Abstract
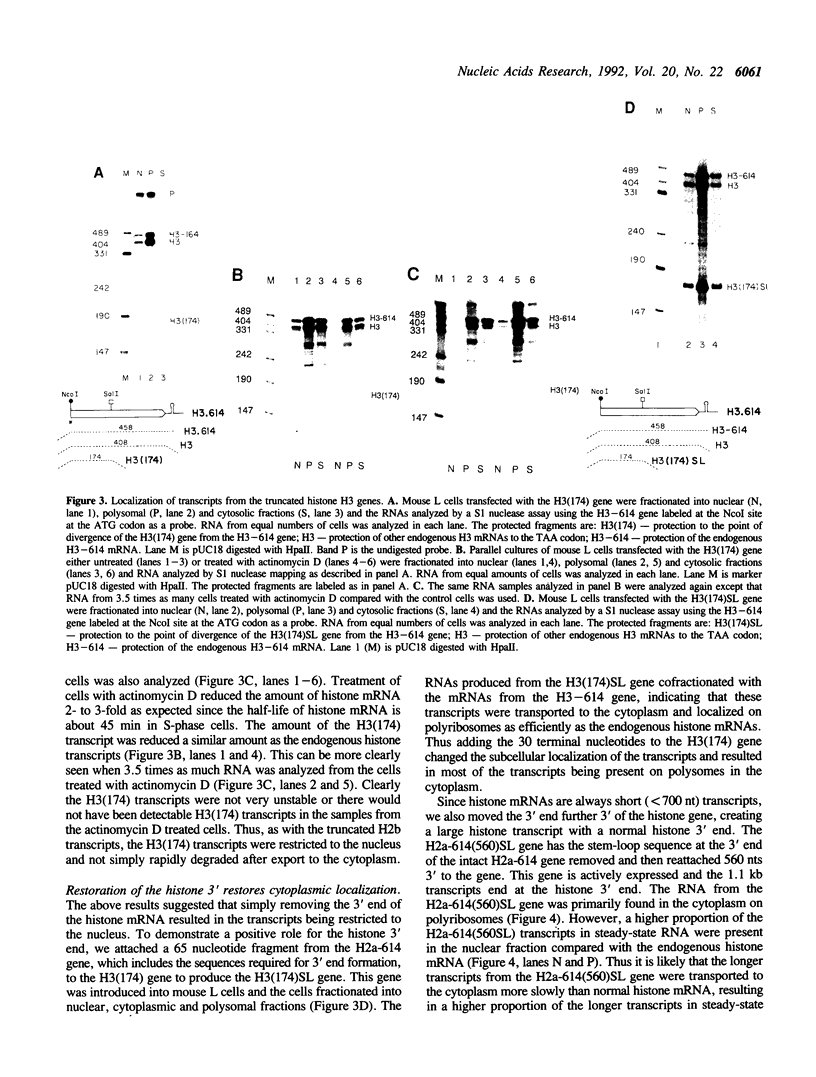
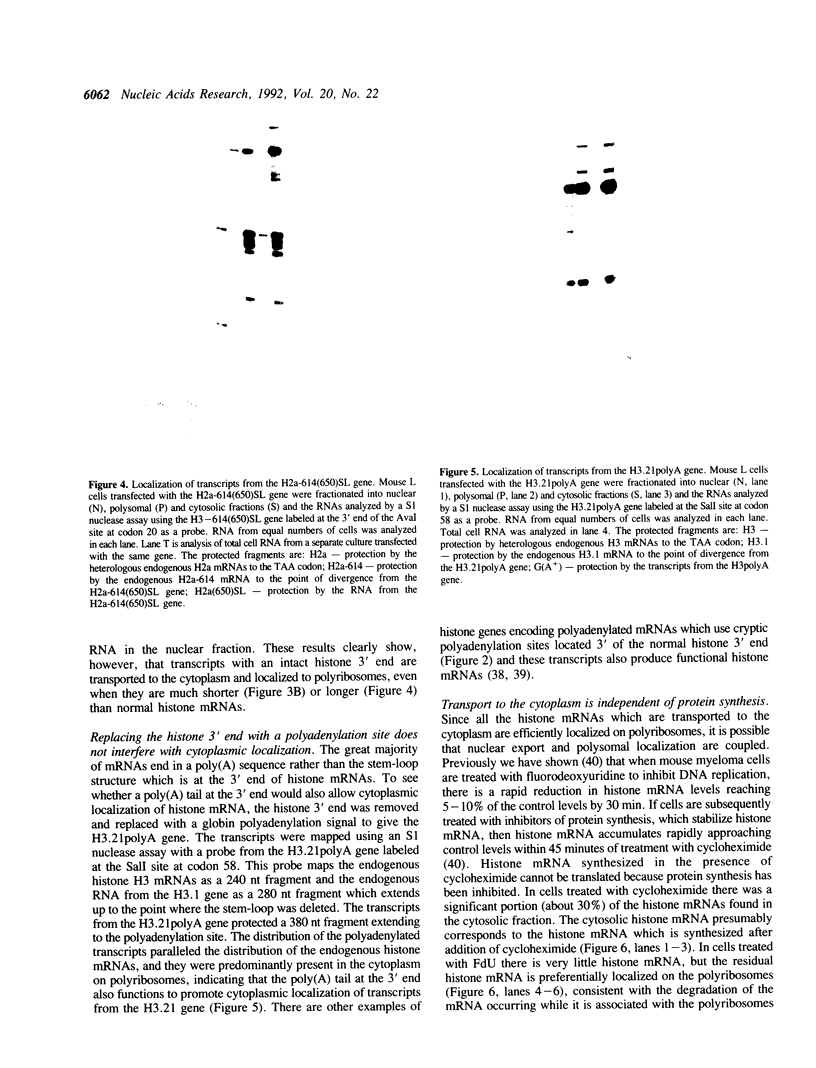
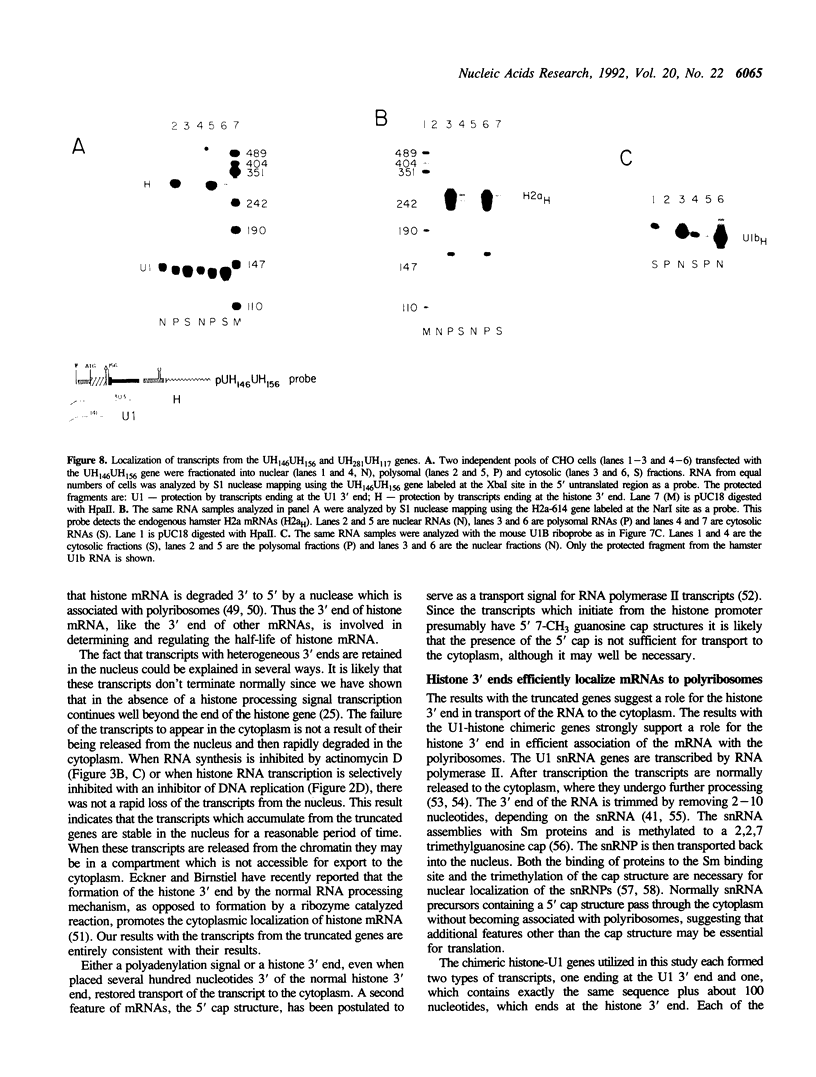
The final step in mRNA biosynthesis is transport of the mRNA from the nucleus to the cytoplasm. Histone genes from which the 3' stem-loop has been deleted are transcribed to give RNAs with heterogeneous 3' ends. These RNAs are localized in the nucleus and are stable. Addition of the histone 3' processing signal either on short (< 250 nts) or long (> 1000 nts) transcripts restores 3' processing and transport of the mRNA to the cytoplasm. In addition chimeric histone-U1 snRNA genes which produced RNAs with either histone or U1 3' ends were analyzed. Transcripts which ended with U1 snRNA 3' ends were not efficiently localized to polyribosomes. However, transcripts containing the same sequences including the snRNA 3' end followed by the histone 3' end were present in the cytoplasm on polyribosomes. Taken together these results suggest that the histone 3' end is required for export of histone mRNA to the cytoplasm and association of the mRNA with polyribosomes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alterman R. B., Sprecher C., Graves R., Marzluff W. F., Skoultchi A. I. Regulated expression of a chimeric histone gene introduced into mouse fibroblasts. Mol Cell Biol. 1985 Sep;5(9):2316–2324. doi: 10.1128/mcb.5.9.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P., Peltz S. W., Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989 Feb;9(2):659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P., Ross J. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem Sci. 1989 Sep;14(9):373–377. doi: 10.1016/0968-0004(89)90011-x. [DOI] [PubMed] [Google Scholar]
- Casey J. L., Koeller D. M., Ramin V. C., Klausner R. D., Harford J. B. Iron regulation of transferrin receptor mRNA levels requires iron-responsive elements and a rapid turnover determinant in the 3' untranslated region of the mRNA. EMBO J. 1989 Dec 1;8(12):3693–3699. doi: 10.1002/j.1460-2075.1989.tb08544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney W. G., Howard D. R., Pollard J. W., Sallustio S., Stanley P. High-frequency transfection of CHO cells using polybrene. Somat Cell Mol Genet. 1986 May;12(3):237–244. doi: 10.1007/BF01570782. [DOI] [PubMed] [Google Scholar]
- Cheng G. H., Nandi A., Clerk S., Skoultchi A. I. Different 3'-end processing produces two independently regulated mRNAs from a single H1 histone gene. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7002–7006. doi: 10.1073/pnas.86.18.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodchoy N., Levine B. J., Sprecher C., Skoultchi A. I., Marzluff W. F. Expression of mouse histone genes: transcription into 3' intergenic DNA and cryptic processing sites downstream from the 3' end of the H3 gene. Mol Cell Biol. 1987 Mar;7(3):1039–1047. doi: 10.1128/mcb.7.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodchoy N., Pandey N. B., Marzluff W. F. An intact histone 3'-processing site is required for transcription termination in a mouse histone H2a gene. Mol Cell Biol. 1991 Jan;11(1):497–509. doi: 10.1128/mcb.11.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Philipson L., Mattaj I. W. Ribonucleoprotein particles in cellular processes. J Cell Biol. 1988 May;106(5):1419–1425. doi: 10.1083/jcb.106.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R., Ellmeier W., Birnstiel M. L. Mature mRNA 3' end formation stimulates RNA export from the nucleus. EMBO J. 1991 Nov;10(11):3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Lührmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990 Aug 17;249(4970):786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- Fox C. A., Wickens M. Poly(A) removal during oocyte maturation: a default reaction selectively prevented by specific sequences in the 3' UTR of certain maternal mRNAs. Genes Dev. 1990 Dec;4(12B):2287–2298. doi: 10.1101/gad.4.12b.2287. [DOI] [PubMed] [Google Scholar]
- Gallie D. R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991 Nov;5(11):2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Gick O., Krämer A., Keller W., Birnstiel M. L. Generation of histone mRNA 3' ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986 Jun;5(6):1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Marzluff W. F. Rapid reversible changes in the rate of histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol Cell Biol. 1984 Feb;4(2):351–357. doi: 10.1128/mcb.4.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Pandey N. B., Chodchoy N., Marzluff W. F. Translation is required for regulation of histone mRNA degradation. Cell. 1987 Feb 27;48(4):615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Graves R. A., Wellman S. E., Chiu I. M., Marzluff W. F. Differential expression of two clusters of mouse histone genes. J Mol Biol. 1985 May 25;183(2):179–194. doi: 10.1016/0022-2836(85)90211-6. [DOI] [PubMed] [Google Scholar]
- Hamm J., Darzynkiewicz E., Tahara S. M., Mattaj I. W. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990 Aug 10;62(3):569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- Hamm J., Mattaj I. W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990 Oct 5;63(1):109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- Harris M. E., Böhni R., Schneiderman M. H., Ramamurthy L., Schümperli D., Marzluff W. F. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991 May;11(5):2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Weiner A. M. Formation of the 3' end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986 Oct 24;47(2):249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Hurt M. M., Chodchoy N., Marzluff W. F. The mouse histone H2a.2 gene from chromosome 3. Nucleic Acids Res. 1989 Nov 11;17(21):8876–8876. doi: 10.1093/nar/17.21.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Standart N. Do the poly(A) tail and 3' untranslated region control mRNA translation? Cell. 1990 Jul 13;62(1):15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt A. M., Pederson T. Accurate and efficient 3' processing of U2 small nuclear RNA precursor in a fractionated cytoplasmic extract. Mol Cell Biol. 1987 Sep;7(9):3131–3137. doi: 10.1128/mcb.7.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird-Offringa I. A., de Wit C. L., Elfferich P., van der Eb A. J. Poly(A) tail shortening is the translation-dependent step in c-myc mRNA degradation. Mol Cell Biol. 1990 Dec;10(12):6132–6140. doi: 10.1128/mcb.10.12.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. J., Chodchoy N., Marzluff W. F., Skoultchi A. I. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem-loop sequence at the 3' end of the mRNA. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6189–6193. doi: 10.1073/pnas.84.17.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. J., Liu T. J., Marzluff W. F., Skoultchi A. I. Differential expression of individual members of the histone multigene family due to sequences in the 5' and 3' regions of the genes. Mol Cell Biol. 1988 May;8(5):1887–1895. doi: 10.1128/mcb.8.5.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. J., Levine B. J., Skoultchi A. I., Marzluff W. F. The efficiency of 3'-end formation contributes to the relative levels of different histone mRNAs. Mol Cell Biol. 1989 Aug;9(8):3499–3508. doi: 10.1128/mcb.9.8.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo S. M., Marzluff W. F., Seufert A. C., Dean W. L., Schultz G. A., Simerly C., Schatten G. Localization and expression of U1 RNA in early mouse embryo development. Dev Biol. 1988 Jun;127(2):349–361. doi: 10.1016/0012-1606(88)90321-1. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Schümperli D. RNA 3' processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987 Jun;6(6):1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madore S. J., Wieben E. D., Pederson T. Intracellular site of U1 small nuclear RNA processing and ribonucleoprotein assembly. J Cell Biol. 1984 Jan;98(1):188–192. doi: 10.1083/jcb.98.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannironi C., Bonner W. M., Hatch C. L. H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3' processing signals. Nucleic Acids Res. 1989 Nov 25;17(22):9113–9126. doi: 10.1093/nar/17.22.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., Brown D. T., Lobo S., Wang S. S. Isolation and characterization of two linked mouse U1b small nuclear RNA genes. Nucleic Acids Res. 1983 Sep 24;11(18):6255–6270. doi: 10.1093/nar/11.18.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., Pandey N. B. Multiple regulatory steps control histone mRNA concentrations. Trends Biochem Sci. 1988 Feb;13(2):49–52. doi: 10.1016/0968-0004(88)90027-8. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986 Sep 12;46(6):905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- McGrew L. L., Richter J. D. Translational control by cytoplasmic polyadenylation during Xenopus oocyte maturation: characterization of cis and trans elements and regulation by cyclin/MPF. EMBO J. 1990 Nov;9(11):3743–3751. doi: 10.1002/j.1460-2075.1990.tb07587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris T. D., Weber L. A., Hickey E., Stein G. S., Stein J. L. Changes in the stability of a human H3 histone mRNA during the HeLa cell cycle. Mol Cell Biol. 1991 Jan;11(1):544–553. doi: 10.1128/mcb.11.1.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa N. M., el-Din A. S., Lobo S. M., Marzluff W. F. A mouse Ulb-2 gene with extensive sequence similarity to a rat Ula gene for 670 nucleotides 5' to the gene. Nucleic Acids Res. 1987 Apr 24;15(8):3622–3622. doi: 10.1093/nar/15.8.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K. L., Oh R., Steitz J. A. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3'-end processing reaction. Mol Cell Biol. 1989 Jul;9(7):3105–3108. doi: 10.1128/mcb.9.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe D., Jacobson A. mRNA poly(A) tail, a 3' enhancer of translational initiation. Mol Cell Biol. 1990 Jul;10(7):3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllner E. W., Kühn L. C. A stem-loop in the 3' untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988 Jun 3;53(5):815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- Neuman de Vegvar H. E., Dahlberg J. E. Nucleocytoplasmic transport and processing of small nuclear RNA precursors. Mol Cell Biol. 1990 Jul;10(7):3365–3375. doi: 10.1128/mcb.10.7.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N. B., Marzluff W. F. The stem-loop structure at the 3' end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987 Dec;7(12):4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N. B., Sun J. H., Marzluff W. F. Different complexes are formed on the 3' end of histone mRNA with nuclear and polyribosomal proteins. Nucleic Acids Res. 1991 Oct 25;19(20):5653–5659. doi: 10.1093/nar/19.20.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris J., Richter J. D. Maturation-specific polyadenylation and translational control: diversity of cytoplasmic polyadenylation elements, influence of poly(A) tail size, and formation of stable polyadenylation complexes. Mol Cell Biol. 1990 Nov;10(11):5634–5645. doi: 10.1128/mcb.10.11.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch D. R., Marzluff W. F. Expression of histone-U1 snRNA chimeric genes: U1 promoters are compatible with histone 3' end formation. Gene Expr. 1991 Apr;1(1):41–53. [PMC free article] [PubMed] [Google Scholar]
- Piper P. W., Aamand J. L. Yeast mutation thought to arrest mRNA transport markedly increases the length of the 3' poly(A) on polyadenylated RNA. J Mol Biol. 1989 Aug 20;208(4):697–700. doi: 10.1016/0022-2836(89)90159-9. [DOI] [PubMed] [Google Scholar]
- Ross J., Kobs G. H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3' terminus and proceeds 3' to 5'. J Mol Biol. 1986 Apr 20;188(4):579–593. doi: 10.1016/s0022-2836(86)80008-0. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Structure of a cluster of mouse histone genes. Nucleic Acids Res. 1983 Oct 11;11(19):6679–6697. doi: 10.1093/nar/11.19.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati D., Schümperli D. Structural and functional characterization of mouse U7 small nuclear RNA active in 3' processing of histone pre-mRNA. Mol Cell Biol. 1988 Apr;8(4):1518–1524. doi: 10.1128/mcb.8.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber C., Schümperli D. 3' processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 1988 Oct 25;16(20):9399–9414. doi: 10.1093/nar/16.20.9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D., Wellman S. E., Marzluff W. F. Sequences of four mouse histone H3 genes: implications for evolution of mouse histone genes. J Mol Evol. 1986;23(3):242–249. doi: 10.1007/BF02115580. [DOI] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]