Abstract
Although the epithelial to mesenchymal transition (EMT) is famous for its role in cancer metastasis, it also is a normal developmental event in which epithelial cells are converted into migratory mesenchymal cells. A prime example of EMT during development occurs when neural crest (NC) cells emigrate from the neural tube thus providing an excellent model to study the principles of EMT in a nonmalignant environment. NC cells start life as neuroepithelial cells intermixed with precursors of the central nervous system. After EMT, they delaminate and begin migrating, often to distant sites in the embryo. While proliferating and maintaining multipotency and cell survival the transitioning neural crest cells lose apicobasal polarity and the basement membrane is broken down. This review discusses how these events are coordinated and regulated, by series of events involving signaling factors, gene regulatory interactions, as well as epigenetic and post-transcriptional modifications. Even though the series of events involved in NC EMT are well known, the sequence in which these steps take place remains a subject of debate, raising the intriguing possibility that, rather than being a single event, neural crest EMT may involve multiple parallel mechanisms.
Keywords: neural crest, EMT, apicobasal polarity, dorsal neural tube, Snail
1. Introduction
During development, most growing cell populations undergo several rounds of transition from their epithelial phenotype into a mesenchymal fate (Epithelial to Mesenchymal Transition, EMT) and/or vice versa (Mesenchymal to Epithelial Transition, MET). Epithelial cells are tightly bound to each other with tight junctions, adherens junctions, desmosomes and gap junctions, which together form coherent apicobasally polarized cell layers. These organized epithelial structures are surrounded with an underlying basement membrane, which physically separates them from other tissues. In contrast, unpolarized mesenchymal cells are not attached by intercellular adhesion complexes. This enables them to respond quickly to environmental cues and to move as individuals throughout the extracellular matrix. Thus, migratory cells display less polarity than epithelial cells, though often communicate with neighboring cells in order to coordinate their movements. EMT enables the construction of various complicated organs with lumens, lobes and separating barriers. However, primary EMT only takes place twice in the embryo, during gastrulation and neural crest formation [1, 2].
The neural crest (NC) provides an excellent model to study the principals of EMT in a nonmalignant environment. Neural crest precursors initially reside within the forming central nervous system but subsequently undergo EMT to migrate to distant locations in the embryo and then differentiate into diverse derivatives. Because neural crest EMT occurs in the early embryo, most knowledge is acquired by using model vertebrate organisms that develop externally such as the frog, chicken and fish.
Upon differentiation, neural crest cells form many derivatives that are prone to malignant transformation, including melanocytes that can form melanomas and glial cells that can form Schwannomas and gliomas. Cancer growth often relies upon reactivation of several developmental cellular programs such as maintenance of the self-renewal and proliferation capacity. In order for the cancer cells to leave the original tumor, EMT machinery is activated, epithelial adhesion is broken down and the metastatic cells escape through the ruptured basement membrane into the circulation. Conversely, MET is needed for the reverse actions at the target site. Since the recognition that EMT is one of the hallmarks of cancer, interest in cancer EMT research has expanded rapidly. Reactivated EMT is also the cause of organ fibrosis, which leads to the loss of functionality of the epithelial structures (e.g. renal tubules) and thus the whole organ [3–5].
2. The neural crest
The neural crest (NC) is a transient embryonic cell population that forms in the dorsal parts of the newly formed neural tube. The NC cells subsequently undergo EMT, and begin to migrate through the mesenchyme of the developing embryo to give rise to the peripheral nervous system, cranial bone and cartilage, melanocytes and parts of the adrenal medulla. The gene regulatory network (GRN) that orchestrates NC induction and specification is evolutionarily conserved in vertebrates [6–10].
Briefly, induction of NC starts during the gastrula stage [11, 12]. A combination of BMP, Wnt and FGF signaling is important for setting up the neural plate border territory. At these stages, BMP (BMP 4 and 7 in the chicken) secreted by the epidermal ectoderm promotes dorsalization and antagonizes Shh from the ventral midline [13, 14]. Wnt-signals, also secreted by the epidermal ectoderm (Wnt8 in fish, frog and lamprey, Wnt6 in chicken) are also critical for NC formation [6, 15–17], as is FGF signaling secreted from the paraxial mesoderm [18].
These extracellular signals regulate expression of a group of essential transcription factors called the neural plate border specifier genes (or dorsal patterning genes). In the frog for example, Msx1 induces expression of Pax3 and Zic1 [19], which together are sufficient to induce NC specifier genes [19–21]. In the chick Pax7, which is expressed in the neural plate border beginning at gastrula stages, is required for specification of NC fate and seems to have a more critical role than Pax3 [11]. In some species like mouse, Pax3 and Pax7 may exhibit functional redundancy but have later defects in neural crest derivatives [22, 23]. The neural plate border specifiers in turn activate the transcription of the NC specification genes (e.g. Snail1/2, SOXE, FoxD3, AP-2 and c-Myc) in the dorsal neural folds [9, 20, 24, 25].
These neural crest specifier genes are thought to switch on the EMT program in a subset of cells in the dorsal neural tube. These detach from the neural epithelium and become pluripotent migratory mesenchymal cells. There are likely to be many additional regulators in the NC GRN, that together orchestrate a complex series of cellular events that allow neural crest cells to emigrate from the neural tube. Various mechanisms, ranging from epigenetics to regulatory RNAs and posttranslational protein modifications, are likely to work coordinately to fine tune the activity of key NC players to contribute to the spatiotemporal control of EMT. Finally, the migrating NC cells assume one of many possible fates either during their migration or upon arriving at their final destination and differentiate into their target tissue [9, 26].
3. The Epithelial to Mesenchymal Transition
Traditionally, EMT is thought to consist of a breakdown of the epithelial adhesion components, loss of apicobasal polarity and rupture of the basement membrane, which then leads to mesenchymal motility. However, these functions overlap and the cell also needs to maintain multipotency, survival and overall adhesive integrity of the source tissue [27]. In this review we focus on what is known about the regulation of EMT in the NC. The “basic rules” apply to all EMT from gastrulation to carcinoma [3], although as discussed below, different individual molecular players fulfill the same tasks in different tissues, species and even at different axial levels, such as the cranial versus trunk region of the neural crest even within the same organism.
3.1 Polarity, adhesive junctions and basement membrane during NC EMT
3.1.1 Adherens junctions and cadherin expression during EMT
Adherens junctions (AJ) are cell-cell adhesion complexes that are comprised of classical calcium dependent cadherins and catenins. These include type I cadherins (E-, N, P- and R-cadherin) and type II cadherins (cadherin 5 and beyond), the extracellular domains of which form a homophilic bond with the cadherins on adjacent cells. The intracellular domain forms a complex with the catenin proteins in order to strengthen the adhesion by connecting with the actin cytoskeleton (the cadherin binds p120- and β-catenin and β-catenin binds α-catenin) or microtubule cytoskeleton (β-catenin, p120catenin) or both [28]. A repertoire of actin-associated proteins including vinculin, α-actinin, formins, ZO1 (also associated with tight junctions), and EPLIN are involved in the binding of α-catenin to the actin filaments [28, 29].
The homophilic interactions of cadherins serve as adhesive sorting factors that mediate morphogenesis of different cell types. During the formation of the neural tube, which is a pseudostratified epithelium, the expression of E-cadherin is gradually downregulated and switched to N-cadherin (N-cad) in the neural progenitors of the neural plate. The non-neural ectoderm continues to express E-cadherin, which thus keeps these two structures separated. N-cad expression is most prominent in the apicolateral adherens junctions of the neural tube where it presumably maintains the apicobasal polarity as well as the adhesive integrity of the neural tube. [30–32]. The neuroepithelial cells also express another classical cadherin, Cad6B, that is first transcribed in the whole neural plate (although most intensively dorsally) and later, prior to neural tube closure, restricted only to the cells in the dorsal neural folds/neural tube [14, 33–35]. Both N-cad and Cad6B transcripts are downregulated from the dorsal neural tube prior to emigration of neural crest cells whereas Cad7/11 expression comes on in the migratory crest [33, 34, 36, 37]. In addition to the localization of Cad6B at the apical membrane cell junction complexes, it (as well as β-catenin) is also expressed in a more nonpolarized fashion ubiquitously throughout the cytoplasm in the dorsal neural tube [35].
N-cadherin during EMT
The hallmark of the EMT program during gastrulation as well as in carcinomas is the direct transcriptional repression of E-cadherin by Snail, which leads to a loss of adherens junctions and cell polarity [4, 32]. In the avian NC, N-cad is lost from the apical membrane junctions prior to emigration [36]. Interestingly, as the full length N-cad antagonizes EMT, it is proteolytically cleaved from the cell membrane via a BMP triggered, ADAM10 metalloprotease dependent mechanism in the delaminating NC cells. The truncated soluble cytoplasmic form CTF2 localizes in the nucleus and switches from an EMT inhibitor into an EMT promoting transcription factor [38]. N-cad is thought to be downregulated by the transcription factor Foxd3 [39] and overexpression of N-cad inhibits NC delamination, migraton and BMP signaling [35, 36, 38]. Even though a breakdown of the apicobasal polarity of the neuroepithelium is required for emigration, the migrating NC cells also display polarity in order to orientate in the correct direction. N-cadherin appears to mediate this reduced contact in migrating NC cells, and is involved in the regulation of contact inhibition of locomotion (CIL) [1, 40, 41].
Cadherin 6B during EMT
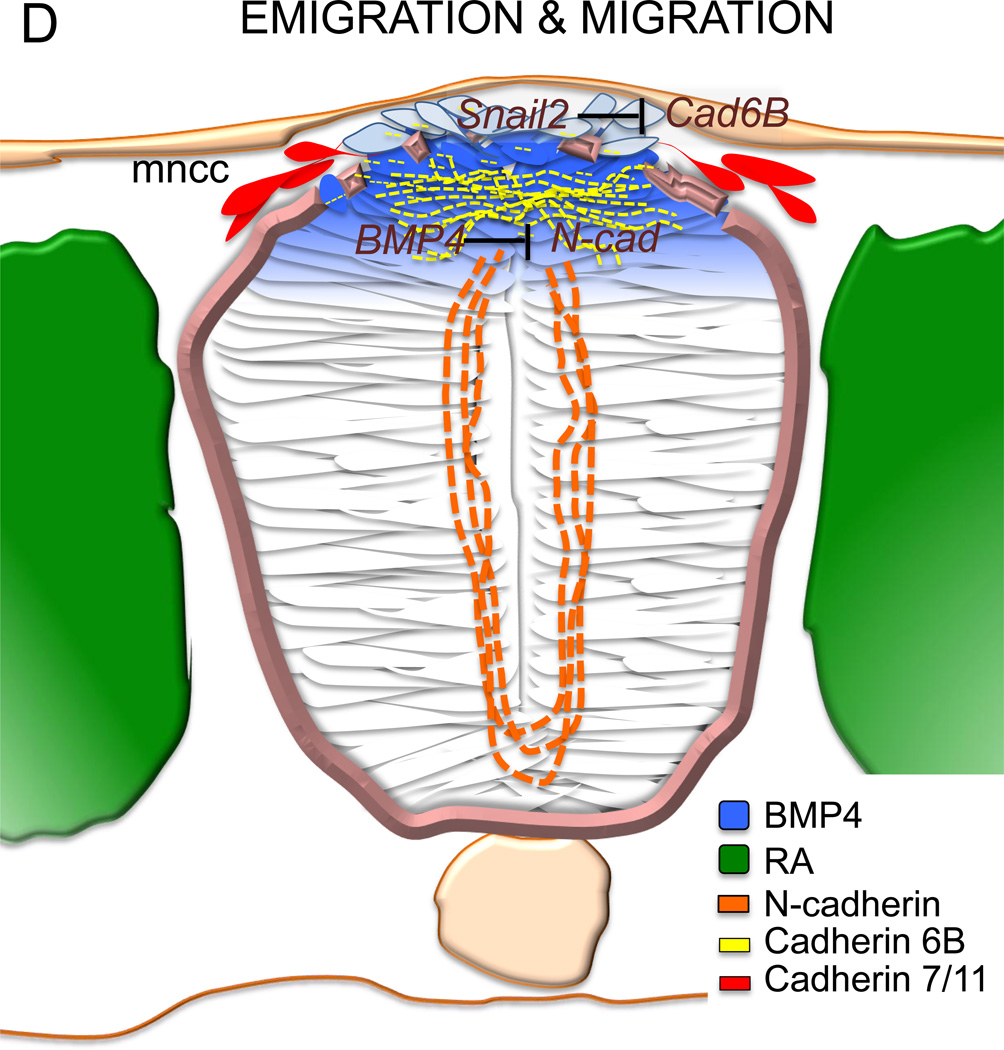
The loss of Cad6B is directly regulated by transcriptional repression of Snail2 prior to migration of NC in the chick [42]. This was thought to regulate the correct timing of NC emigration and delamination from the neural tube without affecting the expression of the migratory proteins of NC [43]. However, a recent study suggests that the avian Cad6b acts via BMP signaling to trigger de-epithelization of the premigratory NC cells without affecting the actual delamination process or breakdown of the basement membrane [35]. Over-expression of Cad6B caused an ectopic loss of polarity, as demonstrated by disruption of ZO-1 expression that lead to a general lack of epithelial integrity. However, the mesenchymal cells were not able to break through the basement membrane but instead accumulated in the lumen of the NT [35]. Accordingly, shRNA mediated knockdown of Cad6B resulted in a lack of depolarization followed by a lack of migration, since the cells had not adopted a mesenchymal phenotype [35]. The seemingly contradictory results from these studies may be due to the different axial differences and timing of performing over-expression and knockdown. For studies of cranial NC [42, 43], translation blocking morpholinos (MO) as well as overexpression constructs were electroporated into the newly formed neural tube at the 2–4 somite stage for Cad6b [43] and the 5–7 somite stage for Snail2 [42]. At this time point the de-epithelization process discovered by Park and Gumbiner (2010) is already strongly ongoing in the cranial premigratory crest (as depolarized Cad6B expression starts in the dorsal neural folds before neural tube closure). Instead, manipulating Cad6b and Snail2 at these later time points likely affects the onset of migration in the cranial NC cells, which indeed operates through Cad6B downregulation. This nicely demonstrates that EMT consists of separately regulated and distinct cellular functions that include NC specification, depolarization, delamination, emigration and migration. Taken the results together, Cad6B appears to be crucial for the loss of polarity in the transition into a mesenchymal cell type and needs to be degraded in order for the proper migration of NC to take place [35, 42]. Further support for this scenario comes from the over-expression phenotype of many different cadherins in the migratory crest [36], all of which block emigration.
A shift from type classical cadherins to more mesenchymal cadherins correlates with the acquisition of cell motility. Premigratory depolarizing NC cells switch N-cad first to Cad6B, which in the migratory cells is again replaced with Cad7 expression in the chick and Cad11 in the frog [26, 33, 44–46].
3.1.2 Rho family of small GTPases control cytoskeleton changes
The members of the Rho family are small G-protein signaling molecules and a subfamily of the Ras superfamily. Rhos participate in various cellular functions including cellular morphology and motility changes by controlling the cytoskeletal rearrangements [47]. In the neurula stage embryo, expression of RhoC is restricted to the notochord. RhoA is ubiquitously expressed in the neural plate and mesoderm but after NT closure it seems to be concentrated in the dorsal lateral part of the NT [14]. RhoB, on the other hand was found as one of the NC genes induced by BMP in the neural plate and its expression is restricted to the pre- and early migratory NC. RhoB null mice do not display any evident developmental defects, which may reflect redundancy between the family members [48]. The avian RhoB expression is turned on after Snail2 and does not appear to affect neural crest cell fate specification (i.e. no alteration in expression of NC specific genes Snail2, Sox9, Cad6B or Foxd3) or migration. Rather, it has been proposed to play an important role in delamination from the neural tube [14, 49], since neural crest cells failed to emigrate from neural plate explants treated with Rho inhibitors [14]. Although originally shown to be essential for NC delamination and thus the loss of epithelial polarity, a more recent paper indicates that both RhoB and RhoA, through ROCK signaling, act to maintain apicobasal polarity and stabilize N-cadherin in a vinculin based manner at the focal adhesion points. The loss of the membrane bound active form of Rho, on the other hand, was associated with delamination and EMT and cytoplasmic nonpolarised RhoB was detected in mesenchymal cells. Both loss and gain of function studies of RhoB and RhoA supported this finding [49]. Moreover, breakdown of RhoA followed by delamination appears to be essential for EMT during gastrulation [50]. The differences in the studies may be due to the more efficient and less toxic reagents used in the more recent study [49]. RhoA in the migratory post-otic NC influences migration rate and filopodia dynamics [51]. Since the inhibitor used by Liu and Jessell [14] inhibits activity of both RhoB and RhoA, it is possible that instead of affecting delamination, the inhibitor was added at a slightly later timepoint and the noted effects were on migration of already delaminated cells. This coupled with possible differences in the axial levels under study may have contributed to the different observations. Also, as RhoB is expressed in the early migratory NC, similar to RhoA, it may have two distinct roles in epithelial versus mesenchymal cells. RhoA negatively regulates the expression of NC genes (including the cranial crest) Snail1/2 and FoxD3 in Xenopus. Interestingly the results with another member of the Rho family, Rac that has been associated with non-canonical Wnt signaling in dendrite morphogenesis [52], were the opposite as its overexpression induced the NC markers [53]. Yet another Rho family protein RAP also has a role in the early NC migration in zebrafish, which is controlled by a RAP mediator RADIL [54].
In addition to cell polarity, RhoB mediated signaling is important for the competence of NC to undergo EMT in the chick. Whereas ectopic expression of Sox9 does not cause EMT, it together with RhoB is sufficient to promote EMT. Ectopic RhoB alone, on the other hand, does not induce HNK-1 positive NC cells but results in disturbed morphology and apoptosis. Interestingly, overexpression of Sox9 and Snail2 induces EMT, suggesting that there is no epistatic relationship between RhoB and the two other NC signaling inducers, respectively [39]. On the other hand, in the avian hindbrain, an increase in expression was reported after Snail2 overexpression [55], which might be due to the differences between cranial and trunk NC or to interesting cross-regulation.
3.1.3 Tight junctions and the protein complexes in the polarized apical plasma membrane
Tight junctions, the most apically located cellular junctions also contribute to the apicobasal polarity of the neural epithelium. Adjacent plasma membranes are sealed together by the transmembrane proteins claudins and occludins. Tight junctions are constantly being remodeled via interactions with the interface protein Zona Occludens ZO-1 and are in contact with Rho GTPases by forming complexes with other junctional adaptor proteins like the portioning defective (PAR), Crumbs and Scribble complexes [56–58]. Tight junctions are prominent in the neural plate but disappear soon after closure of the NT. Loss of tight junctions is associated with an increase in N-cadherin expression as well as maintenance of ZO-I in the apical membranes [57]. Adherens junctions also maintain polarity by communication with the conserved intracellular apical protein complexes which, in Drosophila neuroectoderm, are controlled by Cdc42 and PAR-proteins [59]. The Crumbs complex protein Pals is also expressed in the apical junctions in the avian NT [35]. After loss of tight junctions, maintenance of apical neuroepithelium relies also on tiny cholesterol based prominin-1 containing microdomains (called lipid rafts) of apical membrane protrusions, which also play a major role in the asymmetric division of the neuroepithelial cells [60–62]. Finally, gap junctions replace tight junctions during neural tube closure in the frog [63]. Mouse premigratory NC cells start to express the essential gap junction protein connexin 43, which is particularly important in the cardiac NC [64].
3.1.4 The basement membrane and extracellular matrix
The basement membrane, composed of type IV collagen fibers and different glycoproteins (e.g. laminins, dystroglycans, proteoglycans) separates the neural epithelium form the mesenchyme. The extracellular matrix (ECM) proteins, such as the ADAM proteins and matrix metalloproteinases (MMP), via signaling through integrin and cadherin receptors control the basement membrane and induce its breakdown during EMT to allow migration. MMP-2 and ADAM13 positively correlate with NC delamination and migration [26, 65, 66] and several laminins, proteoglycans and integrin dimers, especially in combination with the β1 integrin subunit, support the migratory NC phenotype [67–72]. Noelin, a secreted glycoprotein that is expressed in the neural folds at the onset of EMT and is maintained all through NC migration has also been reported to positively correlate with the onset of avian NC emigration [73].
3.2 Inductive signals of NC EMT
3.2.1 Notch, BMP, FGF and Wnt signaling
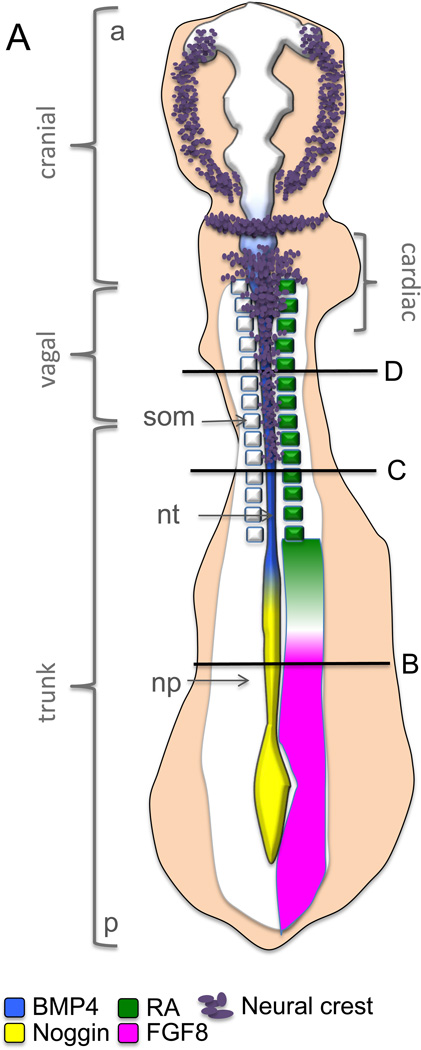
The NC cells are specified at the neural plate border and will obtain a unique cell fate as compared to the prospective cells of the non-neural ectoderm as well as the neural cells, respectively. Notch signaling promotes NC fate in the neural plate in chicken, fish and frog. Induced by the Delta1 ligand expressed by the neighboring endodermal ectoderm cells Notch signaling is needed to induce BMP4 expression in the prospective NC and also to inhibit Snail2 from being expressed too early in the chicken [74–76]. BMP (BMP4 in chicken, BMP2 in mouse), a member of the TGFβ family, secreted in the dorsal NT (as opposed to the BMPs secreted by the epidermal ectoderm needed for NC specification at the neural plate border) induces EMT in the premigratory NC cells and turns on expression of essential NC premigratory genes including, RhoB, Cad6B and Snail2 [14, 38, 77] although the results on Snail2 activation are controversial [78]. Zebrafish mutants of the BMP2 signal transducing pathway display reduction of the NC formation [79] and Smad1 directly binds the chicken Snail2 promoter (which when expressed in the mouse mimics the expression pattern of mouse Snail) and activates EMT [80]. The rostral BMP signal competes with caudally expressed Noggin to establish a rostrocaudal gradient of increasing levels of active BMP in the dorsal NT. In the avian trunk, The BMP/Noggin gradient is controlled by the developing somites, which thus regulate the timing of NC EMT as illustrated in figure 1[78, 81–83]. It is important to note that BMP4 has also been shown to induce apoptosis (via Msx2) in the hindbrain NC [84] and high levels of Snail/Snail2 expression in both mouse and chick hindbrains are sufficient to overcome the BMP induced cell death in the emigrating cells, in which the level of BMP4 expression is again low [85].
Figure 1.
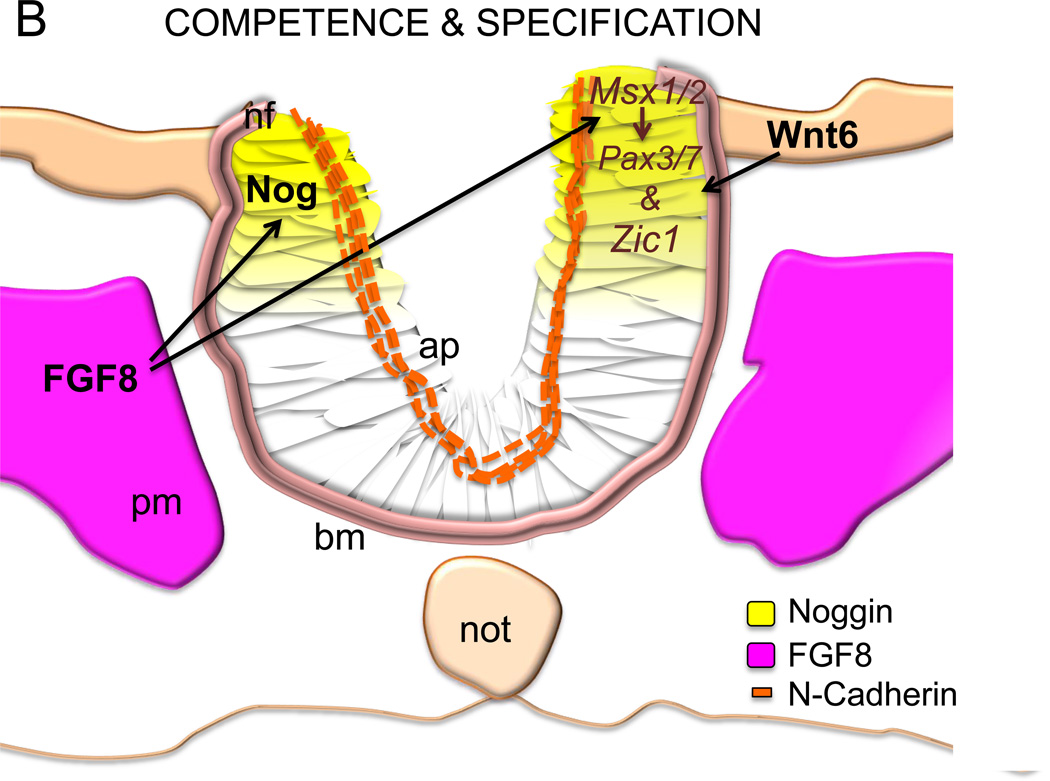
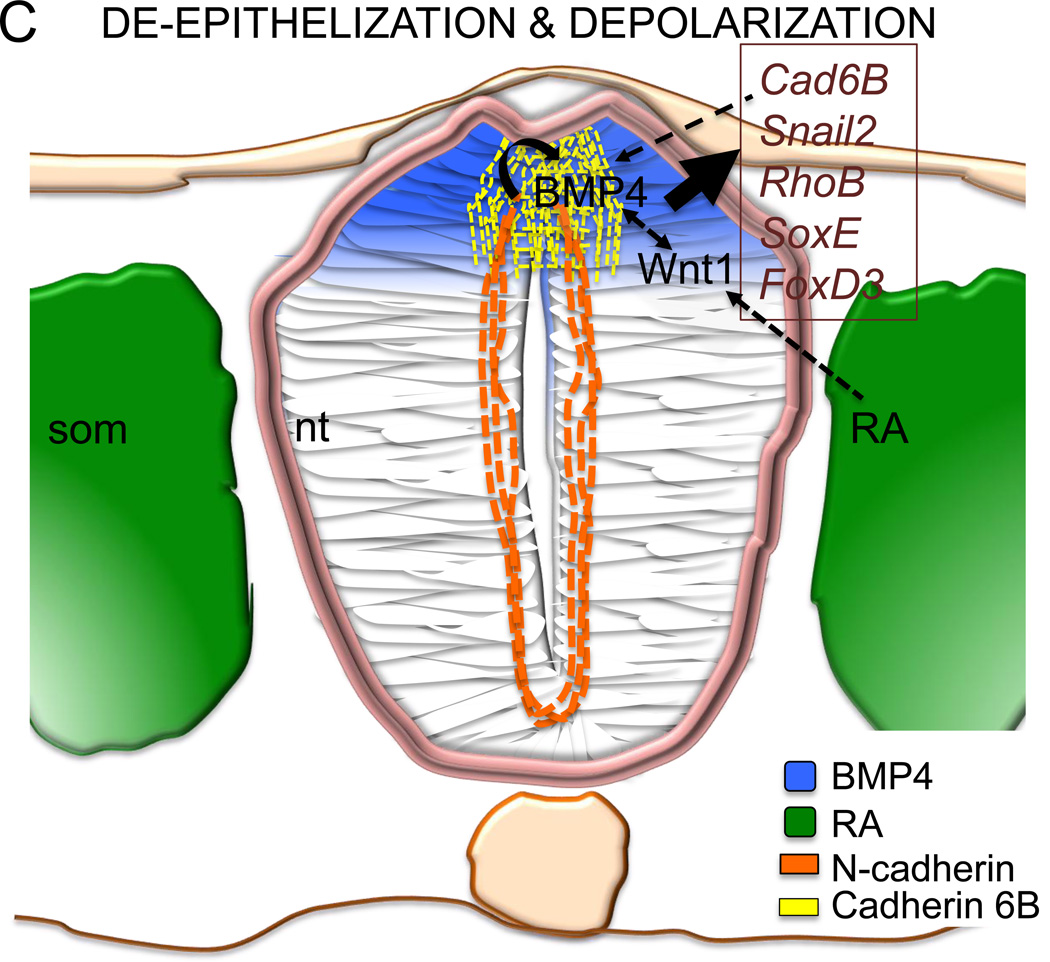
Schematic of neural crest development in the chicken embryo showing that (1A) a rostrocaudal gradient of BMP4 is inhibited by caudally expressed Noggin. The BMP4 gradient in the neural tube, which mediates the timing of neural crest EMT, itself starts rostrally and proceeds caudally as the neural tube closes. FGF8 signaling from the paraxial mesoderm maintains Noggin and is inhibited by retinoic acid signaling (RA) also in a rostrocaudal gradient. Once EMT is complete, mesenchymal neural crest cells emigrate from the neural tube and migrate to their target sites. The black lines mark the axial levels at three transverse levels, representing the gradual progress of EMT (B–D). (1B) FGF secreted from the paraxial mesoderm is required for the specification of the neural crest cells and induces expression of Noggin and Msx1/2 in the dorsal neural folds. These in turn, in the presence of Wnt expressed by the non-neural ectoderm, drives expression of Pax3/7 and Zic1. Apicolaterally bound adherens junction protein N-cadherin maintains the adhesion and polarity of the whole neural epithelium. (1C) RA secreted by the somites induces Wnt1, which induces BMPsignalling and inhibits Noggin. BMP4 is the inducer of the de-epithelization and depolarization factors in a subset of the dorsal neural tube cells that become neural crest. The apicolateral localization of N-cadherin is degraded and replaced by Cadherin 6B in the dorsal neural tube. (1D) The mesenchymal neural crest cells emigrate where the basal lamina is locally ruptured. Cadherin 6B is downregulated by Snail2 and the migratory neural crest cells start to express Cadherin 7/11. som= somite, nt= neural tube, np= neural plate, a= anterior, p= posterior, nne= non-neural ectoderm, pm= paraxial mesoderm, bm= basement membrane, ap= apical side, not= notochord, nf= neural folds, mncc= migratory neural crest cell.
It was recently shown that FGF4/8 secreted by the somites, which signals NC induction at the neural plate border, maintains Noggin expression in the caudal neural tube (as discussed in 3.2.2). Ectopic FGF inhibited expression of NCC specifier genes Snail2 and Sox10. Like the BMP4/Noggin gradient, FGF is also expressed in a rostrocaudal gradient and inhibited by retinoic acid (RA) signaling (Figures. 1A,B). However, RA is not able to directly downregulate Noggin but instead functions through activation of Wnt1, which in turn promotes BMP4 secretion and triggers EMT (Fig. 1C). RA was needed for the onset of NC emigration but not for the expression of NC specification genes such as Snail2 or FoxD3 [86]. Also earlier studies have suggested that BMP induces Wnt1 signalling in the avian trunk NC, which leads to NC delamination through activation of a CyclinD driven transition of the cell cycle from G1 arrest to DNA synthesis (S) phase and Wnt signaling was essential for the avian expression of Cad6B, Msx1 and Pax3 [78]. In line with this, the presence of Wnt1 is required for the transcriptional activation of Snail2 by Pax3 and Zic1 in the Xenopus neural folds in a pathway at least partially mediated by RhoV [19, 87]. Moreover, Wnts and BMPs upregulate transcription of NC genes in explanted chick neural plates [88]. Cv-2 and Cad6B also promote BMP activity in the avian NT [35, 89]. Cad6B links the role of BMP particularly with the onset of de-epithelization of the polar premigratory NC cells and might activate BMP signaling at the receptor level in the absence of ligand [35].
BMP4, which is a member of the TGFβ family, may function in combination with the canonical Wnt-signaling together with N-cadherin. N-cadherin inhibits NC delamination physically by maintaining adhesion junctions but also by storing β-catenin in the adhesive membrane complex [38]. As mentioned earlier, BMP4 triggers cleavage of N-cadherin via the metalloprotease ADAM10. The cleaved intracellular domain, CTF2, activates transcription of Cyclin D and β-catenin, which further enhance de-epithelization by direct transcriptional activation of other EMT genes like Snail2 in the frog [38, 90]. This is similar to events during gastrulation, renal fibrosis and some metastatic cancers, where TGFβ together with Wnts are the main inducers of EMT [4, 91–94]. For example, during gastrulation, Wnt signaling is needed for the competence of the ectoderm to undergo EMT, which is then induced by the TGFβ -superfamily members Nodal and Vg1 [95, 96]. FGF signaling also controls EMT and the maintenance of the mesodermal phenotype by positively regulating Snail, Brachyury and Tbx6 expression during gastrulation [97].
3.2.2 Regulation of genes at the neural plate border before EMT
MAPK-signalling pathway, downstream of FGF8 signalling, is needed for neural crest specification. In the frog neural plate, FGF8 induces Msx1 expression, which in turn, in the presence of Wnt signaling, drives expression of Pax3 and Zic1. In turn, Pax7 is crucial for the NC specification in the chick, see Figure 1B [11, 15, 19, 86]. However, once the premigratory NC cells are specified, FGF8 needs to be downregulated via RA to allow activation of Snail2 and other NC genes e.g. Foxd3, Sox9 and Sox10 and Sox5 (Fig 1A,C). In line with this, the RA synthesizing gene Raldh2 spatiopemporally coincides with the increasing avian Snail2 mRNA [86]. It was recently proposed that the anterior Hox-genes also mediate NC specification and expression of Msx1/2 in a Notch and BMP dependent manner in the cranial NC [98]. In the frog, BMP and Wnt signaling also induce the expression of the transcription factor AP-2, yet another NC specifier that induces expression of Sox9 and Snail2 and marks the NC forming territory at the neural plate [99].
3.3 EMT inducing transcription factors in the neural crest
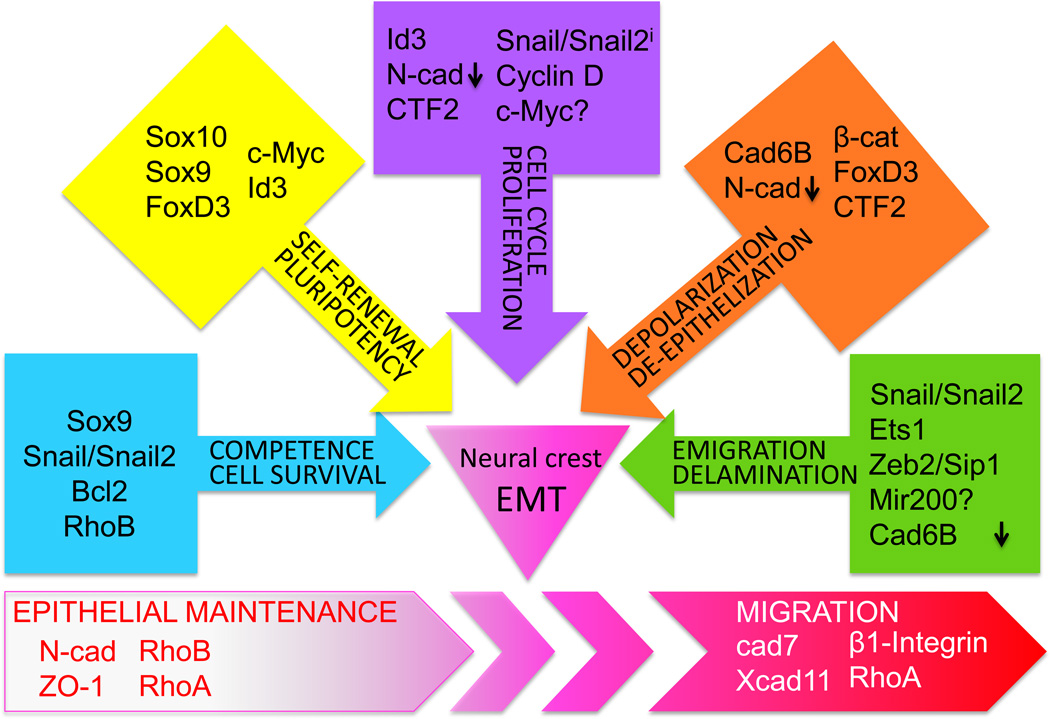
All of the NC specifier genes Snail/Snail2, SoxE and Foxd3 function separately in order for neural crest EMT to occur in the neural folds/NT. Thus, for completion of EMT, the presumptive NC cells need to acquire competence for the pluripotent NC identity, loose the apicobasal polarity and break down the basement membrane [9, 39]. The induction and maintenance of stem cell properties during EMT is crucial during development and cancer and the realization that EMT generates cells with self-renewing properties holds promises of resolving a major problem in cancer biology [100]. Interestingly, the same transcription factors seem to mediate multiple functions during specification, depolarization and migration of NC [39, 101] as illustrated in Figure 2. Because transcriptional and signaling networks often have positive and negative feedback loops, it is often difficult to parse the original order of the events [8, 10].
Figure 2.
The separate subdivisions of EMT that all need to be completed for the proper amount of multipotent neural crest cells to emigrate from the neural tube. This figure outlines the results of multiple studies obtained in several species. i=inhibitory role.
3.3.1 Loss of polarity and delamination
Snail family of transcription factors induce loss of epithelial morphology and migration
Snail/Snail2 has two roles during NC EMT [101]. Snail /Snail2 is the key inducer of EMT marked by the loss of epithelial cell phenotype in NC and the gastrulating embryo [39, 95, 102, 103]. It also has an earlier role during NC specification, which includes maintenance of NC cell survival, which is discussed in 3.3.2. The members of the Snail family of transcription factors all share a similar carboxyl terminal structure that consists of four to six DNA binding zinc finger domains. Snail is the main EMT inducer in mouse premigratory NC whereas only Snail2 (also called Slug) is expressed in the avian dorsal neural folds prior to neural tube closure and remains on during EMT and early migration. The mouse Snail2 expression comes on only in the migratory NC, which thus expresses both Snail and Snail2 [102–104]. The mouse Snail can induce EMT in the avian crest, which indicates functional equality of the two family members [55]. Xenopus Snail1 and Snail2 are both involved in NC specification and EMT. Functional redundancy occurs in frog [105] and mouse NC and thus only the double mutant mice lacking both Snail2 and Snail display a full NC defective phenotype [106]. In zebrafish, Snail1b (a Snail dublicate, previously called Snail2 [107]) expression also defines the NC forming cells [103, 108]. Inhibition of avian Snail2 mRNA (electroporated into the closed NT) blocked emigration from the neuroepithelium, while Snail2 overexpression increased the delamination of premigratory NC as well as the numbers of HNK-1 positive migratory cells [55, 103]. Overexpression of a dominant negative form Snail2 also inhibits NC emigration in Xenopus [101].
Snail proteins repress transcription of target genes by binding to E-boxes in their promoters. For example, in gastrulation, carcinomas and renal fibrosis, Snail induces EMT by directly repressing transcription of E-cadherin, [4, 32, 109, 110]. Snail/Snail2 can also repress other cadherins to allow EMT such as Cad16 in renal fibrosis or Cad6B in the NC [42, 111]. Snail2 also directly represses the adherens junction associated neural N-catenin and MO mediated knockdown of N-catenin increased the number of emigrating cranial NC cells [112]. Snail also directly represses other polarity genes like Claudin, Occludin and Crumbs in in vitro cultured epithelial cells [113, 114]. The Ajuba family of LIM proteins function as corepressors of the Snail family via an interaction with the SNAG domain in the E-cadherin promoter in cell lines. The same corepressors are also required for a proper function of Snail2 in Xenopus NC [115]. Pax3/7 and Zic activate Snail2 in a Wnt dependent manner [11, 19, 20] and also by direct binding the β-cat/ TCF/ Lef complex on the Snail2 enhancer in the frog [90]. The avian Snail2 promoter is also directly activated through Smad1 signalling downstream of BMP4 signalling [80]. Sox9 directly activates Snail2 and also autoregulates its own transcription by directly binding to E-Box2. Similarly, in Xenopus, Snail1 and Snail2 also are needed to maintain each other [105]. In cancer cells Snail2 is required for Twist induced EMT [116].
FoxD3 and Ets1 regulate depolarization and delamination
In all vertebrates, the Forkhead Box family member FoxD3 expression starts in the premigratory NC of the dorsal neural folds, is shut down soon after the beginning of NC migration and reactivated again in the late migratory crest. Finally, it is downregulated as the NC cells reach their final destinations and differentiate [117– 120]. Although knockout models in mice and zebrafish show that lack of Foxd3 does not greatly affect emigration of NC (or expression of RhoB, Snail2 or Cad6B), ectopic FoxD3 affects the transition of epithelial dorsal NT cells into a mesenchymal phenotype. Overexpression of avian Foxd3 in the trunk resulted in total loss of N-cadherin and an increase of β1-integrin as well as other mesenchymal proteins such as Laminin and Cad7 [39, 121–123]. The transcription factor Ets1, which is associated with acquisition of cell mobility and invasiveness during development and cancer, is expressed in the premigratory cranial NC and associated with the breakdown of Laminin and the basement membrane in the avian cranial NC. Ectopic Ets1 did not affect specification of the NC fate or other steps of EMT and alone could not induce expression of FoxD3, Snail2 or Sox10 nor promote transition into the mesenchymal fate [124].
Zeb2
The zinc finger E-box binding homeobox transcriptional repressor family member Zeb2/Sip1 is a known EMT inducer in cancer cells that is activated by Tgfβ/Smad signaling. It can directly repress transcription of E-cadherin as well as other genes involved in epithelial cell junctions and can also function indirectly by repressing miRNAs [125–127]. Xenopus Zeb2 expression starts in the developing neural plate and continues in the premigratory and migratory NC cells [128]. In mouse and frog Zeb2 is thus involved in the specification of the neuroepithelium. It regulates expression of the neural progenitor genes SOX2/3 as well as the neural plate border gene MSX-1 and Zeb2 activity was regulated by BMP/Smad3 as well as FGF signaling [129, 130]. The lack of Zeb2 dramatically affected specification of the vagal NC that forms the enteric nervous system. Mice lacking Zeb2 also displayed ectopic E-cadherin expression throughout the NT and emigration of the cranial NC was inhibited [129, 130].
3.3.2 Neural crest competence, multipotency and cell survival
Sox genes induce NC competence and maintain multipotency
The SoxE genes (Sox8, Sox9 and Sox10) are all expressed in the NC and its derivatives and they exhibit both overlapping and unique functions in NC development. Their expression patterns and several functional studies suggest redundancy between the family members [131, 132] and they are crucial for the early specification as well as for the maintenance of multipotency of premigratory and early migratory NC cells as discussed here. However, SoxE genes also display multiple functions during differentiation of NC [10, 132–136].
In Xenopus Sox8 precedes the expression of Sox9 and Sox10 and its knockdown mainly induces reduction of the expression of Sox10 (but not Sox9 or Snail2) and an overall delay in NC induction [137]. In chicken the timing of the onset of Sox9 expression precedes that of the other premigratory NC markers (Snail2, FoxD3, Sox10 and HNK-1) and its function is crucial for establishing the competence for NC cells to undergo EMT [39, 136, 138]. Wnt and BMP –regulated NC induction in Xenopus and chicken embryos depends on Sox9 transcriptional activator function. Loss of Sox9 in the frog also results in a dramatic loss of NC progenitors particularly affecting the morphology of the craniofacial skeleton [25, 138–140]. Sox9 directly activates Snail2, which in turn activates other NC fate inducing genes and also promotes cell survival in the presumptive NC, which without Sox9 undergo apoptosis and cannot start EMT [39, 138]. In line with this, forced expression of Sox9 increases the territory of NC producing cells at the expense of cells adopting a neural fate but it does not induce emigration of the avian NC [136]. In the cranial but not vagal or trunk premigratory NC, Sox9 directly activates Sox10 although Sox10 levels were reduced in the migratory Sox9−/− trunk NC cells, perhaps representing a secondary effect [39, 141]. Sox9 and Sox10 have also been reported to be able to compensate for the loss of each other without directly activating each other since expression of Sox9 in Sox10 depleted Xenopus embryos rescues NC formation [142]. The knockdown of Sox9 did not affect the levels of FoxD3, suggesting separate regulatory pathways for the key NC specifier genes [39].
Sox10 maintains multipotency of the premigratory and migratory nonmesenchymal (neural, glial and melanocytic) NC progenitors. It seems that Sox10 is especially crucial for the maintenance of glial potential of the NC stem cells [133, 134, 143] even though a high level of Sox10 expression is later needed for the differentiation of NC derived peripheral neurons [133, 143] and melanocytes [144, 145]. Constant overexpression of avian Sox10 induces ectopic NC cell fate despite the absence of dorsal signals in the NT [146]. Knockdown of Xenopus Sox10 causes a loss of premigratory NC precursors affecting both the proliferation and survival of the cells. Also the expression of other NC specific markers was reduced and the timing of Sox10 activation was thought to take place in between the activation of Snail1 and Snail2 [147]. In addition to direct activation by Sox9, Ets1 and c-Myb also directly bind to the Sox10 enhancer and thus directly activate Sox10 [141].
Sox5, a member of the SoxD family, is expressed in the early premigratory NC and its expression seems to be initiated by FoxD3 and Snail2. Ectopic expression of Sox5 resulted in overproduction of NC in the avian cranial crest [150]. In the mouse melanocytic lineage Sox5 inhibits Sox10 transcriptional activity by recruiting chromatin modifiers CTbp2 and HDAC1 to the promoter [150, 151].
Foxd3 maintains NC self-renewal and pluripotency
FoxD3 maintains self-renewal and inhibits differentiation in several stem cell types including e.g. embryonic stem cells In ES cells, inhibition of Foxd3 induced differentiation and it appears to be downstream of the self-renewal inducing factors Nanog, Oct4, c-Myc and Klf. It seems to directly inhibit transcriptional activity of mesoendodermal differentiation genes like Brachyury and Goosecoid [152–154]. FoxD3 also inhibits the cell cycle inhibitor p21 [120]. Foxd3 expression is also critical for the survival and self-renewal of NC. Loss of Foxd3 in neural crest targeted Wnt1-cre mice as well as in a zebra fish mutant (mother superior and Sym1, respectively) causes a devastating loss of NC derivatives [122, 155, 156]. FoxD3-mediated maintenance of self-renewal operates through repression of a mesenchymal as well as melanocytic fate (and FoxD3 thus maintains neural potential) and this lineage specification starts in the delaminating crest [121, 157, 158]. Thus melanocytes, which in chick are the last to emigrate from the NT don’t express Foxd3 [123]. According to several reports, Foxd3 is likely to be activated through distinct pathways from Sox9 and Snail2 [39, 117, 121]. Sox9 deficient premigratory avian NC cells continue to express FoxD3. Furthermore overexpression of Foxd3 gives a distinct phenotype from the ectopic expression of Sox9 and Snail2 [39]. Xenopus FoxD3 is slightly different as it works together with Zic in the early NC to induce Snail2 [159]. Sox10 seems to be downstream of FoxD3 [122], although this may not be a direct connection given the 12 hour induction time shown in the avian overexpression experiments [39, 122] and one study could not see any change in Sox10 or Snail2 levels in the delaminating NC followed by a MO mediated FoxD3 knockdown in zebrafish. The same study suggested that FoxD3 represses itself and Snail1b in the migratory crest [121].
The antiapoptotic Snail/Snail2 is needed for NC specification and cell cycle arrest
Snail/Snail2 (Snail in mouse and fish, Snail2 in chick and both in frog) is expressed in the developing dorsal neural folds and plays a role in the specification of the NC cells [24, 101, 104, 108, 160]. Ectopic overexpression of Snail2 increased the prospective neural crest area in the avian neural plate. However, in explant experiments Snail2 expression alone was not sufficient to induce NC without the presence of Wnt signaling (and to a lesser extent FGF-signaling [17, 39, 101]. In Xenopus the specification of NC in the neural plate is in fact induced Snail, which precedes the expression of Snail2 [24, 161] and overexpression of Snail2 is also sufficient to induce NC [17]. Snail expression in Xenopus thus induces NC specifier genes such as Snail2, Foxd3, Ets1 as well as Twist, which is a yet another known EMT inducer in cancer cells [24, 162] and ectopic Snail2 increases expression of avian RhoB and Pax3 [55].
Another important role of Snail2/Snail in the presumptive NC is to promote cell survival by resisting Msx-mediated apoptosis. Snail2 downregulates the proapoptotic Caspase 9 and promotes activation of the antiapoptotic factor Bcl2. Loss of NC marker expression is rescued by coexpression of BclXL after knockdown of Snail2, thus showing that Snail2 controls NC cell numbers [85, 160, 163]. Snail also induces cell cycle arrest by impairing the transition from early to late G1 by maintaining low levels of CyclinD and it can block the G1/S transition by maintaining high levels of p21. This suggests that Snail expression favors changes in cell shape versus cell division in order to enable EMT [85].
C-Myc and Id3 maintain self-renewal and induce proliferation
The widely studied basic helix-loop-helix transcription factor and oncogene c-Myc maintains self-renewal capacity in several embryonic and adult stem cells as well as cancer stem cells [164]. In the Xenopus, c-Myc controls NC specification at the neural plate border by maintaining multipotency and thus inhibiting premature fate determination, which is directly mediated through Id3 [165, 166]. Id3 also promotes NC proliferation associated with the survival of the NC progenitors at the neural plate border in the frog [167]. On the other hand, according to the avian expression pattern of the Myc-family members in the neural plate border, n-Myc might be responsible for the maintenance of multipotency in early NC since avian c-Myc is turned on only in the migratory NC [168]. c-Myc and n-Myc are known to function similarly during development [169] and they both regulate neural stem cell self-renewal in the mouse embryo [170–172]. The premigratory NC is highly proliferative as compared to the rest the NT and the cyclinD mediated transition (by phosphorylation based inactivation of Rb) of cell cycle regulation point from synthesis to mitosis (the S/G1 phase) has also been associated with the onset of NC emigration [78]. C-myc controls the S/G1 transition by direct activation of cyclins D and E transcription in many cell types [164]. Finally, the tumor suppressor p53 is expressed in the premigratory NC and coordinates growth, suggesting a role in self-renewal and NC specification and p53 null mice also display severe craniofacial defects [173]. Even though more famous for its role ability to induce apoptosis and cellular senescence, p53 also controls self-renewal through repression of c-Myc in neural stem cells and glioma cancer stem cells [174, 175] as well as neural differentiation via cell cycle inhibition by activation of p21 [176, 177]. Likewise, N-myc is associated with neural differentiation of NC derivatives in the ventrolateral pathway. Moderate levels of N-myc protein was detected in all the newly emigrated avian trunk NC cells but the expression was maintained only in the migratory derivatives that assumed a neural fate [178].
3.4. Nontranscriptional regulation of the EMT inducing transcription factors
3.4.1 Epigenetics
Silencing of genes by histone methylation and acetylation plays an important role in transcriptional regulation and phenotype maintenance. The histone demethylase Jumonji (JmjD2A) controls the timing of transcription of NC specifier genes. Occupation of avian JmjD2 on the Sox10 and Snail2 promoters, respectively, keeps them in an inactive H3K9 methylation state and JmjD2A is needed for the switch into the H3K36 methylated active chromatin, which thus allows transcription of the genes at the time of neural tube closure [179]. Similarly, Pax3 expression pattern is also associated with active chromatin H3K4 and H3K36 methylation in mouse fibroblasts [180]. In ES cells Snail recruits components of the repressive Polycomb complex 2 as well as the SIN3A/histone deacetylase to the E-cadherin promoter [181, 182]. Foxd3, more known for its role as a repressor [183], is essential for providing an unmethylated mark at the Alb1 enhancer in ES cells. Overexpression of Foxd3 in embryonic fibroblasts results in a loss of CpG methylation at the Alb1 enhancer and leads to induction of induced pluripotent stem (iPS)cells [184]. C-myc activity correlates with H3-K4 and H3-K79 active methylation marks as well as acetylation of Histone 3 [185]. C-Myc is known for its activation of the transcription of histone acetylases, demethylases and other chromatin modifying complexes as well as for their recruitment to the target promoters [164]. C-Myc induces maintenance of active chromatin in neural stem cells, mediated largely by the histone acetylase GCN5. Loss of n-Myc is associated with a loss of self-renewal capacity and condensed nuclei and terminal differentiation of neurons [186]. Global chromatin condensation has also been associated with onset of migration by perhaps facilitating nuclear movement and reshaping [187].
3.4.2 Posttranslational modifications
The stability of the protein levels of the EMT regulators Snail, Snail2, Twist and Zeb2/Sip1 is regulated by a common mechanism in NC as well as in cancer cells mediated by the F-box protein Partner of paired (Ppa), which serves as the substrate recognition component of the Skp-Cullin-F-box E3 ubiquitin ligase. Ppa thus targets the proteins for degradation and misexpression of Ppa inhibits the formation of neural crest precursors [188–190].
SUMOylation, which interferes with the synergistic interactions of transcription factors to their binding co-factors, is a regulatory mechanism for conferring context-dependent function on widely deployed transcription factors such as the SoxE genes in NC. SUMOylation of Xenopus Sox9 and Sox10 is needed for their activity to induce ear development whereas non-SUMOylated Sox9 and Sox10 are active in NC progenitors and in the differentiating melanocytes [142, 191]. In the premigratory NC, Sox10 is directly activated by Sox9, c-Myb and Ets1, whereas the onset of its expression in the otic placode is driven by a direct binding of a combination of paralogous transcription factors Sox8, c-Myb and Pea3. However, SUMOylation of Sox8 is required for the maintenance of Sox10 expression in the otic placode [141, 192]. Sumoylation of other NC proteins such as Mxs1, have also been reported but the meaning of that in the NC remains unknown [193].
3.4.3 miRNAs and alternative splicing
MicroRNAs have been identified as a new class of EMT regulators, in part owing to their regulation of EMT-inducing transcription factors [27, 194]. The miR-200 family of micro RNAs has an important role in EMT by directly targeting Zeb1/2 and vice versa thus leading to a feedback regulatory loop between EMT and MET [195]. Zeb mediated inhibition of a group of stemness inhibiting miRs (Mir200, Mir203 and miR-183), enhances the activity of self-renewal regulating genes Bmi-1, Sox2 and Klf4, and thus combines stemness and EMT [196]. Stabilization and regulation of splicing by RNA binding proteins are yet additional RNA regulatory steps. Particularly ESRP1/2 have been shown to regulate a whole splicing program that promotes an epithelial phenotype and is thus repressed by EMT [27, 197]. These have yet to be examined in the neural crest system.
3.5 Sequence of events in NC EMT
Even though the series of events involved in EMT are well acknowledged, the sequence in which these steps take place has been under debate and it is not clear if EMT is a single event or can be achieved by multiple parallel mechanisms. The tissue of origin also sets up demands for the order of the EMT steps [198]. Migration studies have also elucidated an important point of the existence of intermediate migratory cells that have undergone EMT (e.g. downregulated E-cadherin) but still maintain a certain amount of polarity [27].
Several hypotheses, which need not be mutually exclusive, have been proposed to explain the onset of NC emigration from the NT. These include the loss of adherens junctions and apicobasal polarity [36, 38] or that the premigratory NC cells may generate a strong enough tractional force to pull themselves away from the adherens junctions without completely downregulating them [199, 200]. The timing of emigration is also regulated by the cell cycle as the G1/S transition has been reported to induce emigration [38, 49, 78, 201]. Interestingly, Snail blocks proliferation in the premigratory NC by arresting the cells in the G0/G1 -phase of the cell cycle. This may allow the cells to change shape as they are not actively dividing. It might also help synchronize cells so that they can all enter the S-phase upon being converted into mesenchymal cells for emigration to occur [85]. The advantage of asymmetrically divided daughter cells that already are detached from the apical membrane was also thought to promote NC fate and emigration [199].
A 4D imaging study of chicken trunk slice cultures shows a surprisingly large variance of the order of events leading to the emigration of NC cells [202]. According to the study, during most of the documented EMTs, the cell was first detached from the apical surface followed by a retraction of the apical tail. After this the cell body translocated out of the epithelium thus suggesting that the downregulation of the apical adhesion complexes was the key step. However there were notable exceptions to this sequence since in around 30% of the cases the cell tail was ruptured while the apicobasal arrangement was still intact (as measured by visualization with α-catenin or actin, respectively) and tail fragments remained behind near the apical surface. Non-apical mitosis, which consisted of only 6% of all documented mitosis in the neuroepithelium, gave rise to 28% of the NC cells in total and in most of the cases both daughter cells became NC. These NC cells had already lost their apicobasal attachments and were rounded up in mitosis when they translocated towards the basal domain of the NT. These and other studies contradict the conclusion that the transition from G1 to S is the trigger of emigration and suggest that the rule may not apply to all NC cells [124, 202]. The cleavage plane of the cells attached to the apical membrane did not play a role in the decision of acquiring a NC fate, as asymmetric and symmetric dividing neuroepithelial cells displayed equal ability to become emigrating neural crest cells. The asymmetrically dividing daughter cells that had lost their contact to the apical membrane also are able to regain it [202]. Another recent live imaging study proposes that the fate of NC (whether to become glia, melanocytes, sensory neurons, peripheral nerves etc.) is already decided according to the location of the cell in the dorsal NT [203]. These two imaging studies also present controversial findings on the exit point of NC from the NT. According to Krispin and collegues, NC cells only exit from the very dorsal tip of the NT whereas Ahlstrom and Ericson report emigrating cells in a more wide range up to a 90° angle [202, 203]. These studies, taken together, highlight the vast heterogeneity in the mechanisms that initiate NC emigration and raise many interesting questions about the mechanisms underlying neural crest EMT. The ultimate microenvironmental cues that influence these events remain to be shown. Clearly, more live imaging studies are needed to combine the knowledge of the fate-determining transcription factors with the morphology and behavior of the emigrating NC cells.
3.6 Mesenchymal to Epithelilal transition
The arrest of neural crest migration is sometimes coupled with a reaggregation process, for example, during formation of peripheral ganglia and craniofacial cartilage. These cells undergo a mesenchymal to epithelial transition (MET). So far, surprisingly little is known about MET in the neural crest derived tissue. Whether the basis of regaining of the epithelial phenotype relies on reversal of the same molecular mechanisms occurring during EMT remains to be studied. Similarities e.g. include re-expression of N-cadherin in sympathetic ganglia, resulting in restoration of cell-cell adhesion and polarity [34, 204, 205]. The role of MET in the reversion of mesenchymal tumor cells, sometimes after long periods of dormancy, to a more epithelial state in distant metastases is a growing field of interest of cancer studies [194, 206, 207] and understanding how neural crest cells switch off their migration program and begin to differentiate may give important clues as to the prevention of tumor dissemination and spreading.
Highlights.
Neural crest (NC) cells undergo EMT to begin migration
The neural crest (NC) is an excellent embryonic model for studying EMT under normal conditions
NC EMT includes several steps from maintenance of multipotency to depolarization and emigration
NC EMT may involve multiple parallel triggering mechanisms
Acknowledgements
This work was funded by grants from Sigrid Juselius Foundation, K Albin Johansson Foundation and Ella and Georg Ehrnrooth Foundation to LK and NIH grants DE017911 and HD037105 to MEB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hay E. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Developmental dynamics. 2005;233:706–720. doi: 10.1002/dvdy.20345. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, Weinberg R. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acloque H, Adams M, Fishwick K, Bronner Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. The Journal of clinical investigation. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiery J, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Boutet A, Esteban M, Maxwell P, Nieto MA. Reactivation of Snail genes in renal fibrosis and carcinomas: a process of reversed embryogenesis? Cell cycle. 2007;6:638–642. doi: 10.4161/cc.6.6.4022. [DOI] [PubMed] [Google Scholar]
- 6.Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Developmental cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Developmental cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling Neural Crest Regulatory Circuits into a Gene Regulatory Network. In: Schekman R, Goldstein L, Lehmann R, editors. Annual Review of Cell and Developmental Biology. Vol 26. 2010. pp. 581–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nature reviews Molecular cell biology. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 10.Nelms B, Labosky PA. Transcriptional Control of neural Crest Development. San Rafael, CA: Morgan & Claypool Life Sciences; 2010. [PubMed] [Google Scholar]
- 11.Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- 12.Ezin A, Fraser S, Bronner-Fraser M. Fate map and morphogenesis of presumptive neural crest and dorsal neural tube. Developmental biology. 2009;330:221–236. doi: 10.1016/j.ydbio.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liem KF, Tremml G, Roelink H, Jessell TM. Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell. 1995;82:969–979. doi: 10.1016/0092-8674(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 14.Liu JP, Jessell TM. A role for rhoB in the delamination of neural crest cells from the dorsal neural tube. Development. 1998;125:5055–5067. doi: 10.1242/dev.125.24.5055. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Castro M, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- 16.Lewis J, Bonner J, Modrell M, Ragland J, Moon R, Dorsky R, et al. Reiterated Wnt signaling during zebrafish neural crest development. Development. 2004;131:1299–1308. doi: 10.1242/dev.01007. [DOI] [PubMed] [Google Scholar]
- 17.LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- 18.Monsoro-Burq A-H, Fletcher R, Harland R. Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development. 2003;130:3111–3124. doi: 10.1242/dev.00531. [DOI] [PubMed] [Google Scholar]
- 19.Monsoro-Burq A-H, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Developmental cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- 21.Hong C-S, Saint-Jeannet J-P. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Molecular biology of the cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansouri A, Stoykova A, Torres M, Gruss P. Dysgenesis of cephalic neural crest derivatives in Pax7−/− mutant mice. Development. 1996;122:831–838. doi: 10.1242/dev.122.3.831. [DOI] [PubMed] [Google Scholar]
- 23.Zhou H-M, Wang J, Rogers R, Conway S. Lineage-specific responses to reduced embryonic Pax3 expression levels. Developmental biology. 2008;315:369–382. doi: 10.1016/j.ydbio.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aybar M, Nieto MA, Mayor R. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130:483–494. doi: 10.1242/dev.00238. [DOI] [PubMed] [Google Scholar]
- 25.Osrio L, Teillet M-A, Palmeirim I, Catala M. Neural crest ontogeny during secondary neurulation: a gene expression pattern study in the chick embryo. The International journal of developmental biology. 2009;53:641–648. doi: 10.1387/ijdb.072517lo. [DOI] [PubMed] [Google Scholar]
- 26.Kuriyama S, Mayor R. Molecular analysis of neural crest migration. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2008;363:1349–1362. doi: 10.1098/rstb.2007.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieto MA. The Ins and Outs of the Epithelial to Mesenchymal Transition in Health and Disease. Annual review of cell and developmental biology. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- 28.Etienne-Manneville S. Control of polarized cell morphology and motility by adherens junctions. Seminars in cell & developmental biology. 2011;22:850–857. doi: 10.1016/j.semcdb.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 29.Yamada S, Pokutta S, Drees F, Weis W, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeichi M. The Cadherins - Cell Cell-Adhesion Molecules Controlling Animal Morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- 31.Hatta K, Takagi S, Fujisawa H, Takeichi M. Spatial And Temporal Expression Pattern Of N-Cadherin Cell-Adhesion Molecules Correlated With Morphogenetic Processes Of Chicken Embryos. Developmental Biology. 1987;120:215–227. doi: 10.1016/0012-1606(87)90119-9. [DOI] [PubMed] [Google Scholar]
- 32.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nature Cell Biology. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa S, Takeichi M. Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development. 1995;121:1321–1332. doi: 10.1242/dev.121.5.1321. [DOI] [PubMed] [Google Scholar]
- 34.Akitaya T, Bronner-Fraser M. Expression of cell adhesion molecules during initiation and cessation of neural crest cell migration. Developmental dynamics. 1992;194:12–20. doi: 10.1002/aja.1001940103. [DOI] [PubMed] [Google Scholar]
- 35.Park KS, Gumbiner BM. Cadherin 6B induces BMP signaling and deepithelialization during the epithelial mesenchymal transition of the neural crest. Development. 2010;137:2691–2701. doi: 10.1242/dev.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa S, Takeichi M. Neural crest emigration from the neural tube depends on regulated cadherin expression. Development. 1998;125:2963–2971. doi: 10.1242/dev.125.15.2963. [DOI] [PubMed] [Google Scholar]
- 37.Inoue T, Chisaka O, Matsunami H, Takeichi M. Cadherin-6 expression transiently delineates specific rhombomeres, other neural tube subdivisions, and neural crest subpopulations in mouse embryos. Developmental biology. 1997;183:183–194. doi: 10.1006/dbio.1996.8501. [DOI] [PubMed] [Google Scholar]
- 38.Shoval I, Ludwig A, Kalcheim C. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 2007;134:491–501. doi: 10.1242/dev.02742. [DOI] [PubMed] [Google Scholar]
- 39.Cheung M, Chaboissier M-C, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Developmental cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 40.Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, et al. Collective Chemotaxis Requires Contact-Dependent Cell Polarity. Developmental Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monier Gavelle F, Duband JL. Cross talk between adhesion molecules: control of N-cadherin activity by intracellular signals elicited by beta1 and beta3 integrins in migrating neural crest cells. The Journal of cell biology. 1997;137:1663–1681. doi: 10.1083/jcb.137.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134:1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coles EG, Taneyhill LA, Bronner-Fraser M. A critical role for Cadherin6B in regulating avian neural crest emigration. Developmental Biology. 2007;312:533–544. doi: 10.1016/j.ydbio.2007.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hadeball B, Borchers A, Wedlich D. Xenopus cadherin-11 (Xcadherin-11) expression requires the Wg/Wnt signal. Mechanisms of development. 1998;72:101–113. doi: 10.1016/s0925-4773(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 45.Borchers A, David R, Wedlich D. Xenopus cadherin-11 restrains cranial neural crest migration and influences neural crest specification. Development. 2001;128:3049–3060. doi: 10.1242/dev.128.16.3049. [DOI] [PubMed] [Google Scholar]
- 46.Vallin J, Girault JM, Thiery JP, Broders F. Xenopus cadherin-11 is expressed in different populations of migrating neural crest cells. Mechanisms of development. 1998;75:171–174. doi: 10.1016/s0925-4773(98)00099-9. [DOI] [PubMed] [Google Scholar]
- 47.Wheeler A, Ridley A. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Experimental cell research. 2004;301:43–49. doi: 10.1016/j.yexcr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 48.Liu AX, Rane N, Liu JP, Prendergast GC. RhoB is dispensable for mouse development, but it modifies susceptibility to tumor formation as well as cell adhesion and growth factor signaling in transformed cells. Molecular and cellular biology. 2001;21:6906–6912. doi: 10.1128/MCB.21.20.6906-6912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Groysman M, Shoval I, Kalcheim C. A negative modulatory role for rho and rho-associated kinase signaling in delamination of neural crest cells. Neural Development. 2008;3 doi: 10.1186/1749-8104-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakaya Y, Sukowati EW, Wu Y, Sheng G. RhoA and microtubule dynamics control cell-basement membrane interaction in EMT during gastrulation. Nature cell biology. 2008;10:765–775. doi: 10.1038/ncb1739. [DOI] [PubMed] [Google Scholar]
- 51.Rupp P, Kulesa P. A role for RhoA in the two-phase migratory pattern of postotic neural crest cells. Developmental biology. 2007;311:159–171. doi: 10.1016/j.ydbio.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 52.Rosso S, Sussman D, Wynshaw Boris A, Salinas P. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nature neuroscience. 2005;8:34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- 53.Broders-Bondon F, Chesneau A, Romero-Oliva F, Mazabraud A, Mayor R, Thiery JP. Regulation of XSnail2 expression by rho GTPases. Developmental Dynamics. 2007;236:2555–2566. doi: 10.1002/dvdy.21273. [DOI] [PubMed] [Google Scholar]
- 54.Smolen G, Schott B, Stewart R, Diederichs S, Muir B, Provencher H, et al. A Rap GTPase interactor, RADIL, mediates migration of neural crest precursors. Genes & development. 2007;21:2131–2136. doi: 10.1101/gad.1561507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.del Barrio M, Nieto MA. Overexpression of Snail family members highlights their ability to promote chick neural crest formation. Development. 2002;129:1583–1593. doi: 10.1242/dev.129.7.1583. [DOI] [PubMed] [Google Scholar]
- 56.Simard A, Di Pietro E, Ryan A. Gene expression pattern of Claudin-1 during chick embryogenesis. Gene expression patterns. 2005;5:553–560. doi: 10.1016/j.modgep.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 57.Aaku Saraste E, Hellwig A, Huttner WB. Loss of occludin and functional tight junctions, but not ZO-1, during neural tube closure--remodeling of the neuroepithelium prior to neurogenesis. Developmental biology. 1996;180:664–679. doi: 10.1006/dbio.1996.0336. [DOI] [PubMed] [Google Scholar]
- 58.Steed E, Balda M, Matter K. Dynamics and functions of tight junctions. Trends in cell biology. 2010;20:142–149. doi: 10.1016/j.tcb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 59.Harris K, Tepass U. Cdc42 and Par proteins stabilize dynamic adherens junctions in the Drosophila neuroectoderm through regulation of apical endocytosis. The Journal of cell biology. 2008;183:1129–1143. doi: 10.1083/jcb.200807020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corbeil D, Marzesco A-M, Wilsch-Bruninger M, Huttner W. The intriguing links between prominin-1 (CD133), cholesterol-based membrane microdomains, remodeling of apical plasma membrane protrusions, extracellular membrane particles, and (neuro)epithelial cell differentiation. FEBS letters. 2010;584:1659–1664. doi: 10.1016/j.febslet.2010.01.050. [DOI] [PubMed] [Google Scholar]
- 61.Marzesco A-M, Janich P, Wilsch-Bräuninger M, Dubreuil V, Langenfeld K, Corbeil D, et al. Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. Journal of cell science. 2005;118:2849–2858. doi: 10.1242/jcs.02439. [DOI] [PubMed] [Google Scholar]
- 62.Röper K, Corbeil D, Huttner WB. Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nature Cell Biology. 2000;2:582–592. doi: 10.1038/35023524. [DOI] [PubMed] [Google Scholar]
- 63.Decker RS, Friend DS. Assembly Of Gap Junctions During Amphibian Neurulation. The Journal of Cell Biology. 1974;62:32–47. doi: 10.1083/jcb.62.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lo CW, Cohen MF, Huang GY, Lazatin BO, Patel N, Sullivan R, et al. Cx43 gap junction gene expression and gap junctional communication in mouse neural crest cells. Developmental genetics. 1997;20:119–132. doi: 10.1002/(SICI)1520-6408(1997)20:2<119::AID-DVG5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 65.Cai DH, Brauer PR. Synthetic matrix metalloproteinase inhibitor decreases early cardiac neural crest migration in chicken embryos. Developmental dynamics. 2002;224:441–449. doi: 10.1002/dvdy.10129. [DOI] [PubMed] [Google Scholar]
- 66.Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribre T, et al. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Current biology. 2001;11:918–930. doi: 10.1016/s0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- 67.Kee Y, Hwang B, Sternberg P, Bronner-Fraser M. Evolutionary conservation of cell migration genes: from nematode neurons to vertebrate neural crest. Genes & development. 2007;21:391–396. doi: 10.1101/gad.1509307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lallier T, Bronner-Fraser M. Alpha 1 beta 1 integrin on neural crest cells recognizes some laminin substrata in a Ca(2+)-independent manner. The Journal of cell biology. 1992;119:1335–1345. doi: 10.1083/jcb.119.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strachan L, Condic M. Cranial neural crest recycle surface integrins in a substratum-dependent manner to promote rapid motility. The Journal of cell biology. 2004;167:545–554. doi: 10.1083/jcb.200405024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alfandari D, Cousin H, Gaultier A, Hoffstrom B, DeSimone D. Integrin alpha5beta1 supports the migration of Xenopus cranial neural crest on fibronectin. Developmental biology. 2003;260:449–464. doi: 10.1016/s0012-1606(03)00277-x. [DOI] [PubMed] [Google Scholar]
- 71.Breau M, Pietri T, Eder O, Blanche M, Brakebusch C, Fässler R, et al. Lack of beta1 integrins in enteric neural crest cells leads to a Hirschsprung-like phenotype. Development. 2006;133:1725–1734. doi: 10.1242/dev.02346. [DOI] [PubMed] [Google Scholar]
- 72.Perris R, Perissinotto D. Role of the extracellular matrix during neural crest cell migration. Mechanisms of development. 2000;95:3–21. doi: 10.1016/s0925-4773(00)00365-8. [DOI] [PubMed] [Google Scholar]
- 73.Barembaum M, Moreno TA, LaBonne C, Sechrist J, Bronner-Fraser M. Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nature Cell Biology. 2000;2:219–225. doi: 10.1038/35008643. [DOI] [PubMed] [Google Scholar]
- 74.Endo Y, Osumi N, Wakamatsu Y. Bimodal functions of Notch-mediated signaling are involved in neural crest formation during avian ectoderm development. Development. 2002;129:863–873. doi: 10.1242/dev.129.4.863. [DOI] [PubMed] [Google Scholar]
- 75.Coffman CR, Skoglund P, Harris WA, Kintner CR. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell. 1993;73:659–671. doi: 10.1016/0092-8674(93)90247-n. [DOI] [PubMed] [Google Scholar]
- 76.Cornell R, Eisen J. Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development. 2002;129:2639–2648. doi: 10.1242/dev.129.11.2639. [DOI] [PubMed] [Google Scholar]
- 77.Kanzler B, Foreman RK, Labosky PA, Mallo M. BMP signaling is essential for development of skeletogenic and neurogenic cranial neural crest. Development. 2000;127:1095–1104. doi: 10.1242/dev.127.5.1095. [DOI] [PubMed] [Google Scholar]
- 78.Burstyn-Cohen T, Stanleigh J, Sela-Donenfeld D, Kalcheim C. Canonical Wnt activity regulates trunk neural crest delamination linking BMP/noggin signaling with G1/S transition. Development. 2004;131:5327–5339. doi: 10.1242/dev.01424. [DOI] [PubMed] [Google Scholar]
- 79.Nguyen VH, Schmid B, Trout J, Connors SA, Ekker M, Mullins MC. Ventral and lateral regions of the zebrafish gastrula, including the neural crest progenitors, are established by a bmp2b/swirl pathway of genes. Developmental biology. 1998;199:93–110. doi: 10.1006/dbio.1998.8927. [DOI] [PubMed] [Google Scholar]
- 80.Sakai D, Tanaka Y, Endo Y, Osumi N, Okamoto H, Wakamatsu Y. Regulation of Slug transcription in embryonic ectoderm by beta-catenin-Lef/Tcf and BMP-Smad signaling. Development, growth & differentiation. 2005;47:471–482. doi: 10.1111/j.1440-169X.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- 81.Steventon B, Araya C, Linker C, Kuriyama S, Mayor R. Differential requirements of BMP and Wnt signalling during gastrulation and neurulation define two steps in neural crest induction. Development. 2009;136:771–779. doi: 10.1242/dev.029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sela-Donenfeld D, Kalcheim C. Inhibition of noggin expression in the dorsal neural tube by somitogenesis: a mechanism for coordinating the timing of neural crest emigration. Development. 2000;127:4845–4854. doi: 10.1242/dev.127.22.4845. [DOI] [PubMed] [Google Scholar]
- 83.Sela-Donenfeld D, Kalcheim C. Regulation of the onset of neural crest migration by coordinated activity of BMP4 and Noggin in the dorsal neural tube. Development. 1999;126:4749–4762. doi: 10.1242/dev.126.21.4749. [DOI] [PubMed] [Google Scholar]
- 84.Graham A, Francis West P, Brickell P, Lumsden A. The signalling molecule BMP4 mediates apoptosis in the rhombencephalic neural crest. Nature. 1994;372:684–686. doi: 10.1038/372684a0. [DOI] [PubMed] [Google Scholar]
- 85.Vega S, Morales A, Ocaa O, Valds F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes & development. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martinez-Morales P, Diez Del Corral R, Olivera-Martnez I, Quiroga A, Das R, Barbas J, et al. FGF and retinoic acid activity gradients control the timing of neural crest cell emigration in the trunk. The Journal of cell biology. 2011;194:489–503. doi: 10.1083/jcb.201011077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guemar L, de Santa Barbara P, Vignal E, Maurel B, Fort P, Faure S. The small GTPase RhoV is an essential regulator of neural crest induction in Xenopus. Developmental biology. 2007;310:113–128. doi: 10.1016/j.ydbio.2007.07.031. [DOI] [PubMed] [Google Scholar]
- 88.Taneyhill L, Bronner-Fraser M. Dynamic alterations in gene expression after Wnt-mediated induction of avian neural crest. Molecular biology of the cell. 2005;16:5283–5293. doi: 10.1091/mbc.E05-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coles E, Christiansen J, Economou A, Bronner-Fraser M, Wilkinson D. A vertebrate crossveinless 2 homologue modulates BMP activity and neural crest cell migration. Development. 2004;131:5309–5317. doi: 10.1242/dev.01419. [DOI] [PubMed] [Google Scholar]
- 90.Vallin J, Thuret R, Giacomello E, Faraldo MM, Thiery JP, Broders F. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. Journal of biological chemistry. 2001;276:30350–30358. doi: 10.1074/jbc.M103167200. [DOI] [PubMed] [Google Scholar]
- 91.Safina A, Varga A, Bianchi A, Zheng Q, Kunnev D, Liang P, et al. Ras alters epithelial-mesenchymal transition in response to TGFbeta by reducing actin fibers and cell-matrix adhesion. Cell cycle. 2009;8:284–298. doi: 10.4161/cc.8.2.7590. [DOI] [PubMed] [Google Scholar]
- 92.Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes & development. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 93.Sato M, Muragaki Y, Saika S, Roberts A, Ooshima A. Targeted disruption of TGF-beta1/ Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. The Journal of clinical investigation. 2003;112:1486–1494. doi: 10.1172/JCI19270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scheel C, Eaton E, Li S, Chaffer C, Reinhardt F, Kah K-J, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nature reviews Molecular cell biology. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 96.Chea H, Wright C, Swalla B. Nodal signaling and the evolution of deuterostome gastrulation. Developmental dynamics. 2005;234:269–278. doi: 10.1002/dvdy.20549. [DOI] [PubMed] [Google Scholar]
- 97.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Developmental cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 98.Gouti M, Briscoe J, Gavalas A. Anterior Hox genes interact with components of the neural crest specification network to induce neural crest fates. Stem cells. 2011;29:858–870. doi: 10.1002/stem.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luo T, Lee Y-H, Saint-Jeannet J-P, Sargent T. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mani S, Guo W, Liao M-J, Eaton E, Ayyanan A, Zhou A, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Developmental biology. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- 102.Nieto MA, Bennett MF, Sargent MG, Wilkinson DG. Cloning and developmental expression of Sna, a murine homologue of the Drosophila snail gene. Development. 1992;116:227–237. doi: 10.1242/dev.116.1.227. [DOI] [PubMed] [Google Scholar]
- 103.Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 104.Sefton M, Sanchez S, Nieto MA. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- 105.Carl TF, Dufton C, Hanken J, Klymkowsky MW. Inhibition of neural crest migration in Xenopus using antisense slug RNA. Developmental biology. 1999;213:101–115. doi: 10.1006/dbio.1999.9320. [DOI] [PubMed] [Google Scholar]
- 106.Murray S, Oram K, Gridley T. Multiple functions of Snail family genes during palate development in mice. Development. 2007;134:1789–1797. doi: 10.1242/dev.02837. [DOI] [PubMed] [Google Scholar]
- 107.Blanco M, Barrallo Gimeno A, Acloque H, Reyes A, Tada M, Allende M, et al. Snail1a and Snail1b cooperate in the anterior migration of the axial mesendoderm in the zebrafish embryo. Development. 2007;134:4073–4081. doi: 10.1242/dev.006858. [DOI] [PubMed] [Google Scholar]
- 108.Thisse C, Thisse B, Postlethwait JH. Expression of snail2, a second member of the zebrafish snail family, in cephalic mesendoderm and presumptive neural crest of wild-type and spadetail mutant embryos. Developmental biology. 1995;172:86–99. doi: 10.1006/dbio.1995.0007. [DOI] [PubMed] [Google Scholar]
- 109.Bolos V, Peinado H, Perez-Moreno M, Fraga M, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. Journal of cell science. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 110.Batlle E, Sancho E, Franc C, Dominguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature Cell Biology. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 111.Boutet A, De Frutos C, Maxwell P, Mayol MJ, Romero J, Nieto MA. Snail activation disrupts tissue homeostasis and induces fibrosis in the adult kidney. EMBO journal. 2006;25:5603–5613. doi: 10.1038/sj.emboj.7601421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jhingory S, Wu CY, Taneyhill LA. Novel insight into the function and regulation of alphaN-catenin by Snail2 during chick neural crest cell migration. Developmental biology. 2010;344:896–910. doi: 10.1016/j.ydbio.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. Journal of cell science. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 114.Whiteman EL, Liu CJ, Fearon ER, Margolis B. The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene. 2008;27:3875–3879. doi: 10.1038/onc.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Langer E, Feng Y, Zhaoyuan H, Rauscher F, Kroll K, Longmore G. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Developmental cell. 2008;14:424–436. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Casas E, Kim J, Bendesky A, Ohno Machado L, Wolfe C, Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer research. 2011;71:245–254. doi: 10.1158/0008-5472.CAN-10-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–4138. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- 118.Yamagata M, Noda M. The winged-helix transcription factor CWH-3 is expressed in developing neural crest cells. Neuroscience Letters. 1998;249:33–36. doi: 10.1016/s0304-3940(98)00385-1. [DOI] [PubMed] [Google Scholar]
- 119.Labosky PA, Kaestner KH. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mechanisms of development. 1998;76:185–190. doi: 10.1016/s0925-4773(98)00105-1. [DOI] [PubMed] [Google Scholar]
- 120.Hromas R, Ye H, Spinella M, Dmitrovsky E, Xu D, Costa RH. Genesis, a Winged Helix transcriptional repressor, has embryonic expression limited to the neural crest, and stimulates proliferation in vitro in a neural development model. Cell and tissue research. 1999;297:371–382. doi: 10.1007/s004410051365. [DOI] [PubMed] [Google Scholar]
- 121.Lister J, Cooper C, Nguyen K, Modrell M, Grant K, Raible D. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Developmental biology. 2006;290:92–104. doi: 10.1016/j.ydbio.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 122.Stewart R, Arduini B, Berghmans S, George R, Kanki J, Henion P, et al. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Developmental biology. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 123.Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–1479. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- 124.Theveneau E, Duband J-L, Altabef M. Ets-1 confers cranial features on neural crest delamination. PLoS ONE. 2007;2:e1142-e. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Molecular cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 126.Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic acids research. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bracken C, Gregory P, Kolesnikoff N, Bert A, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer research. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 128.van Grunsven LA, Papin C, Avalosse B, Opdecamp K, Huylebroeck D, Smith JC, et al. XSIP1, a Xenopus zinc finger/homeodomain encoding gene highly expressed during early neural development. Mechanisms of development. 2000;94:189–193. doi: 10.1016/s0925-4773(00)00318-x. [DOI] [PubMed] [Google Scholar]
- 129.van Grunsven L, Taelman V, Michiels C, Opdecamp K, Huylebroeck D, Bellefroid E. deltaEF1 and SIP1 are differentially expressed and have overlapping activities during Xenopus embryogenesis. Developmental dynamics. 2006;235:1491–1500. doi: 10.1002/dvdy.20727. [DOI] [PubMed] [Google Scholar]
- 130.Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, et al. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. American journal of human genetics. 2003;72:465–470. doi: 10.1086/346092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maka M, Stolt CC, Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Developmental biology. 2005;277:155–169. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 132.Reiprich S, Stolt CC, Schreiner S, Parlato R, Wegner M. SoxE proteins are differentially required in mouse adrenal gland development. Molecular biology of the cell. 2008;19:1575–1586. doi: 10.1091/mbc.E07-08-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kelsh RN. Sorting out Sox10 functions in neural crest development. BioEssays : news and reviews in molecular, cellular and developmental biology. 2006;28:788–798. doi: 10.1002/bies.20445. [DOI] [PubMed] [Google Scholar]
- 134.Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, et al. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes & development. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mori-Akiyama Y, Akiyama H, Rowitch D, de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- 137.O'Donnell M, Hong C-S, Huang X, Delnicki R, Saint-Jeannet J-P. Functional analysis of Sox8 during neural crest development in Xenopus. Development. 2006;133:3817–3826. doi: 10.1242/dev.02558. [DOI] [PubMed] [Google Scholar]
- 138.Sakai D, Suzuki T, Osumi N, Wakamatsu Y. Cooperative action of Sox9, Snail2 and PKA signaling in early neural crest development. Development. 2006;133:1323–1333. doi: 10.1242/dev.02297. [DOI] [PubMed] [Google Scholar]
- 139.Lee Y-H, Aoki Y, Hong C-S, Saint Germain N, Credidio C, Saint Jeannet J-P. Early requirement of the transcriptional activator Sox9 for neural crest specification in Xenopus. Developmental biology. 2004;275:93–103. doi: 10.1016/j.ydbio.2004.07.036. [DOI] [PubMed] [Google Scholar]
- 140.Spokony R, Aoki Y, Saint Germain N, Magner Fink E, Saint-Jeannet J-P. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development. 2002;129:421–432. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- 141.Betancur P, Bronner-Fraser M, Sauka-Spengler T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3570–3575. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Taylor KM, LaBonne C. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Developmental Cell. 2005;9:593–603. doi: 10.1016/j.devcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 143.Kim J, Lo L, Dormand E, Anderson D. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 144.Elworthy S, Lister JA, Carney TJ, Raible DW, Kelsh RN. Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development. 2003;130:2809–2818. doi: 10.1242/dev.00461. [DOI] [PubMed] [Google Scholar]
- 145.Aoki Y, Saint Germain N, Gyda M, Magner Fink E, Lee YH. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Developmental biology. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- 146.McKeown S, Lee V, Bronner-Fraser M, Newgreen D, Farlie P. Sox10 overexpression induces neural crest-like cells from all dorsoventral levels of the neural tube but inhibits differentiation. Developmental dynamics. 2005;233:430–444. doi: 10.1002/dvdy.20341. [DOI] [PubMed] [Google Scholar]
- 147.Honore S, Aybar M, Mayor R. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Developmental biology. 2003;260:79–96. doi: 10.1016/s0012-1606(03)00247-1. [DOI] [PubMed] [Google Scholar]
- 148.Nekrep N, Wang J, Miyatsuka T, German M. Signals from the neural crest regulate beta-cell mass in the pancreas. Development. 2008;135:2151–2160. doi: 10.1242/dev.015859. [DOI] [PubMed] [Google Scholar]
- 149.Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 150.Perez-Alcala S, Nieto MA, Barbas J. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–4465. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- 151.Stolt CC, Lommes P, Hillgrtner S, Wegner M. The transcription factor Sox5 modulates Sox10 function during melanocyte development. Nucleic acids research. 2008;36:5427–5440. doi: 10.1093/nar/gkn527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu Y, Labosky P. Regulation of embryonic stem cell self-renewal and pluripotency by Foxd3. Stem cells. 2008;26:2475–2484. doi: 10.1634/stemcells.2008-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hanna L, Foreman R, Tarasenko I, Kessler D, Labosky P. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes & development. 2002;16:2650–2661. doi: 10.1101/gad.1020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tompers D, Foreman R, Wang Q, Kumanova M, Labosky P. Foxd3 is required in the trophoblast progenitor cell lineage of the mouse embryo. Developmental biology. 2005;285:126–137. doi: 10.1016/j.ydbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 155.Teng L, Mundell N, Frist A, Wang Q, Labosky P. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615–1624. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Montero-Balaguer M, Lang M, Sachdev S, Knappmeyer C, Stewart R, De La Guardia A, et al. The mother superior mutation ablates foxd3 activity in neural crest progenitor cells and depletes neural crest derivatives in zebrafish. Developmental dynamics. 2006;235:3199–3212. doi: 10.1002/dvdy.20959. [DOI] [PubMed] [Google Scholar]
- 157.Mundell N, Labosky P. Neural crest stem cell multipotency requires Foxd3 to maintain neural potential and repress mesenchymal fates. Development. 2011;138:641–652. doi: 10.1242/dev.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Curran K, Lister J, Kunkel G, Prendergast A, Parichy D, Raible D. Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Developmental biology. 2010;344:107–118. doi: 10.1016/j.ydbio.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sasai N, Mizuseki K, Sasai Y. Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development. 2001;128:2525–2536. doi: 10.1242/dev.128.13.2525. [DOI] [PubMed] [Google Scholar]
- 160.Zhang C, Carl TF, Trudeau ED, Simmet T, Klymkowsky MW. An NF-kappa B and Slug Regulatory Loop Active in Early Vertebrate Mesoderm. Plos One. 2006;1 doi: 10.1371/journal.pone.0000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Linker C, Bronner Fraser M, Mayor R. Relationship between gene expression domains of Xsnail, Xslug, and Xtwist and cell movement in the prospective neural crest of Xenopus. Developmental biology. 2000;224:215–225. doi: 10.1006/dbio.2000.9723. [DOI] [PubMed] [Google Scholar]
- 162.Ansieau S, Morel AP, Hinkal G, Bastid J, Puisieux A. TWISTing an embryonic transcription factor into an oncoprotein. Oncogene. 2010;29:3173–3184. doi: 10.1038/onc.2010.92. [DOI] [PubMed] [Google Scholar]
- 163.Tribulo C, Aybar M, Sanchez S, Mayor R. A balance between the anti-apoptotic activity of Slug and the apoptotic activity of msx1 is required for the proper development of the neural crest. Developmental biology. 2004;275:325–342. doi: 10.1016/j.ydbio.2004.07.041. [DOI] [PubMed] [Google Scholar]
- 164.Eilers M, Eisenman R. Myc's broad reach. Genes & development. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in xenopus. Developmental cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 166.Light W, Vernon A, Lasorella A, Iavarone A, LaBonne C. Xenopus Id3 is required downstream of Myc for the formation of multipotent neural crest progenitor cells. Development. 2005;132:1831–1841. doi: 10.1242/dev.01734. [DOI] [PubMed] [Google Scholar]
- 167.Kee Y, Bronner-Fraser M. To proliferate or to die: role of Id3 in cell cycle progression and survival of neural crest progenitors. Genes & development. 2005;19:744–755. doi: 10.1101/gad.1257405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Khudyakov J, Bronner-Fraser M. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Developmental dynamics. 2009;238:716–723. doi: 10.1002/dvdy.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Malynn BA, de Alboran IM, O'Hagan RC, Bronson R, Davidson L, DePinho RA, et al. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes & development. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 170.Kerosuo L, Piltti K, Fox H, Angers-Loustau A, Häyry V, Eilers M, et al. Myc increases self-renewal in neural progenitor cells through Miz-1. Journal of cell science. 2008;121:3941–3950. doi: 10.1242/jcs.024802. [DOI] [PubMed] [Google Scholar]
- 171.Nagao M, Campbell K, Burns K, Kuan C-Y, Trumpp A, Nakafuku M. Coordinated control of self-renewal and differentiation of neural stem cells by Myc and the p19ARF-p53 pathway. The Journal of cell biology. 2008;183:1243–1257. doi: 10.1083/jcb.200807130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Knoepfler P, Cheng P, Eisenman R. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes & development. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rinon A, Molchadsky A, Nathan E, Yovel G, Rotter V, Sarig R, et al. p53 coordinates cranial neural crest cell growth and epithelial-mesenchymal transition/delamination processes. Development. 2011;138:1827–1838. doi: 10.1242/dev.053645. [DOI] [PubMed] [Google Scholar]
- 174.Piltti K, Kerosuo L, Hakanen J, Eriksson M, Angers-Loustau A, Lepp S, et al. E6/E7 oncogenes increase and tumor suppressors decrease the proportion of self-renewing neural progenitor cells. Oncogene. 2006;25:4880–4889. doi: 10.1038/sj.onc.1209492. [DOI] [PubMed] [Google Scholar]
- 175.Zheng H, Ying H, Yan H, Kimmelman A, Hiller D, Chen A-J, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chylicki K, Ehinger M, Svedberg H, Gullberg U. Characterization of the molecular mechanisms for p53-mediated differentiation. Cell growth & differentiation. 2000;11:561–571. [PubMed] [Google Scholar]
- 177.Galderisi U, Jori F, Giordano A. Cell cycle regulation and neural differentiation. Oncogene. 2003;22:5208–5219. doi: 10.1038/sj.onc.1206558. [DOI] [PubMed] [Google Scholar]
- 178.Wakamatsu Y, Watanabe Y, Nakamura H, Kondoh H. Regulation of the neural crest cell fate by N-myc: promotion of ventral migration and neuronal differentiation. Development. 1997;124:1953–1962. doi: 10.1242/dev.124.10.1953. [DOI] [PubMed] [Google Scholar]
- 179.Strobl-Mazzulla PH, Sauka-Spengler T, Bronner-Fraser M. Histone Demethylase JmjD2A Regulates Neural Crest Specification. Developmental Cell. 2010;19:460–468. doi: 10.1016/j.devcel.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Corry G, Hendzel M, Underhill DA. Subnuclear localization and mobility are key indicators of PAX3 dysfunction in Waardenburg syndrome. Human molecular genetics. 2008;17:1825–1837. doi: 10.1093/hmg/ddn076. [DOI] [PubMed] [Google Scholar]
- 181.Herranz N, Pasini D, Daz V, Franc C, Gutierrez A, Dave N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Molecular and cellular biology. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Molecular and cellular biology. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yaklichkin S, Steiner A, Lu Q, Kessler D. FoxD3 and Grg4 physically interact to repress transcription and induce mesoderm in Xenopus. Journal of biological chemistry. 2007;282:2548–2557. doi: 10.1074/jbc.M607412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Xu J, Watts J, Pope S, Gadue P, Kamps M, Plath K, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes & development. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nature Cell Biology. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 186.Knoepfler P, Zhang X-y, Cheng P, Gafken P, McMahon S, Eisenman R. Myc influences global chromatin structure. EMBO journal. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gerlitz G, Bustin M. Efficient cell migration requires global chromatin condensation. Journal of cell science. 2010;123:2207–2217. doi: 10.1242/jcs.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lander R, Nordin K, Labonne C. The F-box protein Ppa is a common regulator of core EMT factors Twist, Snail, Slug, and Sip1. J Cell Biol. 2011;194:17–25. doi: 10.1083/jcb.201012085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Vernon A, LaBonne C. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development. 2006;133:3359–3370. doi: 10.1242/dev.02504. [DOI] [PubMed] [Google Scholar]
- 190.Vinas-Castells R, Beltran M, Valls G, Gomez I, Garcia J, Montserrat-Sents B, et al. The hypoxia-controlled FBXL14 ubiquitin ligase targets SNAIL1 for proteasome degradation. Journal of biological chemistry. 2010;285:3794–3805. doi: 10.1074/jbc.M109.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Girard M, Goossens M. Sumoylation of the SOX10 transcription factor regulates its transcriptional activity. FEBS letters. 2006;580:1635–1641. doi: 10.1016/j.febslet.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 192.Betancur P, Sauka Spengler T, Bronner M. A Sox10 enhancer element common to the otic placode and neural crest is activated by tissue-specific paralogs. Development. 2011;138:3689–3698. doi: 10.1242/dev.057836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gupta V, Bei M. Modification of Msx1 by SUMO-1. Biochemical and biophysical research communications. 2006;345:74–77. doi: 10.1016/j.bbrc.2006.03.232. [DOI] [PubMed] [Google Scholar]
- 194.Polyak K, Weinberg R. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nature Reviews Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 195.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO reports. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wellner U, Schubert J, Burk U, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature Cell Biology. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 197.Warzecha C, Jiang P, Amirikian K, Dittmar K, Lu H, Shen S, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO journal. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nakaya Y, Sheng G. An amicable separation: Chick's way of doing EMT. Cell adhesion & migration. 2009;3:160–163. doi: 10.4161/cam.3.2.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Erickson CA, Reedy MV. Neural crest development: The interplay between morphogenesis and cell differentiation. Current Topics in Developmental Biology, Vol 40. 1998;40:177–209. doi: 10.1016/s0070-2153(08)60367-1. [DOI] [PubMed] [Google Scholar]
- 200.Bilozur ME, Hay ED. Cell migration into neural tube lumen provides evidence for the "fixed cortex" theory of cell motility. Cell motility and the cytoskeleton. 1989;14:469–484. doi: 10.1002/cm.970140405. [DOI] [PubMed] [Google Scholar]
- 201.Burstyn-Cohen T, Kalcheim C. Association between the cell cycle and neural crest delamination through specific regulation of G1/S transition. Developmental cell. 2002;3:383–395. doi: 10.1016/s1534-5807(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 202.Ahlstrom JD, Erickson CA. The neural crest epithelial-mesenchymal transition in 4D: a 'tail' of multiple non-obligatory cellular mechanisms. Development. 2009;136:1801–1812. doi: 10.1242/dev.034785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Krispin S, Nitzan E, Kassem Y, Kalcheim C. Evidence for a dynamic spatiotemporal fate map and early fate restrictions of premigratory avian neural crest. Development. 2010;137:585–595. doi: 10.1242/dev.041509. [DOI] [PubMed] [Google Scholar]
- 204.Kasemeier-Kulesa J, Bradley R, Pasquale E, Lefcort F, Kulesa P. Eph/ephrins and N-cadherin coordinate to control the pattern of sympathetic ganglia. Development. 2006;133:4839–4847. doi: 10.1242/dev.02662. [DOI] [PubMed] [Google Scholar]
- 205.Duband JL, Tucker GC, Poole TJ, Vincent M, Aoyama H, Thiery JP. How do the migratory and adhesive properties of the neural crest govern ganglia formation in the avian peripheral nervous system? Journal of cellular biochemistry. 1985;27:189–203. doi: 10.1002/jcb.240270302. [DOI] [PubMed] [Google Scholar]
- 206.Nguyen D, Bos P, Massague J. Metastasis: from dissemination to organ-specific colonization. Nature Reviews Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 207.Wells A, Yates C, Shepard C. E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clinical & experimental metastasis. 2008;25:621–628. doi: 10.1007/s10585-008-9167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]