Abstract
Gene-to-gene coexpression analysis provides fundamental information and is a promising approach for predicting unknown gene functions in plants. We investigated various associations in the gene expression of tomato (Solanum lycopersicum) to predict unknown gene functions in an unbiased manner. We obtained more than 300 microarrays from publicly available databases and our own hybridizations, and here, we present tomato coexpression networks and coexpression modules. The topological characteristics of the networks were highly heterogenous. We extracted 465 total coexpression modules from the data set by graph clustering, which allows users to divide a graph effectively into a set of clusters. Of these, 88% were assigned systematically by Gene Ontology terms. Our approaches revealed functional modules in the tomato transcriptome data; the predominant functions of coexpression modules were biologically relevant. We also investigated differential coexpression among data sets consisting of leaf, fruit, and root samples to gain further insights into the tomato transcriptome. We now demonstrate that (1) duplicated genes, as well as metabolic genes, exhibit a small but significant number of differential coexpressions, and (2) a reversal of gene coexpression occurred in two metabolic pathways involved in lycopene and flavonoid biosynthesis. Independent experimental verification of the findings for six selected genes was done using quantitative real-time polymerase chain reaction. Our findings suggest that differential coexpression may assist in the investigation of key regulatory steps in metabolic pathways. The approaches and results reported here will be useful to prioritize candidate genes for further functional genomics studies of tomato metabolism.
One of the major challenges of plant systems biology is in understanding genotype-phenotype associations. In that context, biological networks can increase our understanding of how biomolecules interact to function in plants (Fukushima et al., 2009b; Stitt et al., 2010). Large-scale data from genome-wide gene expression profiling with DNA microarrays are publicly available for many species, including Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), poplar (Populus spp.), and some crops (Ogata et al., 2010; Tohge and Fernie, 2010). These data make it possible to use gene coexpression analyses to predict unknown gene functions (Aoki et al., 2007; Usadel et al., 2009). Using pairwise measures (e.g. Pearson’s correlation coefficient), it is possible to generate a coexpression network in which nodes represent genes and edges represent significant correlations between expression patterns. Network representation facilitates the prioritization of candidate genes for further functional genomics studies based on the so-called “guilt-by-association” principle (Saito et al., 2008).
Generally, graph clustering algorithms include hierarchical clustering, density-based and local searches, and other optimization-based clustering, as summarized by Wang et al. (2010). In Arabidopsis and rice microarray data sets, such algorithms, including Markov clustering (Van Dongen, 2000) and DPClus (Altaf-Ul-Amin et al., 2006), were applied to find coexpression modules, which are clusters consisting of densely connected coexpressed genes (Ma et al., 2007; Mentzen and Wurtele, 2008; Fukushima et al., 2009a; Mao et al., 2009). These types of network-module-based approaches are now widely used in attempts to predict new genes involved in biological processes (Saito et al., 2008; Usadel et al., 2009; Mutwil et al., 2011). Other network-based approaches have been applied to annotate unknown genes (Horan et al., 2008), to explore possible genes involved in carbon/nitrogen-responsive machineries (Gutiérrez et al., 2007), and to prioritize candidate genes for a wide variety of traits (Lee et al., 2010).
Subsets of highly coexpressed genes in tomato (Solanum lycopersicum) have been studied. Miozzi et al. (2010), who investigated conserved coexpression in the Solanaceae family, including tomato, tobacco (Nicotiana tabacum), and potato (Solanum tuberosum), used ESTs for tomato and compared transcriptional presence/absence patterns with those of other species to explore functional relationships between genes. Aoki and colleagues (Ozaki et al., 2010) demonstrated that coexpression modules extracted by network-based clustering across various developmental stages and organs facilitated the functional analysis of genes encoding flavonoid biosynthesis in tomato.
Coexpression patterns also change based on different conditions, such as the genotype and tissue type; this has been termed differential coexpression (Choi et al., 2005; Gillis and Pavlidis, 2009). Differential coexpression identifies the rewired edges of coexpression networks and may reflect changes in transcriptome organization (Watson, 2006; Chia and Karuturi, 2010). For example, this feature has been used to identify disease-specific networks (de la Fuente, 2010). Differential metabolomic correlations were utilized in some metabolomics studies (Weckwerth et al., 2004; Morgenthal et al., 2006; Fukushima et al., 2011; Kusano et al., 2011). While the differential expression of genes and gene coexpression in plants have been addressed in earlier microarray studies, their differential coexpression has not been addressed.
Here, we present tomato coexpression modules extracted by graph clustering from over 300 microarrays obtained from publicly available databases and 20 of our own hybridizations in our efforts to unravel associations between gene expression patterns in the tomato. We assessed the clustered modules systematically by Gene Ontology (GO) enrichment analysis. We also investigated differential coexpression among data sets of leaf, fruit, and root samples, based on Fisher’s Z-transformation, to gain insight into the tomato transcriptome. The aims of our study were to (1) characterize coexpression modules in tomato by using graph clustering and GO enrichment analysis, (2) identify significant differential coexpression between organs, and (3) provide clues for elucidating key regulatory networks in a given metabolism. The expression levels of the obtained gene pairs were validated by quantitative real-time (qRT)-PCR.
RESULTS
Construction of a Gene Coexpression Network in Tomato
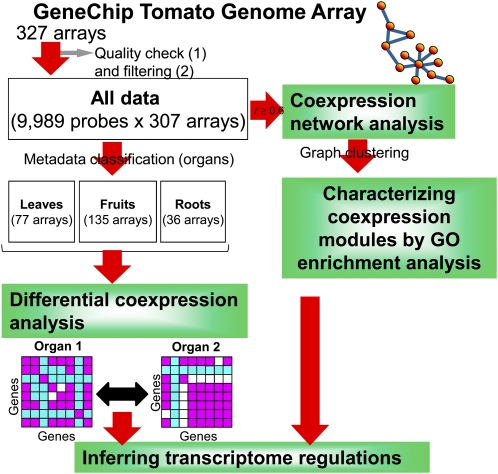
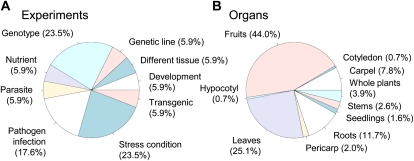
We collected 307 tomato GeneChips from publicly available databases and 20 of our own microarray data sets. A statistical quality check of all microarrays (see “Materials and Methods”) detected 307 high-quality GeneChips; these were used for further analyses (Fig. 1). After normalization, we manually classified them according to meta-data (Supplemental Data S1); 307 arrays included 17 experiments, with 23.5% involving a stress condition, 23.5% a genotype, and 17.6% a pathogen (Fig. 2A). They also corresponded to different organs, including fruits (44%), leaves (25.1%), and roots (11.7%; Fig. 2B). Based on the organ type in the meta-data, we divided the original expression data matrix (307 arrays × 9,989 probe sets, defined as “all data”) into three submatrices that we called “leaves,” “fruits,” and “roots.” We used these matrices, as well as all data, in further analyses (Fig. 1). The distribution of the three expression matrices appeared normal (Supplemental Fig. S1A). To construct the coexpression networks, we calculated correlation matrices using Pearson’s correlation coefficients. The correlation coefficients for each organ were also in normal distribution (Supplemental Fig. S1B).
Figure 1.
Work flow for extracting coexpression modules and for differential coexpression analyses among organs. Coexpression modules for each gene were generated by graph clustering without regard to functional properties. 1, Quality checks of microarrays were performed with robust regression techniques and the Kolmogorov-Smirnov goodness-of-fit statistic D (see “Materials and Methods”). We discarded 20 arrays with low-quality scores (D ≥ 0.15). Probe sets with the prefix AFFX and RPTR were excluded. 2, We rejected 220 probes with the detection call “absent” across all samples.
Figure 2.
Pie charts with a classification of the experiments and organs collected in this study. GeneChips (n = 327) were from publicly available databases including the GEO, ArrayExpress, TFGD, and our own data (see “Materials and Methods”). The 17 experiments contained in the data set were classified into nine experimental (A) and 10 organ (B) categories. [See online article for color version of this figure.]
The Topological Characteristics of Tomato Coexpression Networks Are Highly Heterogenous
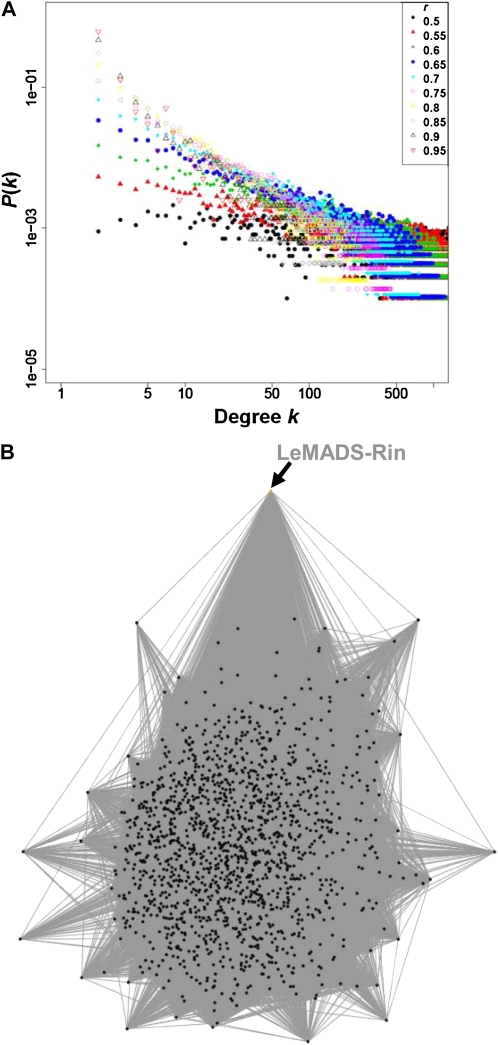
We next constructed tomato coexpression networks. To evaluate whether they manifested the common properties of a complex network, such as power-law degree distribution, we investigated the topological characteristics of the network (e.g. degree distribution and average path length; Table I). We found that the degree distribution of the coexpression network followed a power law (Fig. 3A). In the case of the coexpression network with r ≥ 0.6 (P < 2.1e-31, Pearson’s correlation statistical test), the degree distribution of the all-data network followed a power law with a degree exponent of γ = 1.67. Here, P(k) ∼ k−γ, where γ represents the degree exponent. The average path length and the average clustering coefficient of this network were 2.65 and 0.45, respectively, implying small-world properties and high modularity in the network (for review, see Barabási and Oltvai, 2004). Figure 3B shows a partial coexpression network generated using all data; it was based on the list of first-order genes that neighbored and were coexpressed with the LeMADS-Rin gene, which encodes a MADS box transcriptional factor regulating fruit ripening-related genes (Giovannoni et al., 1995; Vrebalov et al., 2002). The network shows how the neighboring genes of LeMADS-Rin correlate with each other.
Table I. Topological statistics of tomato coexpression networks (r ≥ 0.6).
| Data Sets | No. of Nodes | No. of Edges | Average Path Length | Clustering Coefficient | Average Degree | Degree Exponent |
| Leaves | 9,952 | 2,644,732 | 2.81 | 0.59 | 531.5 | 2.37 |
| Fruits | 9,867 | 1,782,674 | 2.92 | 0.50 | 361.3 | 2.07 |
| Roots | 9,924 | 5,997,644 | 2.62 | 0.70 | 1208.7 | 2.29 |
| All data | 9,797 | 1,754,361 | 2.65 | 0.45 | 358.1 | 1.67 |
Figure 3.
Topological overview of the tomato coexpression network. A, Degree distribution of the network P(k) at various correlation thresholds (r ranging from 0.5 to 0.95); k indicates connectivity, and P(k) indicates the connectivity distribution. B, Partial coexpression network (r ≥ 0.6, P < 2.1e-31) in all data. The network shows how genes neighboring LeMADS-Rin (orange circle) correlate with each other. This undirected graph consists of nodes (black circles) and links (gray edges), indicating genes and positive correlations between genes.
Graph Clustering and GO Enrichment Analyses Reveal Functional Modules in the Tomato Transcriptome Data Set
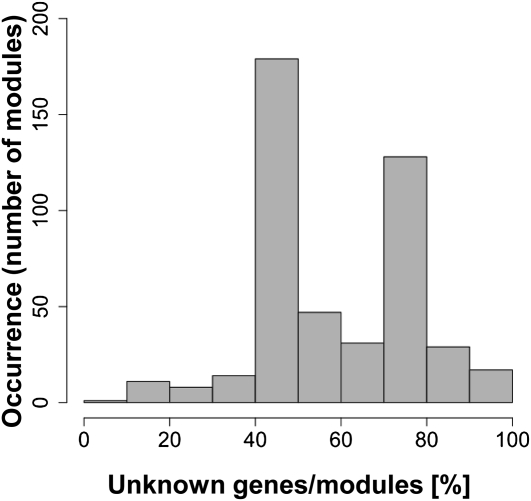
To efficiently identify densely connected nodes in the coexpression network (i.e. a coexpression module), we used the graph clustering algorithm IPCA (Li et al., 2008). We detected 465 modules with at least five gene members in the tomato coexpression network constructed from all data, consisting of 9,797 nodes and 1,754,361 edges; they ranged in size from five to 68 genes (Table II; Supplemental Table S1). We counted the number of unknown genes per module (Fig. 4; Supplemental Table S2). The distribution of the percentage of unknown genes within a module showed a bimodal curve at 50% and 70%.
Table II. Postulated physiological functions of coexpression modules.
We show 15 selected modules with five or more genes. Annotations are based on GO functional categories (FDR < 0.05). See also Supplemental Table S1.
| Module No. | No. of Genes | Predominant Function in Biological Process (Best P Value) |
| 1 | 63 | DNA endoreduplication (2.9e-2) |
| 4 | 54 | Photosynthesis (1.6e-3) |
| 52 | 65 | Response to organic substance (2.8e-2) |
| 90 | 54 | Protein metabolic process (2.2e-2) |
| 107 | 57 | Response to GA stimulus (1.3e-2) |
| 126 | 36 | Cell cycle process (9.2e-3) |
| 174 | 64 | Pathogenesis (4.7e-2) |
| 219 | 47 | Protein farnesylation (4.6e-2) |
| 291 | 49 | Response to organic substance (3.6e-2) |
| 309 | 19 | Apoptosis (3.0e-2) |
| 340 | 7 | Jasmonic acid metabolic process (4.1e-3) |
| 354 | 37 | Response to ethylene stimulus (1.1e-2) |
| 435 | 33 | Response to cold (4.3e-2) |
| 441 | 5 | Primary root development (1.2e-4) |
| 457 | 8 | Jasmonic acid metabolic process (8.7e-3) |
Figure 4.
Distribution of the number of unknown genes within a module.
We then subjected the modules to GO enrichment analysis; for simplicity, we selected a GO term with the best P value within the Biological Process (BP), Cellular Component (CC), and Molecular Function (MF) domains. For overrepresented GO terms, Table III lists the top 10 modules repeatedly assigned by the same functional term (Supplemental Tables S1 and S3). Of the modules extracted, 88% had significantly enriched GO terms in BP, CC, and MF. The distribution of GO terms for each module in tomato is shown in Supplemental Table S3. There were 150, 110, and 51 modules in the specific functional categories of “negative regulation of ligase activity,” “DNA endoreduplication,” and “photosynthesis,” respectively.
Table III. Distribution of GO terms for each module in the tomato data set.
Significance levels are set at FDR < 0.05. This list shows only the top 10 occurrences in this study. Frequency represents occurrences of the modules with specific GO terms. Detailed information is shown in Supplemental Table S3.
| BP Terms | Frequency | CC Terms | Frequency | MF Terms | Frequency |
| Negative regulation of ligase activity | 150 | Chloroplast thylakoid membrane | 59 | Water channel activity | 33 |
| DNA endoreduplication | 111 | PSI | 5 | Peptidase activity | 21 |
| Photosynthesis, light harvesting | 51 | Chloroplast stroma | 4 | Two-component sensor activity | 10 |
| Pathogenesis | 5 | Plant-type cell wall | 3 | Protein binding, bridging | 9 |
| Response to organic substance | 4 | Cytosolic part | 3 | Nucleocytoplasmic transporter activity | 8 |
| Generation of precursor metabolites and energy | 4 | Cytosolic ribosome | 2 | RNA glycosylase activity | 8 |
| Cellular protein metabolic process | 3 | Chloroplast part | 2 | Glc-1-P adenylyltransferase activity | 6 |
| Response to symbiont | 2 | Chloroplast | 2 | 4-Hydroxyphenylacetaldehyde oxime monooxygenase activity | 4 |
| Response to GA stimulus | 2 | Telomeric heterochromatin | 1 | rRNA binding | 3 |
| Response to cold | 2 | Protein farnesyltransferase complex | 1 | Water transmembrane transporter activity | 2 |
Predominant Functions of Coexpression Modules Show Biological Relevance
Below, we focus on the details of the extracted modules in Table II. The modules selected for our analysis follow (Supplemental Table S1).
DNA Endoreduplication
Module 1, consisting of 63 genes, was involved in DNA endoreduplication (false discovery rate [FDR] = 3.1e-2). It contained genes encoding cyclin A1 and A2, histone H4, B1-type cyclin-dependent kinase, ripening-regulated protein (differential display tomato fruit ripening 18), copper transport protein 1-like protein kinase 2, jasmonic acid 1 and 2, cytosolic Gln synthetase 1, and 12-oxophytodienoate reductase 3.
Photosynthesis
Module 4 (54 genes) was associated with photosynthesis (FDR = 1.5e-2). It included genes encoding the PSI subunit II protein precursor, PSII 23-kD protein, chlorophyll a/b-binding protein precursor, ribulose-1,5-bisphosphate carboxylase, ADP-Glc pyrophosphorylase large subunit, PSI reaction center protein subunit 2, and zeaxanthin epoxidase, an enzyme important in the xanthophyll cycle and in abscisic acid biosynthesis.
Response to Cold
Module 435, consisting of 33 genes, was related to “response to cold” (FDR = 4.3e-2). This module included genes encoding cytosolic ascorbate peroxidase 2 and dehydroascorbate reductase 2. These genes are involved in the ascorbate-glutathione cycle that scavenges reactive oxygen species, particularly hydrogen peroxide.
Jasmonic Acid Metabolic Process
Module 457 (eight genes) was involved in the “jasmonic acid metabolic process” (FDR = 8.7e-3). It contained the LeMTS1 gene encoding tomato monoterpene synthase. A homeobox knotted-1-like gene, LeT6 (Chen et al., 1997), was also included in this module. The LeT6 gene is essential for meristem maintenance and the process of leaf initiation (see “Discussion”).
Comparative Analyses of Topological Characteristics in Tomato Transcriptome Coexpression in Different Organ Data Sets
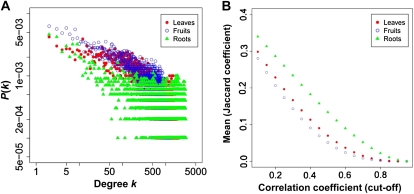
To obtain further insight into the tomato transcriptome, we employed a comparative approach to organ-specific coexpression. Using three subdata sets, leaves, fruits, and roots, we first assessed the topological characteristics of each coexpression network. As shown in Figure 5A, the degree distribution of each network (r ≥ 0.6) was indicative of a typical power-law distribution in a wide range of values for the degree k. We also calculated several graph-theoretic statistics, including the number of edges, average path length, and clustering coefficient (Barabási and Oltvai, 2004; Table I). In the roots data set, we observed that the highest number of edges and the highest clustering coefficient were 5,997,644 and 0.70, respectively. The average path length in three organs was smaller than three, indicating that these networks are characterized by a small-network property. To assess the degree of node overlap among the three data sets, we investigated similarity scores for nodes based on their connection partners using the Jaccard coefficient, which assesses the interaction between two graphs by assessing the tendency that links are present simultaneously in both graphs. Figure 5B shows the relationship between the average of the Jaccard coefficient and the correlation coefficient, and it indicates that the larger the cutoff of the correlation coefficient, the smaller the average similarity. In this graph, we observed that the roots data set had the highest Jaccard coefficient in all ranges.
Figure 5.
Topological overview of organ-specific coexpression networks. A, Degree distribution of the coexpression network (r ≥ 0.6) for three organs: leaves, fruits, and roots. B, Relationship between the average of the Jaccard coefficient and the correlation coefficient. The former can measure the degree overlap between two networks as the ratio of the intersection to the union of the networks.
Direct Measures of Differential Coexpression in the Tomato Transcriptome
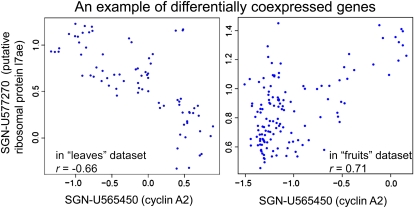
We next focused on changes in coexpression patterns between gene expression levels. We posited that a significant coexpression between given genes may be found under one condition but not another and that the changes elicited under one condition may be reversed under the other condition. An example of changed coexpression is shown in Figure 6. The correlation coefficient between Les.107.1.S1_at (SGN-U565450; cyclin A2) and Les.1081.1.S1_at (SGN-U577270; putative ribosomal protein) was negative (r = −0.66) in leaves but positive (r = 0.71) in fruits. To compare coexpression patterns among organs, we first visualized the three correlation matrices using pseudo-heat maps (Supplemental Fig. S2). While genes involved in photosynthesis were highly coexpressed in leaves and fruits, in roots they showed no remarkable coexpression within the pathway. For other pathways, root data displayed overall positive coexpression.
Figure 6.
A typical example of differentially coexpressed genes in different data sets. The correlation between the expression levels of two genes was significantly different in leaves and fruits (FDR = 9.27e-14). The axes represent relative gene expression. Note that differential coexpression was completely different from differential expression, and the mean level of given genes was significantly different between the two organs (see “Materials and Methods”). [See online article for color version of this figure.]
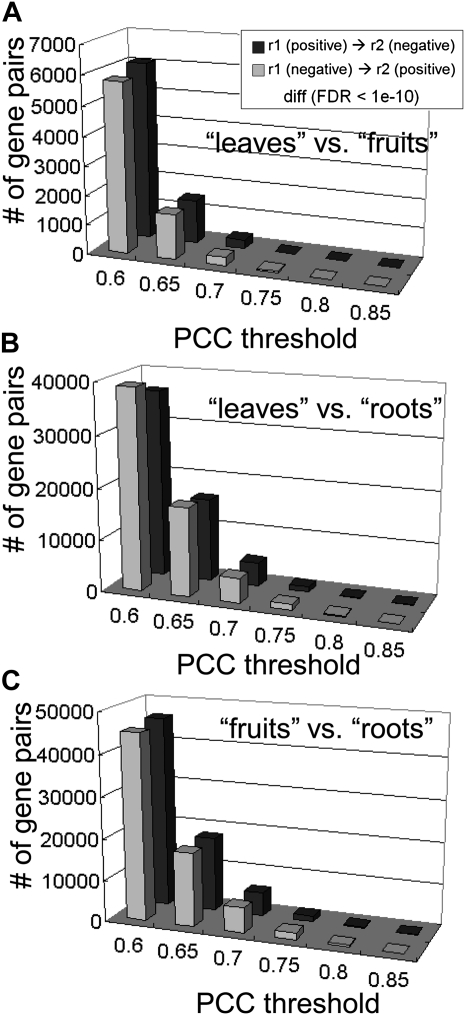
To obtain the difference between coexpression in the three organs, we used Fisher’s Z-transformation. This approach can test directly for differential coexpression by testing, for example, the null hypothesis H0: rL = rF. Here, rL and rF indicate that coexpression is calculated over the leaf and fruit samples, respectively (see “Materials and Methods”). In three comparisons, fruits versus roots, leaves versus fruits, and leaves versus roots, the numbers of significantly different correlation pairs were 753,133, 826,190, and 395,814, respectively (FDR < 1e-10; Fig. 7). Full lists of the differential coexpressions are shown in Supplemental Data S2. Using GO terms, we characterized the gene pairs with significant differential coexpression (Table IV; Supplemental Table S4). For example, in the transition from r > 0.7 in leaves to r < −0.7 in fruits, the differential coexpression included genes associated with cell wall modifications (FDR = 7.37e-3), while in the opposite transition (r < −0.7 in leaves to r > 0.7 in fruits), it included genes involved in flower development (FDR = 5.32e-4). In the case of the GO term “flower development,” we observed 17 annotated probes including LeMADS-Rin and other genes encoding the MADS box protein (Supplemental Table S4).
Figure 7.
Distribution of significantly differentially coexpressed genes between leaves and fruits (A), leaves and roots (B), and fruits and roots (C). This calculation was based on Fisher’s Z-transformation (see “Materials and Methods”). Thresholds correspond to Pearson’s correlation coefficient (PCC). Black and gray bars show the number of transitions from positive to negative correlations and from negative to positive correlations, respectively.
Table IV. Significantly overrepresented GO terms associated with differential coexpression among organs (FDR < 0.05) when the threshold is r = 0.7.
Numbers in parentheses indicate FDR. Detailed information is shown in Supplemental Table S4. n.s., Not significant.
| Domain | Leaves versus Fruits | Leaves versus Roots | Fruits versus Roots |
| Positive to negative | |||
| BP | Cell wall modification (7.37e-3) | n.s. | n.s. |
| CC | n.s. | n.s. | n.s. |
| MF | Enzyme inhibitor activity (4.03e-7) | Calcium ion binding (3.15e-2) | n.s. |
| Negative to positive | |||
| BP | Flower development (5.32e-4) | n.s. | n.s. |
| CC | n.s. | Thylakoid (6.28e-3) | n.s. |
| MF | Enzyme inhibitor activity (4.21e-4) | n.s. | n.s. |
Duplicated Genes Show a Small but Significant Number of Differential Coexpressions
We further investigated whether differential coexpression exists between duplicated genes. We first classified the all-probe-set “target” sequences into similarity clusters (referred to as gene families; see “Materials and Methods”). Consequently, 1,677 obtained clusters were regarded as gene families in this analysis. For 1,677 gene families including duplicated genes, we calculated the number of significantly different coexpressions (FDR < 1e-10). The numbers of duplicated genes that were differentially coexpressed among organs is listed in Table V and Supplemental Table S5. Overall, we were able to observe a small but significant number of differential coexpression relationships (Table V). For example, a gene family, Markov Cluster (MC) 100, was characterized by the GO terms “photosynthesis, light harvesting (FRD = 6.1e-5)” and “protein-chromophore linkage (FDR = 6.1e-5)”. In MC100, there were 19 differentially coexpressed pairs between leaves and roots, while there were 41 pairs between fruits and roots.
Table V. Number of coexpressions of duplicated genes with significant differences among organs.
See also Supplemental Table S5. n.s., Not significant.
| No. of Significantly Different Pairs | No. of Genes within Duplicated Gene Cluster (MC) | MC No. | GO Terms Biological Process (P Value) |
| Leaves versus fruits | |||
| 9 | 12 | MC665 | n.s. |
| 5 | 21 | MC7 | Cellular amino acid derivative biosynthetic process (1.7e-12) |
| 4 | 12 | MC893 | Regulation of transporter activity (3.3e-2) |
| 4 | 9 | MC141 | Brassinosteroid metabolic process (2.0e-6) |
| 4 | 16 | MC584 | Cellular response to auxin stimulus (6.7e-28); auxin-mediated signaling pathway (6.7e-28) |
| 4 | 10 | MC533 | Cellular cell wall organization (1.1e-15) |
| 3 | 5 | MC1117 | Oxidation reduction (1.6e-4) |
| 3 | 13 | MC813 | Ciliary or flagellar motility (1.6e-5) |
| 3 | 7 | MC310 | n.s. |
| Leaves versus roots | |||
| 19 | 16 | MC100 | Photosynthesis, light harvesting (6.1e-5); protein-chromophore linkage (6.1e-5) |
| 6 | 21 | MC7 | Cellular amino acid derivative biosynthetic process (1.7e-12) |
| 4 | 8 | MC10 | Cellular response to auxin stimulus (2.4e-9); auxin-mediated signaling pathway (2.4e-9) |
| 3 | 7 | MC596 | Response to cold (2.0e-3) |
| 3 | 6 | MC748 | Regulation of epidermal cell division (1.8e-2) |
| 3 | 7 | MC1622 | Ethylene metabolic process (4.5e-13) |
| 3 | 12 | MC786 | Phenylpropanoid biosynthetic process (1.3e-4) |
| 3 | 15 | MC493 | Regulation of protein metabolic process (1.6e-5) |
| 3 | 6 | MC19 | Cellular response to auxin stimulus (1.2e-6); auxin-mediated signaling pathway (1.2e-6) |
| Fruits versus roots | |||
| 41 | 16 | MC100 | Photosynthesis, light harvesting (6.1e-5); protein-chromophore linkage (6.1e-5) |
| 8 | 21 | MC7 | Cellular amino acid derivative biosynthetic process (1.7e-12) |
| 7 | 15 | MC493 | Regulation of protein metabolic process (1.6e-5) |
| 5 | 5 | MC134 | Ubiquitin-dependent protein catabolic process (5.0e-3) |
| 4 | 14 | MC296 | Response to chitin (3.9e-15) |
| 4 | 12 | MC181 | Cellular response to hydrogen peroxide (4.4e-21); hydrogen peroxide catabolic process (4.4e-21) |
| 4 | 9 | MC141 | Brassinosteroid metabolic process (2.0e-6) |
| 4 | 9 | MC189 | Gluconeogenesis (4.2e-3) |
| 4 | 6 | MC19 | Cellular response to auxin stimulus (1.2e-6); auxin-mediated signaling pathway (1.2e-6) |
| 3 | 7 | MC1622 | Ethylene metabolic process (4.5e-13) |
| 3 | 8 | MC641 | Gluconeogenesis (1.9e-3) |
To gain further insights into genes with significant differential coexpression, we investigated the number of significant differential coexpressions (FDR < 1e-10) for each of the 256 metabolic pathways from the LycoCyc (Bombarely et al., 2011; Table VI; Supplemental Table S6). For example, there were 15 differential coexpressions in “glycolysis IV (plant cytosol)” between fruits and roots but only three differential coexpressions between leaves and roots. The coexpression patterns in biosynthetic pathways associated with the Calvin cycle, sugar degradation, photorespiration, and the tricarboxylic acid (TCA) cycle, as well as glycolysis, were usually different among organs. In comparisons between leaves and fruits, but not in other comparisons, there were three differentially coexpressed pairs in anthocyanin biosynthesis. On the other hand, gene pairs in flavonoid biosynthesis were different when compared with roots. We also studied duplicated genes encoding isozymes that were primarily derived from LycoCyc. Most of these genes were inferred from computational analysis without any human curation. As shown in Supplemental Table S7, we observed quite a few pairs with significantly different coexpression (FDR < 1e-10) between duplicated genes (isoforms). For example, the number of significantly different coexpression genes encoding β-fructofuranosidase was four of 528 possible gene pairs.
Table VI. Number of coexpressed pairs with significant differences between leaves and fruits in metabolic pathway-related genes.
FDR < 1e-10. We used the LycoCyc database for classification of genes. See also Supplemental Table S6.
| No. of Pairs | No. of Pathway Genes | Pathway |
| 19 | 134 | Suc degradation to ethanol and lactate (anaerobic) |
| 15 | 100 | Glycolysis IV (plant cytosol) |
| 14 | 105 | Fru degradation to pyruvate and lactate (anaerobic) |
| 13 | 103 | Calvin cycle |
| 12 | 93 | Glycolysis I |
| 12 | 90 | Glycolysis V |
| 8 | 71 | Gluconeogenesis |
| 8 | 78 | Glc heterofermentation to lactate I |
| 7 | 78 | Glycolysis II |
| 6 | 71 | Glc fermentation to lactate II |
| 6 | 62 | Photorespiration |
| 5 | 35 | Entner-Doudoroff pathway II nonphosphorylative |
| 5 | 35 | Entner-Doudoroff pathway III semiphosphorylative |
| 5 | 38 | UDP-Glc conversion |
| 4 | 15 | Folate transformations |
| 4 | 26 | FormylTHF biosynthesis I |
| 4 | 38 | FormylTHF biosynthesis II |
| 4 | 55 | Salvage pathways of purine and pyrimidine nucleotides |
| 4 | 28 | TCA cycle |
| 4 | 25 | TCA cycle variation VIII |
| 3 | 20 | Asn degradation I |
| 3 | 46 | Flavonoid biosynthesis |
| 3 | 69 | tRNA charging pathway |
| 2 | 37 | Chlorophyllide a biosynthesis |
| 2 | 11 | Gly degradation I |
| 2 | 41 | Jasmonic acid biosynthesis |
| 2 | 18 | Phe degradation I |
| 2 | 15 | Reductive TCA cycle |
| 2 | 15 | Rib degradation |
| 2 | 38 | Triacylglycerol degradation |
| 2 | 14 | Tyr degradation |
| 1 | 4 | γ-Glutamyl cycle |
| 1 | 24 | Arg biosynthesis II (acetyl cycle) |
| 1 | 27 | Carotenoid biosynthesis |
| 1 | 16 | Cys biosynthesis I |
| 1 | 19 | dTDP-l-Rha biosynthesis I |
| 1 | 18 | Folate polyglutamylation I |
| 1 | 17 | Formaldehyde assimilation I (Ser pathway) |
| 1 | 4 | Glutathione biosynthesis |
| 1 | 10 | Gly betaine degradation |
| 1 | 11 | Gly biosynthesis I |
| 1 | 38 | Lipoxygenase pathway |
| 1 | 9 | Orn biosynthesis |
| 1 | 15 | Phenylpropanoid biosynthesis |
| 1 | 72 | Purine nucleotides de novo biosynthesis I |
| 1 | 39 | Respiration (anaerobic) |
| 1 | 12 | Salvage pathways of purine nucleosides |
| 1 | 16 | Salvage pathways of purine nucleosides II, plant |
| 1 | 39 | Suberin biosynthesis |
| 1 | 39 | Suc biosynthesis |
| 1 | 50 | Suc degradation I |
| 1 | 33 | Sulfate assimilation III |
| 1 | 13 | Superpathway of Ser and Gly biosynthesis II |
Differential Coexpression Provides Clues of Key Regulatory Steps in Metabolic Pathways
To interpret differential coexpression in metabolic pathways, we investigated, in detail, two biosynthetic pathways, lycopene and flavonoid, in a proof-of-concept study.
Lycopene Biosynthesis
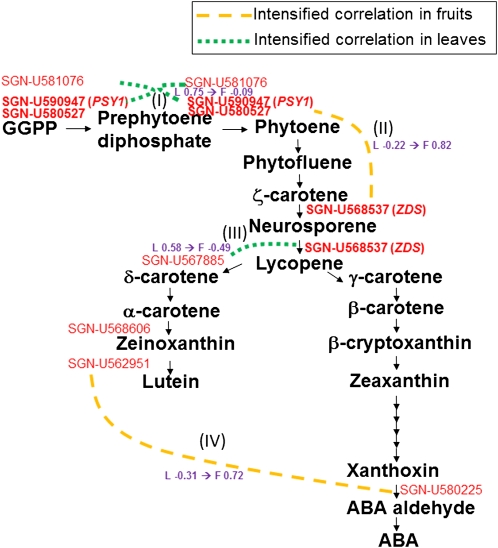
The changes in coexpression between organs involved in carotenoid biosynthesis are of particular interest because this pathway contributes to the production of lycopene and β-carotene, which can be utilized as indicators of tomato quality. The mapping of differential coexpression between leaves and fruits onto a lycopene biosynthesis pathway (Fig. 8) revealed significantly different coexpression in this pathway. The coexpression of genes encoding phytoene synthase (PSY1; Les.3171.4.A1_at; SGN-U580527) and abscisic acid stress ripening 1 (Les.4930.1.A1_at; SGN-U581076) was significantly different between leaves and fruits (Fig 8, I). In leaves, their expression was positively correlated (r = 0.75); there was no correlation in fruits (r = −0.09). The expression of the PSY1 gene (Les.3171.4.A1_at; SGN-U580527) and that of the gene encoding ζ-carotene desaturase (ZDS; Les.20.1.S1_a; SGN-U568537) exhibited a highly positive correlation (r = 0.82) in fruits and a weak negative correlation in leaves (r = −0.22; Fig. 8, II). The ZDS gene (Les.20.1.S1_at; SGN-U568537) and the gene encoding LELYCOCYC lycopene ε-cyclase (Les.3771.1.S1_at; SGN-U567885) were mildly coexpressed in leaves (r = 0.58); in fruits, the coexpression was mildly negative (r = −0.49; Fig. 8, III). We also observed differential coexpression in genes associated with lutein biosynthesis and abscisic acid biosynthesis pathways (Fig. 8, IV). The coexpression between the gene encoding ε-ring hydroxylase (Les.4915.1.S1_at; SGN-U562951) and the short-chain dehydrogenase/reductase (SDR) homolog (LesAffx.68802.1.S1_at; SGN-U580225) was highly positive (r = 0.72) in fruits and mildly negative (r = −0.31) in leaves.
Figure 8.
Differential coexpressions mapped onto the lycopene biosynthesis pathway. Solid arrows show the reaction steps. Orange dashed lines and green broken lines represent intensified correlations in fruits (F) and leaves (L), respectively.
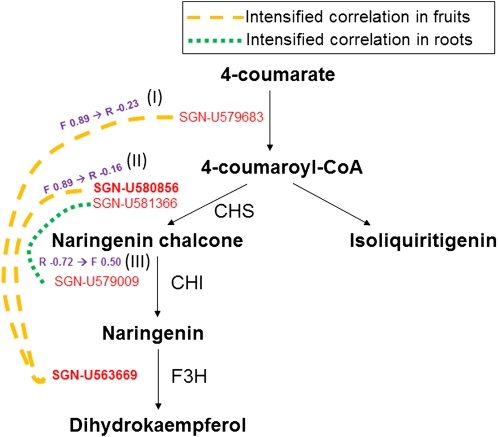
Flavonoid Biosynthesis
There were three differential coexpression patterns in flavonoid biosynthesis (Fig. 9). The gene encoding flavanone 3-hydroxylase (Les.2278.1.S1_at; SGN-U563669) and the gene encoding 4-coumarate-CoA ligase (Les.5848.1.A1_at; SGN-U579683) exhibited a highly positive correlation (r = 0.89) in fruits and a weak negative correlation in roots (r = −0.23; Fig. 9, I). Similarly, the gene encoding flavanone 3-hydroxylase (Les.2278.1.S1_at; SGN-U563669) and the gene for naringenin-chalcone synthase (Les.3649.1.S1_at; SGN-U580856) showed a highly positive correlation (r = 0.89) in fruits and a weak negative correlation in roots (r = −0.16; Fig. 9, II). The last pair manifested a pattern that was in direct contrast to the one observed between the gene encoding chalcone isomerase (Les.3218.1.S1_at; SGN-U579009) and putative naringenin-chalcone synthase (Les.4911.1.S1_at; SGN-U581366), which showed a positive correlation (r = 0.50) in fruits and a highly negative correlation (r = −0.72) in roots (Fig. 9, III).
Figure 9.
Differential coexpression mapping to the flavonoid biosynthesis pathway. Solid arrows represent reaction steps. Orange dashed lines and green broken lines indicate intensified correlations in fruits (F) and roots (R), respectively. CHS, Chalcone synthase; CHI, chalcone isomerase; F3H, flavanone-3-hydroxylase.
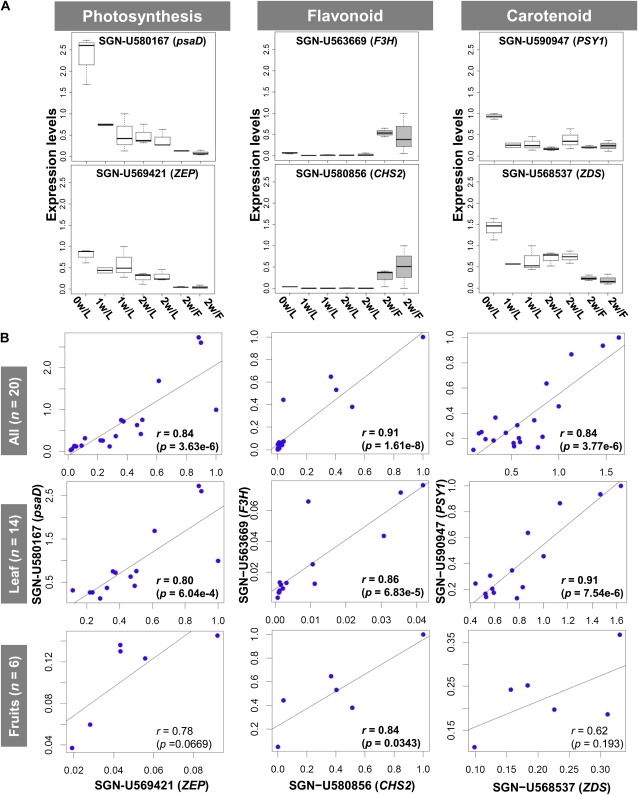
Independent Experimental Verification of the Microarray Data Sets Using qRT-PCR
To independently confirm the expression profiling data, six expressed genes identified in our microarray results were assayed with qRT-PCR at seven experimental conditions using the same RNA sample that was used for our original experiment consisting of 20 microarrays (see “Materials and Methods”). We chose six genes that were involved in photosynthesis, flavonoid biosynthesis, and carotenoid biosynthesis, which corresponded with three types of expression pattern: (1) leaf specific, (2) fruit specific, and (3) moderate (Fig. 10A). We verified that the expression patterns were consistent between microarray and qRT-PCR analyses and showed very good reproducibility (Supplemental Fig. S3). Thus, we can state the following: (1) the significant coexpression in module 4, which includes the genes ZEP and psaD, was reproducible (r = 0.84, P = 3.63e-6; Fig. 10), and (2) regarding flavonoid genes, the calculation based on fruit samples resulted in significant coexpression (r = 0.84, P = 0.0343). This result supports, at least in part, our approach (Fig. 9). These verification results suggest that the resulting coexpressions derived from 307 collected data sets, as well as each subdata set (leaves, fruits, and roots), are relevant, although we must note coexpression between genes with low-level expression.
Figure 10.
qRT-PCR assessment of microarray results. A, Box plots of expression values measured by qRT-PCR across seven experimental conditions, including leaf (L) and fruit (F) samples. Six genes associated with biosynthetic pathways were involved in photosynthesis, photosystem 1 reaction center protein subunit 2 (psaD; SGN-U580167) and zeaxanthin epoxidase (ZEP; SGN-U569421); flavonoids, flavanone 3-hydroxylase (F3H; SGN-U563669) and chalcone synthase 2 (CHS2; SGN−U580856); and carotenoids, phytoene synthase 1 (PSY1; SGN−U590947) and ζ-carotene desaturase (ZDS; SGN-U568537). B, Coexpression patterns between the genes, as determined by qRT-PCR (blue circles). Correlation between qRT-PCR and microarray analyses was also verified as shown in Supplemental Figure S3. w, Week. [See online article for color version of this figure.]
DISCUSSION
We present a comprehensive coexpression network analysis based on over 300 tomato microarrays and investigate the topological properties of the network. The degree distribution of the coexpression network followed the heterogeneous power law. The subdata sets from three organs, leaf, fruit, and root samples, also exhibited high heterogeneity and modularity (e.g. power-law degree distributions and small-world properties). This result indicates that the properties of the network are consistent with earlier reports on typical coexpression networks (Stuart et al., 2003). We constructed genome-wide tomato coexpression networks with a correlation cutoff value of r ≥ 0.6 (P < 2.1e-31, Pearson’s correlation statistical test) using all data. Our threshold selection was supported, at least partially, by earlier studies. Aoki et al. (2007) showed that the Arabidopsis coexpression network had a minimal network density at a cutoff r value ranging from 0.55 to 0.60. Using simulated data sets, Elo et al. (2007) demonstrated that a cutoff value of r = 0.6 yielded both low error and high reproducibility.
We clustered densely coexpressed genes by graph clustering, a nontargeted approach that divides the tomato coexpression network into gene modules. We detected 465 coexpression modules and assessed them by GO term enrichment analysis. A coexpression module-based approach applied to 67 microarrays from 24 different tissues in tomato provided strong predictions for gene functions, such as the pathways involved in flavonoid biosynthesis (Ozaki et al., 2010). Based on the evolutionary conservation of coexpression in the Solanaceae family, Miozzi et al. (2010) developed the data-mining tool, ORTom, which predicts functional relationships among tomato genes. The Tomato Functional Genomics Database (TFGD) offers a Web-based retrieval tool for coexpressed genes (Fei et al., 2011). There are several differences between our approach and these previous studies. First, our study involved a much larger number of microarrays (more than 300) and covered wider experimental conditions, including various stress experiments. Second, we introduced differential coexpression into our plant transcriptome study. In addition, the extracted modules carried many unknown genes (Fig. 4). Finally, the expression profiles of six selected genes were independently verified by qRT-PCR (Fig. 10; Supplemental Fig. S3).
The coexpression module-based approach by graph clustering is an important approach to facilitate predictions (Mentzen and Wurtele, 2008; Atias et al., 2009; Fukushima et al., 2009a; Mao et al., 2009). In our previous works (Fukushima et al., 2009a, 2011), we used DPClus-based graph clustering to characterize gene coexpression networks and metabolomic correlation networks. Although we believe that DPClus is also useful for gene coexpression networks, it has a limitation on the number of nodes (limitation: n < 5,000). To this end, we chose IPCA (Li et al., 2008), which enables us to perform the graph clustering for larger scale networks in this study. We demonstrated that 88% of all coexpression modules in the tomato were assigned by GO terms, although the modules to which we assigned GO terms overlapped markedly. The tomato genome sequencing project and enhanced genomic annotations in tomato (Barone et al., 2008; Bombarely et al., 2011) will improve such characterizations of coexpression modules. We highlighted four modules, module 1, module 4, module 435, and module 457 (Table II). The coexpression between genes encoding cyclin B1 and histone H4 in module 1 (GO term DNA endoreduplication) has been observed in Arabidopsis (see the ATTED-II database; Obayashi et al., 2011). Module 4 (GO term photosynthesis) is consistent with previous reports in Arabidopsis and rice (Mentzen and Wurtele, 2008; Fukushima et al., 2009a). Genes in module 435 (GO term response to cold) are reasonable in the sense that abiotic stresses, including cold stress, are related to oxidative stress that perturbs almost all functions in a plant cell. Module 457 (GO term jasmonic acid metabolic process) was also relevant because it included the LeMTS1 gene. According to van Schie et al. (2007), the expression of the gene in tomato leaves was induced by jasmonic acid treatment, and this gene encoded a linalool synthase. We also observed the LeT6 gene (Chen et al., 1997) in this module. Although both genes were highly coexpressed, their relationship is currently unclear. Using the arf6arf8 double mutant, Tabata et al. (2010) showed that Arabidopsis auxin response factors 6 and 8 regulate jasmonic acid biosynthesis and floral organ development via class 1 KNOX genes. This mutant is similar to that found in plants harboring a mutation associated with the biosynthetic pathway of jasmonic acid, which causes morphologic abnormality in floral organ development. Our clustering results may suggest a yet unknown transcriptional coordination between jasmonic acid biosynthesis and KNOX genes in tomato. The predominant functions of the coexpression modules detected here show biological relevance. Our top-down approach revealed good candidates for functional modules in the tomato transcriptome. That is, when using the guilt-by-association principle, such a coexpression module-based approach can help to predict the function of unknown genes within a module, although coexpressed genes are not necessarily involved in the same biological process.
We studied differential coexpression among tomato leaves, fruits, and roots using Fisher’s Z-transformation. Direct measurements showed that duplicated genes as well as metabolic genes exhibit a small but significant number of differential coexpressions. Gene clusters assigned the GO term flower development as an example of switching coexpression included LeMADS-Rin (Table IV; Supplemental Table S4). Regarding the link between the identification of differential coexpression and the identification of coexpressed gene modules, we describe this as a complementary relationship. The reason for this statement is 2-fold. Because gene coexpression can be interpreted as a “fingerprint” of the underlying transcriptional network, it can be used to compare two physiological states of cellular systems. That is, the differential coexpression approach leads to a systematic difference in transcriptomic levels among such organs. Second, the comparative coexpression study here provides a way to use the observed coexpression to find additional information about the plant transcriptome. It is obvious that identifying coexpressed gene modules is not sufficient, because differentially coexpressed gene groups also reflect the changes in underlying transcriptional regulation.
We also demonstrated the existence of several pairs within the LycoCyc pathway (enzymatic isoforms) that had significantly different coexpression between duplicated genes. In our proof-of-concept study, we investigated various patterns of differential coexpression in two biosynthetic pathways associated with lycopene and flavonoid. In the lycopene biosynthesis pathway, we identified four significant differences in coexpression between leaves and fruits (Fig. 8). Of the genes involved, PSY encodes a key regulatory enzyme (Cazzonelli and Pogson, 2010). Positive coexpression of the PSY and ZDS genes in tomato fruits is highly likely from a proof-of-concept point of view regarding differential coexpression approaches. The differential coexpression between the two isoforms of the PSY gene may reflect organ-specific regulation at the transcription level. We found that two genes encoding the ε-ring hydroxylase and the SDR homolog exhibited markedly different coexpression patterns in leaves and fruits. In fruits, the high coexpression (r = 0.73) between the genes encoding ε-ring hydroxylase and SDR was partly consistent with the observation that two abscisic acid-deficient mutants, sitiens and flacca, exhibit a decrease in lutein levels (Galpaz et al., 2008). Our approach suggests that the correlation between abscisic acid and lutein levels in tomato plants is attributable to their transcriptional coordination.
Based on transcriptome coexpression analysis, several flavonoid biosynthetic genes in Arabidopsis were characterized (Luo et al., 2007; Yonekura-Sakakibara et al., 2007, 2008, 2012). Flavonoids in plants play significant roles in many biological processes, such as protecting plants against ultraviolet light, providing pigmentation in fruits and flowers, and protecting against diseases and pests in higher plants (Buer et al., 2010). In the tomato, the total flavonoid concentration in roots is lower than that in leaves and fruits (Zornoza and Esteban, 1984); among the three organs, tomato fruits manifest the highest concentration of total flavonoids. Earlier studies had detected up to 70 flavonoids, including naringenin chalcone, kaempferol, and quercetin, in tomato fruits (cv Micro-Tom; Moco et al., 2006; Iijima et al., 2008). Our results showed strong coexpression patterns among genes encoding chalcone synthase, chalcone isomerase, and flavanone 3-hydroxylase in fruits (Fig. 9). However, there was no coexpression between these genes in roots. In addition, we found a reversal of coexpression between genes encoding chalcone synthase and chalcone isomerase. This suggests a pronounced change in the underlying transcriptional regulation of the flavonoid pathway. Taken together, these observations suggest that at least in the lycopene and flavonoid pathways, these differential coexpressions reflect a key regulatory mechanism among organs.
Because a marked change in coexpression patterns among organs may reflect underlying transcriptional changes directly or indirectly, the genes related to these differential coexpressions may act distinctly in those organs. Differential coexpression analysis represents a good data-mining approach to prioritizing candidate genes in further functional genomics studies on computationally assigned genes encoding an enzyme, for example. Additional work is needed to evaluate the conservation of differential coexpression patterns among species, and more efficient tools for their assessment in plants would be helpful. For example, like Kappa-view 4 (Sakurai et al., 2011), the mapping of differential coexpressions onto metabolic pathways is highly useful in the field of plant research. We offer our extensive analysis of the coexpression networks to aid researchers in the selection and prioritization of candidate genes in studies on tomato functional genomics.
MATERIALS AND METHODS
Plant Material and Growth Conditions
We developed a plant irradiation system based on applying light-emitting diodes to a leaf. We used this system to investigate changes in transcript profiles of leaves and fruits grown under different light conditions. Seeds from tomato (Solanum lycopersicum ‘Reiyo’) were sown in 72-cell plant trays (Takii Seed) and grown in a commercial soil mix (Napura Soil Mixes) for 2 weeks in a growth chamber (MKV DREAM) at 25°C/20°C (light/dark) and 900 μL L−1 CO2 concentration with a light/dark cycle of 16 h/8 h for 2 weeks at Chiba University. Then, seedlings were transferred to pots one by one (final size, 2.4 L) and grown in a growth chamber (Asahi Kogyosha). The photosynthetic photon flux level in the growth chamber was adjusted to 450 to 500 μmol m−2 s−1 when we measured at the meristem of each tomato plant (light source, ceramic metal halide lamps). After plant flowering in summer 2010, we removed leaves and trusses from each plant, with the exception of the second truss, a leaf below the second truss, and the meristem. Light-emitting diode irradiation at 0, 200, and 1,000 μmol m−2 s−1 was directly applied to the leaf using a plant irradiation system (humidity, 70%; CO2 concentration, 900 μL L−1). Plant material was harvested after 0, 1, and 2 weeks. Twenty samples were analyzed (14 leaf samples and six pericarp samples), and two to three biological replicates were used.
GeneChip Microarray Analysis
Analysis was performed using the Affymetrix GeneChip Tomato Genome Array according to the manufacturer’s instructions. This microarray includes more than 9,000 transcripts and does not cover all of the approximately 35,000 genes encoded in the tomato genome, which are located largely in euchromatic regions (Van der Hoeven et al., 2002). RNA was extracted using the standard procedure of Affymetrix. Our own data have been deposited in the Gene Expression Omnibus (GEO; Barrett et al., 2011) and are accessible through the GEO series accession number GSE35020.
qRT-PCR Analysis
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). Reverse transcription of each total DNase-treated (Qiagen) RNA sample was performed using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). qRT-PCR with the first-strand cDNA using the Fast SYBR Green Master Mix (Applied Biosystems) was performed on the ABI StepOnePlus Real Time PCR system (Applied Biosystems). qRT-PCR primers used in this study are listed in Supplemental Table S8.
Data Collection and Preprocessing
We collected 307 GeneChips from publicly available databases including GEO, ArrayExpress (Parkinson et al., 2011), and TFGD (Fei et al., 2011) and included 20 of our own hybridizations (GEO accession no. GSE35020), resulting in a total of 327 arrays. The collected data set contains 17 experiments (Fig. 2; Supplemental Data S1). The raw CEL data were normalized by the robust multichip average (Irizarry et al., 2003) with Bioconductor (Gentleman et al., 2004). The resulting values were normalized to the same range by means-based scale normalization. Probe sets with the prefix RPTR or AFFX were removed. Using the detection call (present/absent) from the MAS5 algorithm (http://www.affymetrix.com/support/technical/whitepapers/sadd_whitepaper.pdf) with default settings, we excluded all probe sets with absent calls in all samples. For the visual inspection of the quality of microarrays based on chip pseudoimages of residual weights and signed residuals, we used the R packages affy (Gautier et al., 2004) and affyPLM (Gentleman et al., 2005). We next performed a quality check of the microarrays based on the Kolmogorov-Smirnov goodness-of-fit statistic D (Persson et al., 2005) and discarded all GeneChips with D ≥ 0.15, resulting in 307 high-quality data sets for further analyses. This calculation was done with HDBStat! (Trivedi et al., 2005).
Constructing Coexpression Networks and Topological Analyses
Using Pearson’s correlation coefficient (r), we calculated correlation matrices of subdata sets classified by organ names (leaves, fruits, and roots) as well as all data (Fig. 1). The number of arrays corresponding to the leaves, fruits, and roots were 77, 135, and 36, respectively. A correlation matrix was calculated for the remaining 9,989 probe sets. All topological analyses, such as calculation of the Jaccard coefficient, were performed in R with the igraph package (Csardi and Nepusz, 2006). Coexpression networks were visualized using the Cytoscape software program (Shannon et al., 2003).
Graph Clustering and Functional Enrichment Analysis
To extract coexpression modules, we utilized the graph clustering algorithm IPCA (Li et al., 2008), an extension of the original DPClus algorithm (Altaf-Ul-Amin et al., 2006). DPClus is based on the combined periphery and density of graphs to extract dense subgraphs. The parameters we used were as follows: modules with sizes smaller than five (S = 5), a shortest path length of two (P = 2), and a threshold parameter Tin of 0.6 (T = 0.6).
We used BiNGO (Maere et al., 2005), a tool to assess overrepresentation or underrepresentation in a set of genes, for the analysis of significantly overrepresented GO categories among coexpression modules detected by graph clustering. We selected a GO term with the best P value within the BP, CC, and MF domains. The Benjamini and Hochberg (1995) correction for FDR was applied in functional enrichment analysis.
Calculating Differential Coexpression
We used the term “differential coexpression” to describe significant correlations between given genes found under one, but not another, condition. This definition also included instances in which the correlations were changed to the opposite direction under two conditions (Fig. 6). It provided a means to identify associations that were dramatically changed by mutations, organs, or treatments. For example, the correlation between gene expressions was calculated over the leaf sample (rL) and over the fruit sample (rF). In this case, the differential coexpression could be elucidated by testing the null hypothesis H0: rL = rF. To test whether two coexpressions were significantly different from one another, the correlations were transformed by Fisher’s Z-transformation. If there were two correlations with sample sizes n1 and n2, they were each transformed into Fisher’s Z values, Z = 1/2[ln(1 + r)/(1 − r)]. Under the null hypothesis that the population correlations are equal, the Z value, 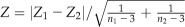 has an approximately normal distribution. We also calculated the local FDR for all correlation tests with the fdrtool package (Strimmer, 2008).
has an approximately normal distribution. We also calculated the local FDR for all correlation tests with the fdrtool package (Strimmer, 2008).
Definition of Duplicated Genes
The tomato target sequences from the Affymetrix Web site (http://www.affymetrix.com/Auth/analysis/downloads/data/Tomato.target.zip) were utilized in an all-against-all BLASTX search (cutoff threshold, E < 1e-5). We used the Markov chain clustering algorithm (http://micans.org/mcl/; Van Dongen, 2000) to assign the target sequences to clusters. The 1,677 obtained clusters were regarded as gene families in this analysis.
Gene Annotation
Annotation information (release 31) for tomato was downloaded from the Affymetrix Web site. Gene ontology of tomato genes was based on information from TFGD (Fei et al., 2011). To assign the Sol Genomics Network (SGN) unigene (Mueller et al., 2005; Bombarely et al., 2011), similarity searches between the Affymetrix probe set identifier and the SGN unigene were performed using BLASTN (Altschul et al., 1990) with a threshold E value of 1e-10. For functional categories of central metabolism, we used the MapMan flat file (Thimm et al., 2004) from the Web site (http://mapman.gabipd.org/web/guest/mapmanstore). We also used the SolCyc (LycoCyc) database (Bombarely et al., 2011) for the classification of metabolic genes.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The distribution of the expression matrix (A) and correlation coefficients (B) for the three organs are based on all probe sets calculated using the robust multichip average algorithm.
Supplemental Figure S2. Correlation heat maps for four data sets: all data, leaves, fruits, and roots.
Supplemental Figure S3. Correlation of expression values measured by qRT-PCR and microarray.
Supplemental Table S1. List of 465 coexpression modules in the tomato all-data data set.
Supplemental Table S2. List of unknown genes for each of the 465 modules in the tomato all-data data set.
Supplemental Table S3. Distribution of GO terms for each module in the tomato all-data data set.
Supplemental Table S4. Significantly overrepresented GO terms associated with differential coexpression among organs (FDR < 0.05).
Supplemental Table S5. Number of coexpressions of duplicated genes with significant differences between leaves and fruits.
Supplemental Table S6. Number of coexpressed pairs with significant differences between organs (FDR < 1e-10).
Supplemental Table S7. Number of coexpressed pairs with significantly different coexpression between duplicated genes (isoforms) within the LycoCyc database version 1.0.
Supplemental Table S8. qRT-PCR primers used in this study.
Supplemental Data S1. Meta-data for the microarrays used in this study.
Supplemental Data S2. List of differential coexpressions between the data sets of leaves and fruits, leaves and roots, and fruits and roots.
Supplementary Material
Acknowledgments
We gratefully acknowledge the tomato research community for providing their microarray data through GEO, ArrayExpress, and TFGD databases. We also thank Tetsuya Sakurai and Kenji Akiyama for computational assistance, Takashi Osanai for technical support for the qRT-PCR instrument, and Ursula Petralia for editing our manuscript.
References
- Altaf-Ul-Amin M, Shinbo Y, Mihara K, Kurokawa K, Kanaya S. (2006) Development and implementation of an algorithm for detection of protein complexes in large interaction networks. BMC Bioinformatics 7: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Aoki K, Ogata Y, Shibata D. (2007) Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol 48: 381–390 [DOI] [PubMed] [Google Scholar]
- Atias O, Chor B, Chamovitz DA. (2009) Large-scale analysis of Arabidopsis transcription reveals a basal co-regulation network. BMC Syst Biol 3: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási AL, Oltvai ZN. (2004) Network biology: understanding the cell’s functional organization. Nat Rev Genet 5: 101–113 [DOI] [PubMed] [Google Scholar]
- Barone A, Chiusano ML, Ercolano MR, Giuliano G, Grandillo S, Frusciante L. (2008) Structural and functional genomics of tomato. Int J Plant Genomics 2008: 820274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. (2011) NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res 39: D1005–D1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 39: D1149–D1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Imin N, Djordjevic MA. (2010) Flavonoids: new roles for old molecules. J Integr Plant Biol 52: 98–111 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson BJ. (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Chen JJ, Janssen BJ, Williams A, Sinha N. (1997) A gene fusion at a homeobox locus: alterations in leaf shape and implications for morphological evolution. Plant Cell 9: 1289–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia BK, Karuturi RK. (2010) Differential co-expression framework to quantify goodness of biclusters and compare biclustering algorithms. Algorithms Mol Biol 5: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Yu U, Yoo OJ, Kim S. (2005) Differential coexpression analysis using microarray data and its application to human cancer. Bioinformatics 21: 4348–4355 [DOI] [PubMed] [Google Scholar]
- Csardi G, Nepusz T. (2006) The igraph software package for complex network research. InterJournal Complex Systems 1695. http://igraph.sf.net [Google Scholar]
- de la Fuente A. (2010) From ‘differential expression’ to ‘differential networking’: identification of dysfunctional regulatory networks in diseases. Trends Genet 26: 326–333 [DOI] [PubMed] [Google Scholar]
- Elo LL, Järvenpää H, Oresic M, Lahesmaa R, Aittokallio T. (2007) Systematic construction of gene coexpression networks with applications to human T helper cell differentiation process. Bioinformatics 23: 2096–2103 [DOI] [PubMed] [Google Scholar]
- Fei Z, Joung JG, Tang X, Zheng Y, Huang M, Lee JM, McQuinn R, Tieman DM, Alba R, Klee HJ, et al. (2011) Tomato Functional Genomics Database: a comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res 39: D1156–D1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A, Kanaya S, Arita M. (2009a) Characterizing gene coexpression modules in Oryza sativa based on a graph-clustering approach. Plant Biotechnol 26: 485–493 [Google Scholar]
- Fukushima A, Kusano M, Redestig H, Arita M, Saito K. (2009b) Integrated omics approaches in plant systems biology. Curr Opin Chem Biol 13: 532–538 [DOI] [PubMed] [Google Scholar]
- Fukushima A, Kusano M, Redestig H, Arita M, Saito K. (2011) Metabolomic correlation-network modules in Arabidopsis based on a graph-clustering approach. BMC Syst Biol 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J. (2008) Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J 53: 717–730 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. (2004) affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W. (2005) Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J, Pavlidis P. (2009) A methodology for the analysis of differential coexpression across the human lifespan. BMC Bioinformatics 10: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ, Noensie EN, Ruezinsky DM, Lu X, Tracy SL, Ganal MW, Martin GB, Pillen K, Alpert K, Tanksley SD. (1995) Molecular genetic analysis of the ripening-inhibitor and non-ripening loci of tomato: a first step in genetic map-based cloning of fruit ripening genes. Mol Gen Genet 248: 195–206 [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. (2007) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, Zhu JK, Cushman JC, Gollery M, Girke T. (2008) Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol 147: 41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y, Nakamura Y, Ogata Y, Tanaka K, Sakurai N, Suda K, Suzuki T, Suzuki H, Okazaki K, Kitayama M, et al. (2008) Metabolite annotations based on the integration of mass spectral information. Plant J 54: 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Kusano M, Tabuchi M, Fukushima A, Funayama K, Diaz C, Kobayashi M, Hayashi N, Tsuchiya YN, Takahashi H, Kamata A, et al. (2011) Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice. Plant J 66: 456–466 [DOI] [PubMed] [Google Scholar]
- Lee I, Ambaru B, Thakkar P, Marcotte EM, Rhee SY. (2010) Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat Biotechnol 28: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen JE, Wang JX, Hu B, Chen G. (2008) Modifying the DPClus algorithm for identifying protein complexes based on new topological structures. BMC Bioinformatics 9: 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P, et al. (2007) Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J 50: 678–695 [DOI] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ. (2007) An Arabidopsis gene network based on the graphical Gaussian model. Genome Res 17: 1614–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Mao L, Van Hemert JL, Dash S, Dickerson JA. (2009) Arabidopsis gene co-expression network and its functional modules. BMC Bioinformatics 10: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentzen WI, Wurtele ES. (2008) Regulon organization of Arabidopsis. BMC Plant Biol 8: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miozzi L, Provero P, Accotto GP. (2010) ORTom: a multi-species approach based on conserved co-expression to identify putative functional relationships among genes in tomato. Plant Mol Biol 73: 519–532 [DOI] [PubMed] [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoort J, de Vos CH. (2006) A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol 141: 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenthal K, Weckwerth W, Steuer R. (2006) Metabolomic networks in plants: transitions from pattern recognition to biological interpretation. Biosystems 83: 108–117 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, et al. (2005) The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol 138: 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, Fernie AR, Usadel B, Nikoloski Z, Persson S. (2011) PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23: 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Nishida K, Kasahara K, Kinoshita K. (2011) ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol 52: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata Y, Suzuki H, Sakurai N, Shibata D. (2010) CoP: a database for characterizing co-expressed gene modules with biological information in plants. Bioinformatics 26: 1267–1268 [DOI] [PubMed] [Google Scholar]
- Ozaki S, Ogata Y, Suda K, Kurabayashi A, Suzuki T, Yamamoto N, Iijima Y, Tsugane T, Fujii T, Konishi C, et al. (2010) Coexpression analysis of tomato genes and experimental verification of coordinated expression of genes found in a functionally enriched coexpression module. DNA Res 17: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, et al. (2011) ArrayExpress update: an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res 39: D1002–D1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K. (2008) Decoding genes with coexpression networks and metabolomics: ‘majority report by precogs.’ Trends Plant Sci 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Sakurai N, Ara T, Ogata Y, Sano R, Ohno T, Sugiyama K, Hiruta A, Yamazaki K, Yano K, Aoki K, et al. (2011) KaPPA-View4: a metabolic pathway database for representation and analysis of correlation networks of gene co-expression and metabolite co-accumulation and omics data. Nucleic Acids Res 39: D677–D684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Sulpice R, Keurentjes J. (2010) Metabolic networks: how to identify key components in the regulation of metabolism and growth. Plant Physiol 152: 428–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K. (2008) fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24: 1461–1462 [DOI] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, Aida M, Tian CE, Ueno Y, Yamamoto KT, Machida Y, Nakamura K, Ishiguro S. (2010) Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51: 164–175 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tohge T, Fernie AR. (2010) Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat Protoc 5: 1210–1227 [DOI] [PubMed] [Google Scholar]
- Trivedi P, Edwards JW, Wang J, Gadbury GL, Srinivasasainagendra V, Zakharkin SO, Kim K, Mehta T, Brand JP, Patki A, et al. (2005) HDBStat!: a platform-independent software suite for statistical analysis of high dimensional biology data. BMC Bioinformatics 6: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Obayashi T, Mutwil M, Giorgi FM, Bassel GW, Tanimoto M, Chow A, Steinhauser D, Persson S, Provart NJ. (2009) Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ 32: 1633–1651 [DOI] [PubMed] [Google Scholar]
- Van der Hoeven R, Ronning C, Giovannoni J, Martin G, Tanksley S. (2002) Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen S. (2000) Graph clustering by flow simulation. PhD thesis. University of Utrecht, Utrecht, The Netherlands [Google Scholar]
- van Schie CC, Haring MA, Schuurink RC. (2007) Tomato linalool synthase is induced in trichomes by jasmonic acid. Plant Mol Biol 64: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang J, Li M, Deng Y, Pan Y. (2010) Recent advances in clustering methods for protein interaction networks. BMC Genomics (Suppl 3) 11: S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. (2006) CoXpress: differential co-expression in gene expression data. BMC Bioinformatics 7: 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W, Loureiro ME, Wenzel K, Fiehn O. (2004) Differential metabolic networks unravel the effects of silent plant phenotypes. Proc Natl Acad Sci USA 101: 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromori T, Ito T, Shinozaki K, et al. (2012) Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J 69: 154–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Niida R, Saito K. (2007) Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J Biol Chem 282: 14932–14941 [DOI] [PubMed] [Google Scholar]
- Zornoza P, Esteban RM. (1984) Flavonoids content of tomato plants for the study of the nutritional status. Plant Soil 82: 269–271 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.