Abstract
Biomimetic nanopores based on membrane-spanning single-walled carbon nanotubes have been designed to include selectivity filters based on combinations of anionic and cationic groups mimicking those present in bacterial porins and in voltage-gated sodium and calcium channels. The ion permeation and selectivity properties of these nanopores when embedded in a phospholipid bilayer have been explored by molecular dynamics simulations and free energy profile calculations. The interactions of the nanopores with sodium, potassium, calcium, and chloride ions have been explored as a function of the number of anionic and cationic groups within the selectivity filter. Unbiased molecular dynamics simulations show that the overall selectivity is largely determined by the net charge of the filter. Analysis of distribution functions reveals considerable structuring of the distribution of ions and water within the nanopores. The distributions of ions along the pore axis reveal local selectivity for cations around filter, even in those nanopores (C0) where the net filter charge is zero. Single ion free energy profiles also reveal clear evidence for cation selectivity, even in the C0 nanopores. Detailed analysis of the interactions of the C0 nanopore with Ca2+ ions reveals that local interactions with the anionic (carboxylate) groups of the selectivity filter lead to (partial) replacement of solvating water as the ion passes through the pore. These studies suggest that a computational biomimetic approach can be used to evaluate our understanding of the design principles of nanopores and channels.
Nanopores in membranes are of both biological and technological importance, the latter including stochastic biosensors, selective water pores for desalination, and biomedical diagnostics (1–3). Nanopores may be designed de novo from nonbiological materials or by reengineering biological nanopores. One may design artificial biomimetic nanopores which reproduce functions of biological systems (4, 5). For example, synthetic nanopores based on the permeation properties of biological channels (6) or which mimic the transport properties of the nuclear pore complex (7) have been designed.
Knowing the structure of ion channels provides possible design principles for biomimetic nanopores. Charged amino acid side chains are important in the selectivity properties of a number of biological channels and pores. For example, bacterial porins have cationic and basic side chains on opposite sides of the pore, with the exact balance pattern of charges governing the cation vs. anion selectivity (8). ELIC, a bacterial pentemeric ligand-gated ion channel, is cation selective and has a ring of five anionic glutamate (E) side chains (9). Rings of anionic side chains have also been suggested to play a key role in the ion selectivity of voltage-gated calcium (Cav) and sodium (Nav) channels (10–12). Thus, Cav (and some Nav) channels have four anionic residues in a ring (an EEEE motif, where E is the amino acid glutamate). The recent determination of the crystal structure of a bacterial Nav channel (NavAb) reveals the structure of a ring of four anionic (glutamate; i.e., an EEEE motif) side chains in the selectivity filter of the channel (13). It is therefore timely to design biomimetic nanopores based on Nav-like and related selectivity filters.
Carbon nanotubes (CNTs) have considerable potential as nanopores. Recent studies have demonstrated ion transport through CNTs linking two aqueous reservoirs (14, 15). In combination with advances in methods for chemical functionalization (16), these studies make CNTs attractive possible templates for biomimetic design of nanopores.
Molecular dynamics (MD) simulations have been used extensively to explore the behavior of water (17–20) and of ions in CNT nanopores (21). Such studies have included simulations of the effects of functionalization to add charges to the walls (22–25) and/or the mouths (26) of CNTs. Effects of functionalization inside CNTs on water behavior have also been studied (27). Ion transport through CNTs has been examined (21), demonstrating that wider nanotubes, even if hydrophobic, can allow passage of ions (28).
Membranes formed by parallel arrays of CNTs may permit water permeation while excluding ions, thus offering possible devices for water desalination (3, 18, 29, 30). These studies have focused on narrower (radius < 0.5 nm) CNTs (3), and have included consideration of the effects of functionalization of narrow (8,8) CNTsat the mouth (31, 32). Other studies, both experimental and computational, have examined how CNT derivatization may be used to promote selective ion permeation of CNTs. Negatively charged functional groups at CNT tips result in entry of cations (33). In one biomimetic design (34), a CNT was derivatized to resemble the selectivity filter of a potassium channel filter with the resultant nanopore and embedded in a graphene-like “membrane” (also see ref. 35).
In the current study, we design a series of CNT-based biomimetic nanopores with selectivity filters derived from those of Nav and Cav channels and of a length capable of spanning a phospholipid bilayer. We use MD simulations to demonstrate and explain the ion selectivity of the resultant nanopores. The results both provide insights into the fundamental “design principles” of biological ion channels and demonstrate how these principles may be mimicked in synthetic nanopores.
Results and Discussion
Model Nanopores.
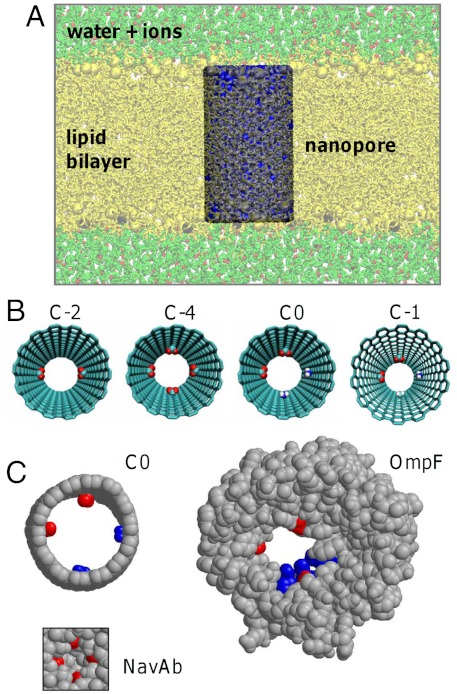
We set out to design CNT-based nanopores comparable in dimension to biological nanopores comparable to those formed by bacterial outer membrane proteins such as porins (8) and OmpG (36). Noting that ionic conductances have been demonstrated through single-walled CNTs of radius ca. 0.5—1 nm (14), we therefore modeled an armchair (14,14) CNT of length 3.61 nm, capable of spanning a phospholipid (dioleoyl phosphatidyl choline, DOPC) bilayer of thickness ca. 3.5 nm and with an internal radius (determined using HOLE; ref. 37) of 0.76 nm (Fig. 1). We aimed at mimicking the charge distributions observed in various bacterial (nano)pores by modifying the pristine CNT nanopore (Fig. 1C). In bacterial porins, the ion selectivity is controlled by the relative numbers of cationic and anionic side chains in the filter region of the pore (8). In calcium channels (CaV) (and indeed in some sodium channels; ref. 13), the selectivity filter is formed by four anionic glutamate side chains, whereas in other sodium channels (NaV) the number of anionic side chains is reduced to two and a cationic side chain is also present (10). Therefore, we designed CNT-based nanopores in which we attached either two or four carboxylate groups to the inner wall of the CNT, or two carboxylates at one side and two protonated amines at the other side of the pore. These were either attached midway along the nanopore (models C-2, C-4, and C0, respectively; Fig. 1B) or at one end of the nanopore (models E-2, E-4, and E0). To these models, we added a model C-1, in which there are two protonated amines at one side and one carboxylate (plus a methyl group in the fourth location) at the other side of the pore. The nanopore radius thus was reduced from 0.76 nm for the pristine CNT to ca. 0.55 nm in the region of the modifications. This constriction should be sufficient to accommodate a solvated cation or anion and is comparable to the radius of some biological nanopores; e.g., ca. 0.7 nm in OmpG (38) and ca. 0.3 nm for the selectivity filter of the recently determined bacterial NavAb channel structure (13).
Fig. 1.
Simulations of derivatized CNTs in a phospholipid bilayer.(A) The simulation system consisting of the CNT nanopore (gray/blue) in a phospholipid bilayer (yellow, DOPC lipid molecules) with water and ions on either side of the membrane. (B) The model nanopores viewed down the pore axis. Either two carboxylate (net charge -2), four carboxylate (net charge -4), two carboxylate plus two amine (net charge 0), or two carboxylate plus one amine (net charge -1) groups were attached to the inner wall of a (14,14) pristine CNT either at the center (C) or end (E; see Table S1) of the nanopore. (C) Comparison of the C0 model nanopore with two biological nanopores (the bacterial outer membrane porin OmpF, and the voltage-gated sodium channel NavAB). In each case, the key anionic and cationic groups of the selectivity filter (residues E62, D113, E117, R42, R82, R132 in OmpF; residues E177 in NavAb) are colored red and blue, respectively.
Two further model nanopores were investigated: C-EEEE and C-DEKA (Table S1). In these, the side chains of four glutamate (E) residues (as in CaV and NavAb channels) or of an aspartate (D), glutamate (E), a lysine (K), and an alanine (A) (as in vertebrate Nav channels) were used to form the centrally located selectivity filter. C-EEEE has a net charge of -4 and C-DEKA of -1; i.e., they are analogous to C-4 and C-1, respectively, but with more flexible functional groups forming the selectivity filter. The radius of these nanopores in the vicinity of the filter was ca. 0.35 nm.
Unbiased MD Simulations.
In order to assess the ion selectivity of the pore models, 30-ns equilibrium MD simulations were performed. The membrane was exposed on both faces to an electrolyte solution, namely 1 M NaCl, 1 M KCl, or 0.5 M CaCl2. We analyzed the number of ions present inside each of the nanopores over the course of the simulations. The pristine CNT nanopore admits, on average, ca. 4 Na+ and 4 Cl- ions when bathed in 1 M NaCl solution (Fig. 2 and Table S1), along with ca. 200 water molecules. Thus, the concentration of ions in the nanopore is equivalent to that in the bulk solution, and the pristine nanopore is not selective for either of the ions present. The total number of ions in the pore fluctuates over time between 0 and 8, but the number of Na+ and Cl- ions inside the pore is always approximately equal; thus, the volume of the CNT is charge neutral, as has also been observed (29) for CNTs not embedded in a membrane. If two carboxylate groups are present (C-2), a small degree of selectivity for Na+ over Cl- ions appears, which becomes more marked for C-4 (with four carboxylates) with a mean excess of 3.3 Na+ over Cl- (Table S1). On average, the excess of cations is such that the overall contents of the nanopore, taking into account the charge on the carboxylates, remains approximately neutral. Again, there is a clear correlation between the number of anions and cations in the pore over time; i.e., neutrality is preserved. The same degree of selectivity is seen for the C-EEEE nanopore (with the more flexible anionic side chains) as for C-4. The C-1 model gives a modest degree of selectivity (a mean excess of 0.5 Na+ over Cl-). This degree of selectivity is the same for C-DEKA, but is increased slightly (to an a mean excess of 0.9 cation over Cl-) in E-1 or in the presence of KCl.
Fig. 2.
Numbers of sodium and chloride ions inside the nanopores during 30-ns equilibrium MD simulations in the presence of 1 M NaCl. These are shown for the pristine and C-2, C-4, and C0 nanopore simulations. (Similar results are seen for the E-2, E-4, and E0 simulations.)
For the C0 systems there is little or no selectivity, so that the overall number of anions and cations in the pore is the same, but the mean number (ca. 5.5 of both Na+ and Cl-) is somewhat greater than the corresponding mean (ca. 4) for the pristine nanotubes. Analysis of the simulations of the E models revealed very similar results in terms of numbers of ions in the pore, suggesting that the selectivity created with by introduction of the carboxylate groups is not dependent on the position of the derivatization (Fig. S1 and Table S1).
We checked the kinetics of entry of ions into the nanopores. Water and ions enter within the first 0.1–0.3 ns. Thus, we are confident that a 30-ns simulation provides an equilibrium picture of ions within the nanopore. Experimentally observed ionic currents through CNTs are of the order of 2 nA (14), which corresponds to a mean passage time of ca. 0.1 ns. Analysis of the diffusion coefficients of ions along the nanopore axis (Table S2) yields ion diffusion coefficients lower than those observed in bulk, as was also observed for simulations of ions in smooth cylindrical channels (39).
Comparable simulations and analysis were performed with 1 M KCl or 0.5 M CaCl2 as the electrolyte. Similar trends in selectivity were seen (Fig. S1 and Table S1). In general, the net number of K+ ions inside all the channels studied is slightly greater than the number of Na+ ions, which perhaps may be explained in terms of a lower dehydration energy for K+ than Na+, although the ions remain largely hydrated when within the pore. For CaCl2, again the presence of carboxylate groups favors the entry of the cation into the nanopore, such that for the C-4 model the number of Ca2+ and Cl- ions within the pore are equal (at ca. 4 of each on average) even though the concentration of Ca2+ in the bulk is half that of Cl-. Thus, electroneutrality of the pore plus contents was maintained. Diffusion coefficients were significantly reduced (by more than an order of magnitude) for Ca2+ ions within the derivatized nanopores reflecting the stronger interaction (see below) of the divalent ion with the selectivity filters of the different nanopores.
Spatial Distributions.
From the equilibrium MD simulations, one can derive the average spatial distributions of the water and ions within the nanopores. There is a significant degree of structuring of water and ions within the nanopores, as seen previously (20). For the pristine nanotubes, the radial distribution profiles reveal the shell structure of the nanopore contents (Fig. 3), water molecules forming three concentric shells consistent with a nanopores radius of ca. 0.75 nm. We note that a similar shell structure has been observed in simulations of ions in smooth cylindrical channels (39). For the pristine CNT in NaCl, the radial distribution profile for Na+ shows two maxima in between (radially) the water maxima. This profile is also seen for the cations in the corresponding KCl 1 M and CaCl2 simulations (Fig. S2). The Cl- distribution is broader.
Fig. 3.
Radial distributions of Na+ ions (black), Cl- ions (red), and water (green) for pristine, C-4, and C-2 nanopores in a DOPC bilayer in a solution of 1 M NaCl. The gray and blue vertical broken lines represent the radius of the CNT nanopore and of the selectivity filter (defined by the minimum radius along the pore axis), respectively.
The introduction of two carboxylate groups in the C-2 nanopore leaves the water and Cl- radial distributions largely unchanged. However, the Na+ distribution changes to include a clear peak at r ∼ 0.5 nm, corresponding to Na+ ions in the selectivity filter region. This peak is even more marked in the C-4 nanopore, for which there is also a shift in the Cl- radial distribution toward the center of the nanopore, reflecting repulsion of the anions from the ring of carboxylate groups. Interestingly, even though the C0 nanopore is overall electroneutral, the C0 and C-1 systems shows radial distribution functions (Fig. S2), which are quite similar to those for C-2, suggesting that their selectivity filters may be more favorable to Na+ than to Cl-.
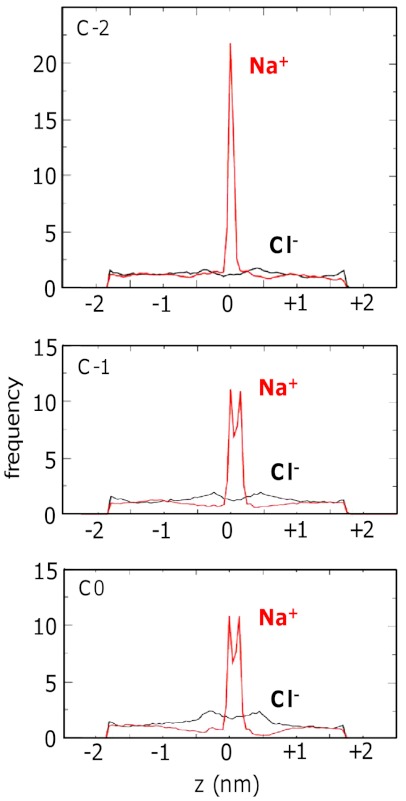
This view is reinforced by the frequency distributions of cations and anions along the pore (z) axis (Fig. 4). From these it can be seen that in the pristine CNT there is perhaps a weak preference for anions (Fig. S3). In marked contrast in all of the derivatized nanotubes (C0 through to C-4) there is a preference for cations over anions in the region of the selectivity filter. Even in C0 the selectivity in this region is about 4∶1 for cations over anions, in spite of the net neutral total charge of this nanopore (and similar behavior is seen for the E0 model, in which case the selectivity filter is close to the mouth of the nanopores; see Fig. S3). This result is of special interest given the selectivity of a DEKA motif (corresponding to C-1) in the selectivity filter of vertebrate Nav channels.
Fig. 4.
Distributions of Na+ ions (red) and Cl- ions (black) along the long axis (z) for the C-2, C-1, and C0 nanopores in a DOPC bilayer in a solution of 1 M NaCl. The selectivity filters are positioned at z ca. 0 nm.
Free Energy Profiles.
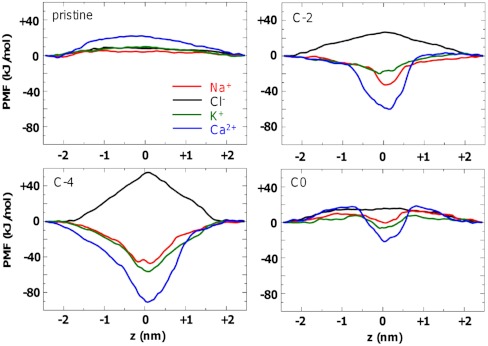
The thermodynamic basis of the observed patterns of ion selectivity may be explored by calculation of potentials of mean force (PMFs) (Fig. 5); i.e., of free energy profiles for a given ion as it is moved along the long (z) axis of the pore. It should be noted that this yields a single ion PMF (i.e., other ions were not present within the pore during the PMF simulations), which results in deeper wells than would be derived for multiion PMFs from the distributions in Fig. 4. For the pristine nanotube it can be seen that all ions experience a barrier for passage along the (hydrophobic) nanopores, and that this is greatest for the divalent Ca2+ ion. This barrier is largely electrostatic in origin, as shown qualitatively by Poisson–Boltzmann (PB) calculations (using the software package APBS, ref. 40; Fig. S4), and has been described in previous studies of model hydrophobic nanopores (28).
Fig. 5.
Potentials of mean force (PMFs) for single ions as a function of position along the z axis of the pore. The bilayer extends from z ca. -1.5 to +1.5 nm, and the CNT from z ca. -1.8 to +1.8 nm. The selectivity filters are positioned at z ca. 0 nm.
The PMF profile for the C-4 system is of special interest in the context of the EEEE motif in the selectivity filter of the bacterial NavAb sodium channel (Fig. 1C) (13) and of the EEEE or EEDD motifs in the filters of Cav channels. It can be seen that there is a clear preference for cations over anions, and also for Ca2+ over the monovalent cations.
The PMF profiles for C-2 also show a clear preference for cations over anions, and an especially deep energy well for Ca2+ ions. Significantly, it can be seen from these PMFs that the profiles for Na+ and Cl- are not simple mirror images of one another, but that there is a more pronounced energy well for cations in the vicinity of the selectivity filter. This pattern matches the results seen for distributions of ions along the long axis of the nanopores (Fig. 4), and suggests a more complex mechanism of selectivity than just electrostatic interactions between the ion and the charge on the wall of the nanopores.
The C0 (Fig. 5) and C-1 PMF profiles (Fig. S5) are very similar. These nanopores are especially relevant in the context of vertebrate Nav channels (which have a DEKA motif, net charge -1) and to porins such as OmpF (Fig. 1C). Again, local interactions in the vicinity of the selectivity filter make the filter region selective for cations over anions, even though the C0 pore is overall electroneutral; this is not seen in PB electrostatics calculations (Fig. S4).
The nature of the “specific” interactions of cations in the vicinity of the selectivity filter is revealed by examining the number of contacts to water and to the filter groups formed by the ions as a function of ion position along the pore axis (Fig. 6 and Fig. S6). For example, in Fig. 6A we show the contacts to water and to carboxylate groups of the filter for a Ca2+ ion moved along the long axis of the C0 nanopore. It can be seen that, in the vicinity of the selectivity filter, 2–3 waters from the cation solvation shell are replaced by interactions of the ion with the filter carboxylates (as is seen in the system snapshots in Fig. 6C). Similar effects are seen for all three cation species in all of the derivatized nanopores studied (Fig. S6). Even in the C0 nanopore there are no significant interactions between the Cl- ion and the  groups of the selectivity filter (Fig. S7).
groups of the selectivity filter (Fig. S7).
Fig. 6.
Calcium ions in the C0 pore system. (A) Numbers of contacts between calcium ions and water molecules (black) or carboxylate side chains (red) as a function of z derived from the PMF simulations of the C0 nanopore system. The charged groups are positions at z ca. 0 nm. (B and C) Two views of the C0 nanopore and Ca2+ ions. (B) A single Ca2+ ion at z ca. 0 nm; (C) snapshots of Ca2+ ion positions for multiple z values showing the clustering about the carboxylate (red) groups.
Conclusions.
By combining a CNT-template with insights from ion channel structure we have designed a set of cation-selective nanopores. The C-2 and C-4 nanopores (the latter mimicking the EEEE motif in the filter of Cav and NavAb channels) provide significant cation selectivity. Although the C-1 (mimicking the DEKA filter motif of vertebrate Nav channels) and C0 nanopores are not globally selective (i.e., in terms of the total number of ions within the nanopore), they show high local selective in the vicinity of the filter. Evaluation of PMFs reveals that Ca2+ is especially favored, in part as a result of the close association of the (partly dehydrated) cation with the carboxylates of the selectivity filter.
Our results suggest the utility of a biomimetic approach to “stress testing” our understanding of design principles of nanopores and channels. As further structural data emerge and more advanced nanopore synthesis becomes possible (41), this may become a more general approach.
Methods
Atomic coordinates for the (14,14) pristine CNT were generated using TubeGen (http://turin.nss.udel.edu/research/tubegenonline.html). This CNT is an example of a (n, n) or armchair nanotube (see ref. 42 for nomenclature), and is such that the carbon–carbon bonds are perpendicular to the tube axis. Parameterization was based on that used in previous studies (17) (see SI Text for further details of all methods). Water (SPC/E)/ion combination parameters were as in AMBER10 (43). The general AMBER force field (GAFF) was used for DOPC lipids (44). After CNT insertion in a preformed bilayer, the complete system was solvated. Water in the inside of the CNT was removed, so that at the start of the simulation the channel was completely dry. The resultant system was ionized using different salt concentrations (NaCl 1 M, KCl 1 M, and CaCl2 0.5 M). The initial size of the unit cell was equal to 18.2 × 17.4 × 7.0 nm3 and contained 560 lipids and approximately 18,000 water molecules.
Simulations were performed using GROMACS4 (www.gromacs.org) (45). All the systems were energy minimized, thermalized, and equilibrated, followed by unrestrained simulations for at least 30 ns (time step of 2 fs) for each system studied. The constant pressure and temperature canonical ensemble was employed with the pressure of 1 bar controlled using a semiisotropic Parrinello–Rahman barostat (46) and the temperature of 300 K imposed by a Berendsen (47) thermostat. The LINCS (48) and Particle Mesh Ewald methods were used (49) (see SI Text for details).
The potential of mean force (PMF) of a given ion moved along the nanopore (z) axis was calculated using umbrella sampling along the z axis from -2.475 nm to +2.475 nm using 100 equidistant windows each of width 0.05 nm and a harmonic force constant of ca. 1,000 kJ mol-1 nm-2. A simulation of length 1 ns was carried out for each window. Extension to 2 ns did not change significantly the PMF profiles. The biased distributions were recombined and unbiased with the Weighted Histogram Analysis Method (50) in Grossfield’s implementation (http://membrane.urmc.rochester.edu/content/wham). The first 0.6 ns of each window run were discarded as equilibration time, leaving a total of 0.4 ns per window. Data were analyzed using GROMACS and locally written code. Molecular graphic images were prepared using VMD (51).
Supplementary Material
Acknowledgments.
We thank all of our colleagues for their interest in this work, especially Jayne Wallace and Oliver Beckstein. This work was supported by the Spanish Ministry of Education (Programa de movilidad José Castillejo) and Xunta de Galicia (Programa postdoctoral ÁngelesAlvariño). The calculations were carried out on the MareNostrum supercomputer at the Barcelona Supercomputer Center and on the National Grid Service. Research in M.S.P.S.’s group is supported by the Biotechnology and Biological Sciences Research Council, the Engineering and Physical Sciences Research Council, and the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119326109/-/DCSupplemental.
References
- 1.Bayley H, Cremer PS. Stochastic sensors inspired by biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 2.Kasianowicz JJ, et al. Nanoscopic porous sensors. Annu Rev Anal Chem. 2008;1:737–766. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 3.Corry B. Designing carbon nanotube membranes for efficient water desalination. J Phys Chem B. 2008;112:1427–1434. doi: 10.1021/jp709845u. [DOI] [PubMed] [Google Scholar]
- 4.Hou X, et al. A biomimetic potassium responsive nanochannel: G-quadruplex DNA conformational switching in a synthetic nanopore. J Am Chem Soc. 2009;131:7800–7805. doi: 10.1021/ja901574c. [DOI] [PubMed] [Google Scholar]
- 5.Hou X, et al. A biomimetic asymmetric responsive single nanochannel. J Am Chem Soc. 2010;132:11736–11742. doi: 10.1021/ja1045082. [DOI] [PubMed] [Google Scholar]
- 6.Dehez F, Tarek M, Chipot C. Energetics of ion transport in a peptide nanotube. J Phys Chem B. 2007;111:10633–10635. doi: 10.1021/jp075308s. [DOI] [PubMed] [Google Scholar]
- 7.Jovanovic-Talisman T, et al. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2009;457:1023–1027. doi: 10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowan SW, et al. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 9.Hilf RJC, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 10.Heinemann SH, et al. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–443. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- 11.Nonner W, Gillespie D, Henderson D, Eisenberg B. Ion accumulation in a biological calcium channel: Effects of solvent and conining pressure. J Phys Chem B. 2001;105:6427–6436. [Google Scholar]
- 12.Boda D, et al. Steric selectivity in Na channels arising from protein polarization and mobile side chains. Biophys J. 2007;93:1960–1980. doi: 10.1529/biophysj.107.105478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, et al. Translocation of single-stranded DNA through single-walled carbon nanotubes. Science. 2010;327:64–67. doi: 10.1126/science.1181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi W, et al. Dynamics of simultaneous, single ion transport through two single-walled carbon nanotubes: Observation of a three-state system. J Am Chem Soc. 2011;133:203–205. doi: 10.1021/ja108011g. [DOI] [PubMed] [Google Scholar]
- 16.Hirsh A. Functionalization of single-walled carbon nanotubes. Angew Chem Int Ed Engl. 2002;41:1853–1859. doi: 10.1002/1521-3773(20020603)41:11<1853::aid-anie1853>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 17.Hummer G, Rasaiah JC, Noworyta JP. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 18.Kalra A, Garde S, Hummer G. Osmotic water transport through carbon nanotube membranes. Proc Natl Acad Sci USA. 2003;100:10175–10180. doi: 10.1073/pnas.1633354100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu F, Schulten K. Water and proton conduction through carbon nanotubes as models for biological channels. Biophys J. 2003;85:236–244. doi: 10.1016/S0006-3495(03)74469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexiadis A, Kassinos S. Molecular simulation of water in carbon nanotubes. Chem Rev. 2008;108:5014–5034. doi: 10.1021/cr078140f. [DOI] [PubMed] [Google Scholar]
- 21.Peter C, Hummer G. Ion transport through membrane-spanning nanopores studied by molecular dynamics simulations and continuum electrostatics calculations. Biophys J. 2005;89:2222–2234. doi: 10.1529/biophysj.105.065946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao R, Aluru NR. Atypical dependence of electroosmotic transport on surface charge in a single-wall carbon nanotube. Nano Lett. 2003;3:1013–1017. [Google Scholar]
- 23.Gong X, et al. A charge-driven molecular water pump. Nat Nanotechnol. 2007;2:709–712. doi: 10.1038/nnano.2007.320. [DOI] [PubMed] [Google Scholar]
- 24.Lu H, Zhou X, Wu F, Xu Y. Effect of charge on water filling/emptying transitions of nanochannel. J Phys Chem B. 2008;112:16777–16781. doi: 10.1021/jp802263v. [DOI] [PubMed] [Google Scholar]
- 25.Zuo G, Shen R, Ma S, Guo W. Transport properties of single-file water molecules inside a carbon nanotube biomimicking water channel. ACS Nano. 2010;4:205–210. doi: 10.1021/nn901334w. [DOI] [PubMed] [Google Scholar]
- 26.Huang L-L, et al. Molecular dynamics simulation study of the structural characteristics of water molecules confined in functionalized carbon nanotubes. J Phys Chem B. 2006;110:25761–25768. doi: 10.1021/jp064676d. [DOI] [PubMed] [Google Scholar]
- 27.Zhu YD, et al. Molecular dynamics study of pore inner wall modification effect in structure of water molecules confined in single-walled carbon nanotubes. J Phys Chem C Nanomater Interfaces. 2009;113:882–889. [Google Scholar]
- 28.Beckstein O, Tai K, Sansom MSP. Not ions alone: Barriers to ion permeation in nanopores and channels. J Am Chem Soc. 2004;126:14694–14695. doi: 10.1021/ja045271e. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Murad S, Jameson CJ. Ion permeation dynamics in carbon nanotubes. J Chem Phys. 2006;125:084713–084726. doi: 10.1063/1.2337289. [DOI] [PubMed] [Google Scholar]
- 30.Fornasiero F, et al. Ion exclusion by sub-2-nm carbon nanotube pores. Proc Natl Acad Sci USA. 2008;105:17250–17255. doi: 10.1073/pnas.0710437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corry B. Water and ion transport through functionalised carbon nanotubes: Implications for desalination technology. Energy Environ Sci. 2011;4:751–759. [Google Scholar]
- 32.Chen Q, et al. Water transport and purification in nanochannels controlled by asymmetric wettability. Small. 2011;7:2225–2231. doi: 10.1002/smll.201100287. [DOI] [PubMed] [Google Scholar]
- 33.Majumder M, Chopra N, Hinds BJ. Effect of tip functionalization on transport through vertically oriented carbon nanotube membranes. J Am Chem Soc. 2005;127:9062–9070. doi: 10.1021/ja043013b. [DOI] [PubMed] [Google Scholar]
- 34.Gong X, et al. A controllable molecular sieve for Na+ and K+ ions. J Am Chem Soc. 2010;132:1873–1877. doi: 10.1021/ja905753p. [DOI] [PubMed] [Google Scholar]
- 35.Zhu YD, et al. Molecular simulation study of the effect of inner wall modified groups on ionic hydration confined in carbon nanotube. Fluid Phase Equilib. 2010;297:215–220. [Google Scholar]
- 36.Yildiz O, Vinothkumar KR, Goswami P, Kühlbrandt W. Structure of the monomeric outer-membrane porinOmpG in the open and closed conformation. EMBO J. 2006;25:3702–3713. doi: 10.1038/sj.emboj.7601237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smart OS, et al. Hole: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graphics. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 38.Chen M, Khalid S, Sansom MSP, Bayley H. Outer membrane protein G: Engineering a quiet pore for biosensing. Proc Natl Acad Sci USA. 2008;105:6272–6277. doi: 10.1073/pnas.0711561105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynden-Bell R, Rasaiah JC. Mobility and solvation of ions in channels. J Chem Phys. 1996;105:9266–9280. [Google Scholar]
- 40.Baker NA, et al. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou X, Guo W, Jiang L. Biomimetic smart nanopores and nanochannels. Chem Soc Rev. 2011;40:2385–2401. doi: 10.1039/c0cs00053a. [DOI] [PubMed] [Google Scholar]
- 42.Dresselhaus MS, Dresselhaus G, Jorio A. Unusual properties and structure of carbon nanotubes. Annu Rev Mater Res. 2004;34:247–278. [Google Scholar]
- 43.Joung IS, Cheatham TE. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siu SWI, Vacha R, Jungwirth P, Böckmann RA. Biomolecular simulations of membranes: Physical properties from different force fields. J Chem Phys. 2008;128:125103. doi: 10.1063/1.2897760. [DOI] [PubMed] [Google Scholar]
- 45.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 46.Parrinello M, Rahman A. Polymorphic transitions in single-crystals—a new molecular-dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 47.Berendsen HJC, et al. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 48.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 49.Essmann U, et al. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 50.Kumar S, et al. The weighted histogram analysis method for free-energy calculations on biomolecules.1. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 51.Humphrey W, Dalke A, Schulten K. VMD—visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.