413–2422. doi: 10.1101/gr.1195703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipniacki T, et al. Mathematical model of NF-kappaB regulatory module. J Theor Biol. 2004;228:195–215. doi: 10.1016/j.jtbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Lipniacki T, et al. Single TNFalpha trimers mediating NF-kappaB activation: stochastic robustness of NF-kappaB signaling. BMC Bioinformatics. 2007;8:376. doi: 10.1186/1471-2105-8-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung MH, et al. Dynamic effect of bortezo-mib on nuclear factor-kappaB activity and gene expression in tumor cells. Mol Pharmacol. 2008;74:1215–1222. doi: 10.1124/mol.108.049114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayot F, Jayaprakash C. NF-kappaB oscillations, cell-to-cell variability. J Theor Biol. 2006;240:583–591. doi: 10.1016/j.jtbi.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Krishna S, Jensen MH, Sneppen K. Minimal model of spiky oscillations in NF-kappaB signaling. Proc Sci Natl Acad USA. 2006;103:10840–10845. doi: 10.1073/pnas.0604085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yde P, et al. Modeling the NF-kappaB mediated inflammatory response predicts cytokine waves in tissue. BMC Syst Biol. 2011;5:115. doi: 10.1186/1752-0509-5-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh G, Lee DY. Mathematical modeling, sensitivity analysis of the integrated TNFal-phamediated apoptotic pathway for identifying key regulators. Comput Biol Med. 2011;41:512–528. doi: 10.1016/j.compbiomed.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Peng H, et al. Drug inhibition profile prediction for NF kappaB pathway in multiple myeloma. PLoS ONE. 2011;6:e14750. doi: 10.1371/journal.pone.0014750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu WH, Wang FS, Chang MS. Sensitivity analysis of dynamic biological systems with time-delays. BMC Bioinformatics. 2010;11(Suppl 7):S12. doi: 10.1186/1471-2105-11-S7-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng SC, et al. Computational modeling with forward and reverse engineering links signaling network and genomic regulatory responses: NF-kappaB signaling-induced gene expression responses in inflammation. BMC Bioinformatics. 2010;11:308. doi: 10.1186/1471-2105-11-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radulescu O, et al. Robust simplifications of multiscale biochemical networks. BMC Syst Biol. 2008;2:86. doi: 10.1186/1752-0509-2-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han L, Zhao Y, Jia X. Mathematical modeling identified c-FLIP as an apoptotic switch in death receptor induced apoptosis. Apoptosis. 2008;13:1198–1204. doi: 10.1007/s10495-008-0252-3. [DOI] [PubMed] [Google Scholar]
- 29.Lipniacki T, Kimmel M. Deterministic, stochastic models of NF-kappaB pathway. Cardiovasc Toxicol. 2007;7:215–234. doi: 10.1007/s12012-007-9003-x. [DOI] [PubMed] [Google Scholar]
- 30.Monk NA. Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Curr Biol. 2003;13:1409–1413. doi: 10.1016/s0960-9822(03)00494-9. [DOI] [PubMed] [Google Scholar]
- 31.Pando MP, Verma IM. Signal-dependent, -independent degradation of free, IkappaBalpha NF-kappa B-bound. J Biol Chem. 2000;275:21278–21286. doi: 10.1074/jbc.M002532200. [DOI] [PubMed] [Google Scholar]
- 32.Mathes E, et al. NF-kappaB dictates the degradation pathway of IkappaBalpha. EMBO J. 2008;27:1357–1367. doi: 10.1038/emboj.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathes E, et al. Flexible regions within I{kappa}B{alpha} create the ubiquitin-independent degradation signal. J Biol Chem. 2010;285:32927–32936. doi: 10.1074/jbc.M110.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kearns JD, et al. IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J Cell Biol. 2006;173:659–664. doi: 10.1083/jcb.200510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paszek P, Jackson DA, White MR. Oscillatory control of signalling molecules. Curr Opin Genet Dev. 2010;20:670–676. doi: 10.1016/j.gde.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Cheong R, et al. Transient IkappaB kinase activity mediates temporal NF-kappaB dynamics in response to a wide range of tumor necrosis factor-alpha doses. J Biol Chem. 2006;281:2945–2950. doi: 10.1074/jbc.M510085200. [DOI] [PubMed] [Google Scholar]
- 37.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 38.Werner SL, et al. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22:2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Basak S, et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih VF, et al. Kinetic control of negative feedback regulators of NF-kappaB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proc Natl Acad Sci USA. 2009;106:9619–9624. doi: 10.1073/pnas.0812367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barken D, et al. Comment on ‘‘Oscillations in NF-kappaB signaling control the dynamics of gene expression’’. Science. 2005;308:52. doi: 10.1126/science.1107904. author reply 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheong R, et al. Information transduction capacity of noisy biochemical signaling networks. Science. 2011;334:354–358. doi: 10.1126/science.1204553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sung MH, et al. Sustained oscillations of NF-kappaB produce distinct genome scanning and gene expression profiles. PLoS ONE. 2009;4:e7163. doi: 10.1371/journal.pone.0007163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson DE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 45.Lee TK, et al. A noisy paracrine signal determines the cellular NF-kappaB response to lipopolysaccharide. Sci Signal. 2009;2:ra65. doi: 10.1126/scisignal.2000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jungnickel B, et al. Clonal deleterious mutations in the IkappaBalpha gene in the malignant cells in Hodgkin’s lymphoma. J Exp Med. 2000;191:395–402. doi: 10.1084/jem.191.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kato M, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 48.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rui L, et al. Malignant pirates of the immune system. Nat Immunol. 2011;12:933–940. doi: 10.1038/ni.2094. [DOI] [PubMed] [Google Scholar]
- 50.Truhlar SM, et al. Pre-folding IkappaBalpha alters control of NF-kappaB signaling. J Mol Biol. 2008;380:67–82. doi: 10.1016/j.jmb.2008.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Dea EL, Kearns JD, Hoffmann A. UV as an amplifier rather than inducer of NF-kappaB activity. Mol Cell. 2008;30:632–641. doi: 10.1016/j.molcel.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolfrum S, et al. The protective effect of A20 on atherosclerosis in apolipoprotein E-deficient mice is associated with reduced expression of NF-kappaB target genes. Proc Natl Acad Sci USA. 2007;104:18601–18606. doi: 10.1073/pnas.0709011104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matmati M, et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat Genet. 2011;43:908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- 54.Ashall L, et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalita MK, et al. Sources of cell-to-cell variability in canonical nuclear factor-{kappa}B (NF-{kappa}B) signaling pathway inferred from single cell dynamic images. J Biol Chem. 2011;286:37741–37757. doi: 10.1074/jbc.M111.280925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giorgetti L, et al. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell. 2010;37:418–428. doi: 10.1016/j.molcel.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 57.Covert MW, et al. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 58.de Wit H, et al. Regulation of p100 (NFKB2) expression in human monocytes in response to inflammatory mediators and lymphokines. Leukemia. 1998;12:363–370. doi: 10.1038/sj.leu.2400950. [DOI] [PubMed] [Google Scholar]
- 59.Legarda-Addison D, Ting AT. Negative regulation of TCR signaling by NF-kappaB2/p100. J Immunol. 2007;178:7767–7778. doi: 10.4049/jimmunol.178.12.7767. [DOI] [PubMed] [Google Scholar]
- 60.Stadanlick JE, et al. Tonic B cell antigen receptor signals supply an NF-kappaB substrate for prosurvival BLyS signaling. Nat Immunol. 2008;9:1379–1387. doi: 10.1038/ni.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Behar M, Hoffmann A. Understanding the temporal codes of intra-cellular signals. Curr Opin Genet Dev. 2010;20:684–693. doi: 10.1016/j.gde.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Futterer A, et al. The lymphotoxin beta receptor controls organogenesis and affinity maturation in peripheral lymphoid tissues. Immunity. 1998;9:59–70. doi: 10.1016/s1074-7613(00)80588-9. [DOI] [PubMed] [Google Scholar]
- 63.Rennert PD, et al. Lymph node genesis is induced by signaling through the lymphotox-in beta receptor. Immunity. 1998;9:71–79. doi: 10.1016/s1074-7613(00)80589-0. [DOI] [PubMed] [Google Scholar]
- 64.Neumann B, et al. Defective Peyer’s patch organogenesis in mice lacking the 55-kD receptor for tumor necrosis factor. J Exp Med. 1996;184:259–264. doi: 10.1084/jem.184.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiu A, et al. Hodgkin lymphoma cells express TACI and BCMA receptors and generate survival and proliferation signals in response to BAFF and APRIL. Blood. 2007;109:729–739. doi: 10.1182/blood-2006-04-015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lawrence T, et al. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 67.Rao P, et al. IkappaBbeta acts to inhibit and activate gene expression during the inflammatory response. Nature. 2010;466:1115–1119. doi: 10.1038/nature09283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Basak S, Shih VF, Hoffmann A. Generation, activation of multiple dimeric transcription factors within the NF-kappaB signaling system. Mol Cell Biol. 2008;28:3139–3150. doi: 10.1128/MCB.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]