Summary
The evolutionary ‘decision’ to store genetic information away from the place of protein synthesis, in a separate compartment, has forced eukaryotic cells to establish a system to transports mRNAs from the nucleus to the cytoplasm for translation. To ensure export to be fast and efficient, cells have evolved a complex molecular interplay that is tightly regulated. Over the last few decades, many of the individual players in this process have been described, starting with the composition of the nuclear pore complex to proteins that modulate co-transcriptional events required to prepare an mRNP for export to the cytoplasm. How the interplay between all the factors and processes results in the efficient and selective export of mRNAs from the nucleus and how the export process itself is executed within cells, however, is still not fully understood. Recent advances in using proteomic and single molecule microscopy approaches have provided important insights into the process and its kinetics. This review summarizes these recent advances and how they led to the current view on how cells orchestrate the export of mRNAs.
1. Introduction
The export of mRNAs is one of the many steps along the gene expression pathway and reflects only a short time period within the lifetime of an mRNA [1]. However, mRNA export cannot be seen as an isolated process, as it has been functionally linked to different upstream and downstream events, in particular the localization of the gene within the nucleus, transcription, mRNA processing and quality control [2]. Disruption of upstream events affects export, but how and which exact steps in the export process are affected is not very well understood. Genetic and biochemical approaches allowed the isolation of what we think may be most of the factors involved in mRNA export, however, exactly how these factors function in the process of mRNA export is still unknown.
As for many biological processes, the use of model organisms was shown to be a very fruitful approach in dissecting the mechanisms of mRNA export. In particular, genetic screens and proteomic approaches in the yeast S.cerevisiae have identified many players involved in mRNA export and it is reasonable to speculate that, at least for yeast, most mRNA maturation factors have been found [3,4]. While a ‘clear’ picture as has become apparent in yeast, it has not yet emerged for higher eukaryotes where many of the yeast proteins are conserved and often additional components have been found, suggesting an even more complex regulation. On the other hand, more is know in higher eukaryotes of how an mRNA is actually exported on a cellular level. As early as in the 1970s electron micrographs have shown mRNAs crossing nuclear pores, and recently single molecule techniques have allowed the monitoring and visualization of individual mRNA molecules during export in real time [5]. Consequently, microscopy has become a key tool to determine how cellular factors act in concert to regulate kinetics along the different steps of the mRNA export pathway [6].
This review tries to combine the dense knowledge about the mechanistic details, mainly observed in yeast, with the insights gained from recent single molecule studies in higher eukaryotes to illustrate the current understanding on how this important part of the gene expression pathway is executed.
2. Getting made and ready to be exported: linking mRNA export with transcription
For an mRNA to be exported, it first has to be processed, folded and assembled into an export-competent mRNP. To allow for a coordinated efficiency of these events, they occur co-transcriptionally and can be mediated in two different ways: i) by RNA maturation factors present on the polymerase complex which are later transferred to the RNA, and, ii) by proteins that are recruited directly to nascent transcripts [7]. Two complexes have so far been implicated in this recruitment, one is the C-terminal domain (CTD) of the large subunit of RNA Polymerase II (Pol II), the other the multi-protein complex THO, which itself is recruited to the elongating polymerase by an unknown mechanism [2]. This orchestrated formation of export-competent mRNPs is moreover controlled by a surveillance mechanism, ensuring only properly assembled and processed mRNPs will be released from the site of transcription into the nucleoplasm, a process that requires the nuclear exosome [8].
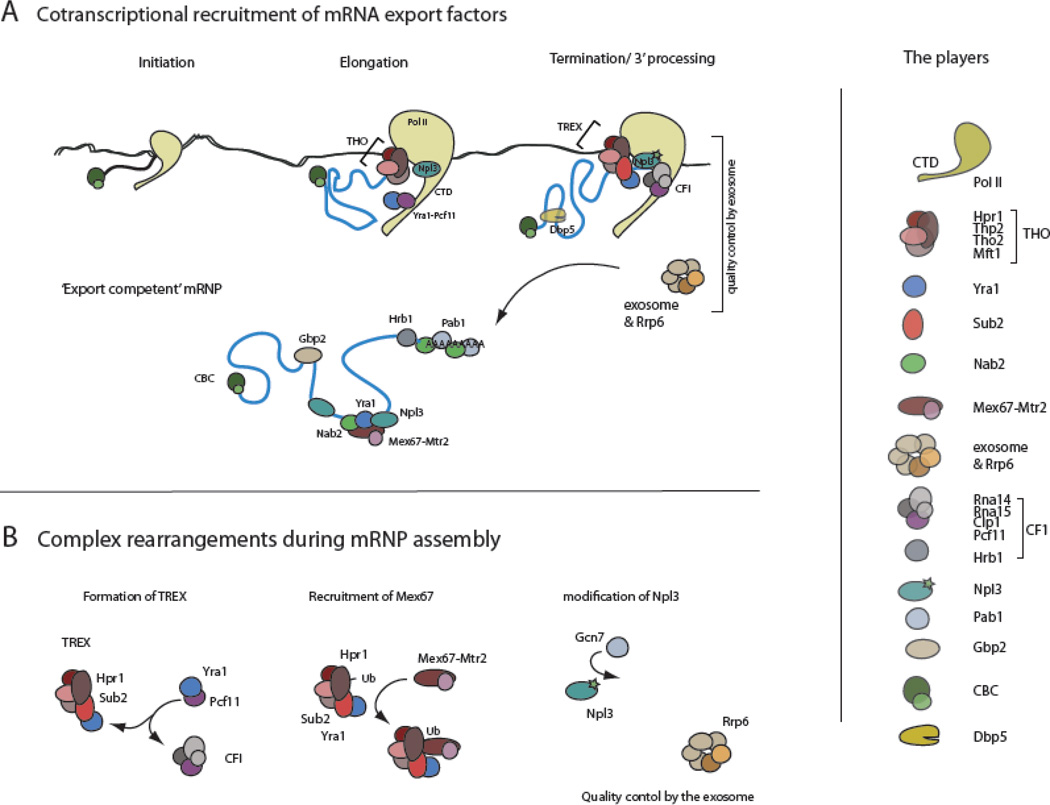
Co-transcriptional recruitment of the mRNA export machinery
It has been known for some years that many of the processes rearranging mRNAs during their maturation occur co-transcriptionally and that a number of them are mediated by the phosphorylation cycle of the CTD. The CTD consists of heptad peptide repeats that are reversibly phosphorylated at Ser2, Ser5 and Ser7. Both phosphorylation and dephosphorylation of the CTD were shown to mediate co-transcriptional mRNA maturation events such as recruitment of the capping enzymes (by Ser5-P) and the 3’ end processing machinery (by Ser2-P) as well as stimulation of 3’end processing of nascent transcripts, respectively [9], [10].
In the last few years several studies showed that the CTD, however, is not just involved in events affecting splicing and 3’ end processing, but has a role in orchestrating the recruiting of the mRNA export machinery onto nascent transcripts. Recent chromatin immuno-precipitation (ChIP) studies from Bentley and coworkers revealed that the mRNA export factor Yra1 is recruited to elongating Pol II through an interaction with the 3’end processing factor Pcf11, which itself binds to Ser2-P CTD (Figure 1) [11,12]. Once transcription reaches the 3’ end of a gene, Yra1 is then competed off by the cleavage and polyadenylation factor (CF1A) component Clp1, resulting in concomitant 3’ end cleavage and transcription termination followed by mRNA release and polyadenylation as well as the transfer of Yra1 to the mRNA and its binding to the ATP-dependent RNA helicase Sub2 [11,12]. These findings supersede the initial believe that Sub2 is responsible for recruiting the export factor Yra1p to mRNPs [13,14].
Figure 1. Co-transcriptional assembly of an export competent mRNP.
The assembly of an export competent mRNP is orchestrated by the coordinated a recruitment of mRNA export factors to the nascent mRNA. The THO complex is recruited to the polymerase complex by an unknown interaction with the polymerase and ensures that the nascent mRNA is not forming DNA-RNA hybrids. It also recruits Sub2. The 3’ processing factor Pcf11 binds to the Ser2 phosphorylated CTD and brings Yra1 to the 3’ end of the gene where it is transferred to Sub2. Npl3 is also deposited on the mRNA during 3’ end formation. Only fully processed mRNPs can leave the site of transcription, and export competency is monitored by the nuclear exosome component Rrp6.
Sub2 itself is part of the Transcription Export (TREX) complex (composed of THO, Tex1, Sub2 and Yra1) and is involved in splicing and mRNA export. It is recruited to elongating transcripts by the THO complex [14], an early mRNA-associating maturation complex. THO (composed of Tho2, Hpr1, Mft1 and Thp2) plays a so far undefined role at the interface between transcription and mRNA export; tho null mutants in S. cerevisiae, while viable, exhibited impaired transcription elongation and RNA export defects [14–17]. One intriguing feature of yeast tho mutants are their transcription-associated hyper-recombination phenotype, where the nascent mRNA is observed to form stretches of DNA:RNA hybrids (R-loops) which can be reduced by RNaseH over-expression [18–20]. This suggests that THO could play a role in compacting the pre-mRNA during transcription elongation to prevent its pairing to chromatin. Although THO components have been found to co-immunopurify with Pol II complexes and the complex has a potential role during elongation, no direct interaction between THO components and the polymerase or the CTD has been described to date [2]. Therefore it remains unclear how THO is recruited to elongating complexes. Abruzzi and colleagues reported that recruitment of the THO component Hpr1 is RNA-independent in yeast, however, recent studies in mammals suggest that recruitment of THO does depend on the nascent transcript [21,22]
However, tho mutants also exhibit mRNA export defects and a number of genetic studies using yra1 and sub2 mRNA export mutants have demonstrated a link between the export machinery and THO, and thus transcription [13,14]. A recent study by Rougemaille and colleagues [23] showed that both THO and Sub2 are required to coordinate mRNA processing and export; they observed that tho/sub2 mutants induced an elongation defect stalling Pol II close to the 3’ end, resulting in the dissociation of CF1A and inhibiting both polyadenylation as well as the release of the mRNA. Instead, complexes accumulated at the 3’ end processing/termination site that contained DNA, RNA as well as proteins and have been dubbed “heavy chromatin” [23,24]. On the other hand, heavy chromatin was not detected in strains lacking 3′ end-processing factors or mRNA export mutants, implying that these factors may play a role in forming heavy chromatin. Interestingly, the authors also found that nuclear pore complex (NPC) components constitute a large portion of heavy chromatin, suggesting that the NPC itself may associate with the 3′ ends of genes during transcription at least for a subset of genes, as this was mostly shown for heat shock-regulated genes. Taken together, and considering recent data by the Bentley lab [11,12], it could be speculated that in these mutants Yra1 cannot associate with Sub2 and remains bound to Pcf11. Subsequently, Clp1 cannot bind to Pcf11, no 3’ end cleavage takes place and the mRNP is not released and exported but rather accumulates as a large complex at the 3’ end. Together, these observations indicate an important role for THO–Sub2 in coordinating a remodeling step at the 3′ end of genes that is critical for the dissociation of CF1A, polyadenylation and transcript release.
Npl3 is another RNA binding protein that participates in linking transcription elongation with 3’ processing and mRNA export. Recruited to the elongating polymerase complex, it promotes elongation, the recruitment of splicing factors and prevents premature termination [25,26]. During elongation, Npl3 is progressively phosphorylated by caseine kinase 2 (Ck2), reducing its anti-termination activity and allowing the recruitment of CF1A to the 3’ end. During 3’ processing, Npl3 is then de-phosphorylated by Glc7, which was suggested to promote its interaction with the mRNA export receptor Mex67 [27]. Indeed it was shown that in a mutant where phosphorylation of Npl3 was eliminated, increased amounts of Mex67, as well as Npl3 itself, were present on polyadenylated mRNAs, indicating that dephosphorylation of Npl3p does promote binding of Mex67p to RNA. Moreover, Mex67 was demonstrated to interact with Npl3 directly in vitro [27]. Npl3, as well as Yra1, Nab2 and Hrp1, is also modified by the arginine methylation [28,29]. These modifications, although not essential for general mRNA export, were suggested to facilitate the release and export of specific transcripts [29].
A further link between transcription and export is the recruitment of the mRNA export adapter Mex67. Recently, Dieppois and coworkers demonstrated the recruitment of Mex67 to elongating Pol II mediated by THO [30]. As demonstrated also by structural studies, this interaction involves the C-terminal ubiquitin-associated (UBA) domain of Mex67 (Mex67-UBA) and Hpr1, which is ubiquitylated by the E3 ubiquitin ligase Rsp5 [30–32] (Figure 1). Mex67 was also shown to be associated with “heavy chromatin” of the HSP104 gene in Δmft1 mutant cells [23]. So far this recruitment of Mex67 to elongating transcripts has only been shown for a subset of genes (GAL2, GAL10 and HSP104) and whether this mechanism is applicable to all genes still has to be investigated.
Co-transcriptional mRNA quality control
Correct transcription, processing and polyadenylation are all required for the assembly of an export-competent mRNP. Eukaryotic cells have evolved a co-transcriptional quality control mechanism to determine whether mRNAs have been transcribed and assembled correctly. Based on that mechanism, the ability of mRNAs to leave the site of transcription is tightly controlled, as mutations within the DNA, errors in transcription, splicing, processing or assembly can all lead to the formation of aberrant pre-mRNPs. Mutations in proteins affecting specific steps of pre-mRNA maturation have been shown to result in the accumulation of immature mRNPs in a tight focus close to or at a site of transcription [2,33]. The mRNAs within these foci were shown to have been cleaved and polyadenylated, hence suggesting that retention of aberrant mRNAs is a post-transcriptional event [34]. The exact mechanism of how this surveillance and quality control is achieved is not know, but genetic studies in yeast have identified the nuclear exosome, and in particular its component Rrp6, as a key player in the recognition and retention of defective transcripts [8,35]. Rrp6 is required for co-transcriptional surveillance of mRNA biogenesis [13,35,36] and its function was shown to be enhanced by the TRAMP (Trf–Air–Mtr4 polyadenylation) complex, which acts as a cofactor for mRNA surveillance [34,37]. However, how exactly this is achieved mechanistically is still unclear. While much of our knowledge about mRNA surveillance comes from studies in yeast, the composition and structure of the exosome are highly conserved from yeast to humans, which suggests that the its functions are also conserved in higher eukaryotes [38,39].
Considering all the above data strongly underlines the important connection and coordination between 3’ processing and the assembly of an export-competent mRNP. Another interesting aspect of linking transcription with export is from a kinetic point of view. As different processes along the gene expression pathway are linked they may provide feedback loops to ensure correct co-transcriptional processing, assembly and release of mRNAs. One example is the transferal of Mex67 and its adaptors Yra1, Npl3 and the poly(A)-binding protein Nab2 from the transcription machinery onto the mRNA. Mutations in Mex67 and Yra1 interfere with correct 3’end formation [35]. This indicates that proper recruitment and transfer of export factors from the transcription machinery onto the mRNA, and thus formation of an export-competent mRNP, may have an impact (or feedback) on optimal THO–Sub2 function and stimulate 3’end processing; improper recruitment or transfer (due to mutations in the protein, for example) will result in targeting the mRNP for degradation [40]. Thus, the entire process of co-transcriptional mRNP assembly converges to guarantee kinetically efficient 3’-end formation, in order to compete against mRNA retention and degradation by the exosome surveillance complex. Finally, it is interesting to note that while many studies have shown that, in yeast, recruitment of THO/TREX is coupled to transcription, in higher eukaryotes, the recruitment of TREX is coupled to splicing. Moreover, recent studies are consistent with the view that in metazoans TREX promotes export through binding and releasing spliced mRNPs from nuclear speckles [41–44].
3. On track: Moving on to the periphery
After the mRNA is transcribed, processed, assembled into an mRNP and checked by a surveillance mechanism, the mRNP is released into the nucleoplasm to find its way to the nuclear periphery (Figure 2A). It is now generally accepted that mRNAs move within the nucleoplasm by random Brownian motion and not by motor driven transport [1,45–53]. However, single molecule experiments in metazoan cells showed that despite moving randomly, mRNAs move in a discontinuous manner; periods of fast diffusion are intermitted by periods of much slower mRNA movement, during which mRNAs diffuse within a small region for an extended period of time (also called ‘corralled movement’) [50,51,53]. As mRNA diffusion slows down considerably when mRNA enters a chromatin-dense region, slower mRNA movement is believed to be mainly caused by a molecular crowding effect rather than by mRNAs directly interacting with any cellular component during corralled movement in chromatin dense regions. Consistent with this theory, single mRNAs never become completely immobile during corralled movement. However, the existence of a higher order organization allowing facilitated movement of large macromolecular complexes within the nucleus cannot completely be excluded.
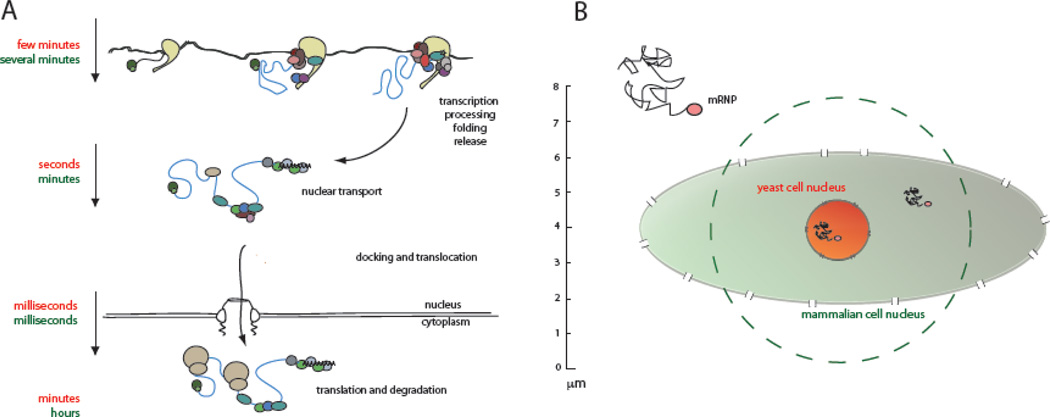
Figure 2. Timescales and distances for different steps of the mRNA export process in yeast and vertebrates.
A. Cartoon showing the gene expression pathway. Transcription, nuclear diffusion and cytoplasmic lifetimes differ significantly between lower and higher eukaryotes, mRNP translocation through the pore, however, is likely to be similar in all organisms. Timescales in yeast are shown in red, timescales in vertebrates in green. B. Comparison of nuclear size in yeast and in vertebrates. Diffusion of an mRNP to the nuclear periphery by Brownian motion will require significantly more time in cells with a larger nucleus.
The actual speed at which mRNAs diffuse within the nucleus is still up for discussion. A number of studies using a variety of imaging approaches resulted in different estimates that vary considerably from each other (from 0.005 µm2/sec up to more than 1 µm2/sec) [1,45,46,48,50–54]. The main reason for these discrepancies is most likely technical. Interestingly, there is a large discrepancy in the values of the estimated diffusion coefficient between studies that used single molecule tracking (SMT) techniques to measure diffusion, but surprisingly little between studies that extracted their values measuring the behavior of total nuclear mRNAs by labeling all mRNAs simultaneously. Measuring the behavior of the whole population of nuclear polyadenylated mRNA using a GFP-tagged nuclear poly(A)binding protein by florescence recovery after photobleaching (FRAP) suggested an mRNA diffusion coefficient of 0.6µm2/sec, similar to measurements by florescence correlation spectroscopy approaches (FCS) that observed the diffusion of dT-labeled nuclear mRNA (1.0µm2/sec) [46,48,52]. However, these numbers were challenged by single molecule tracking experiments that allowed following individual mRNAs in the nucleus and, at least in part, suggested a much slower mRNA diffusion rate. Using different labeling approaches (MS2 system, anti-sense oligos and molecular beacons), these SMT experiments estimated diffusion speeds varying from 0.005µm2/sec for a large dystrophin mRNA to 3.4µm2/sec for a small beta-actin mRNA [1,50–54]. Interestingly, however, the discrepancies might lie less in the size of the molecules than in the technology used to obtain the numbers. Obtaining accurate SMT data relies heavily on the ability not just to see single molecules but also to detect ALL molecules, which can be problematic when molecules diffuse at different speeds. If image acquisition is slow, faster diffusing molecules will not be detected and therefore the behavior of the entire population will be misinterpreted, as only slow moving molecules are observed and measured. Importantly, in experiments that measured slow diffusion constants, data was acquired using much slower frame rates than in experiments revealing faster diffusion times (300ms per frame down to 5ms per frame). Therefore, it might well be possible that data acquired with slow frame rates simply missed fast moving particles, and that diffusion rates for most mRNAs are hence much faster than suggested by many SMT experiments - closer to 1µm2/sec as was also suggested by FRAP and FCS data. This would moreover be consisted with the diffusion constant obtained for mRNP-sized inert dextran molecules. Interestingly, recent experiments from the Kubitcheck lab, using fast acquisition SMT as well as FCS, measured a diffusion constant that also fits within this range [52]. Future experiments will have to further resolve these discrepancies.
So how long does it take an mRNA (or mRNP) to reach the nuclear periphery? Considering that mRNAs move by Brownian motion, the transit time of an mRNP across a typical mammalian cell nucleus (7µm in diameter) was calculated to be in the range of 2–6 minutes [55]. However, the recent observation that mRNPs can spend about 50% of their time ‘corralling’, might significantly increase their transit time [1,50–54]. On the other hand, many nuclei are not perfect spheres but rather ellipsoids, which substantially reduces the distance an mRNA has to travel to encounter the periphery in one dimension (Figure 2B). Ultimately it depends on where a gene is located within the nucleus; if it is found close to the periphery, the probability of quickly finding a pore will be higher. Furthermore, these calculations only estimate the time an mRNA requires to reach the nuclear periphery. It is likely, however, that mRNPs do not always automatically encounter a nuclear pore and that finding an NPC might be equally difficult a task as finding its way to the periphery in the first place. Measurements using a number of different microscopy approaches showed that mRNAs reach the cytoplasm within a timeframe anywhere from a few minutes to up to 30 minutes after being transcribed or injected into a nucleus, suggesting that mRNPs reach the nuclear periphery within a few minutes [56].
However, even with the recent development of fast microscopy techniques, it has not yet been possible to follow a single mRNA all the way from its site of transcription to and through a nuclear pore and thus to get a precise timeframe for these steps. This is mostly due to the speed of mRNP diffusion, which is too fast to follow an mRNP in all four dimensions and over long periods of time using current imaging technologies. Due to their fast diffusion rates, mRNPs will not stay in a single imaging plane for longer than a few hundred milliseconds, and those that do are not likely to reflect the average mRNP population. Once an mRNP is lost from the imaging plane, it is almost impossible to find the same mRNA again, as nuclei often contain multiple copies that cannot be distinguished from one another. To ensure that an individual mRNP remains in focus and tracked, it would have to be followed in all three dimensions as well as over time (4D), with frame rates per imaging plane in the tenths of milliseconds and in multiple planes simultaneously; a feat that is very difficult to achieve with currently available technologies. In addition, many single molecule studies use fluorescent proteins to label mRNAs which limits the time mRNPs can be imaged due to photo-stability and photo-toxicity. As export occurs minutes after an mRNP has been released into the nucleoplasm, it will be very challenging to image this entire process using fluorescent proteins and alternative labeling strategies will have to be used that, in combination with improvements in fast 4D image acquisition, will allow the tracking of nuclear mRNPs over longer time frames.
4. Where to next: Finding a pore
Reaching the nuclear periphery is the first step on the way out of the nucleus. Once mRNPs have reached the nuclear periphery, they have to encounter an NPC, interact with its structure and get access to the pore. Thus encountering a nuclear pore is the next step. The surface of the inner nuclear membrane is densely packed with nuclear pore complexes. Cyro-EM studies in yeast have shown that a G1 cell nucleus contains about 90 NPCs [57]. Considering a typical nuclear surface area of 7.4µm2 and a diameter of an NPC of 100µm, one can calculate that NPCs account for about 10% of the total surface of the inner nuclear membrane [58]. In higher eukaryotes, NPCs are found at a similar density, although it varies considerably between different cell types [59,60]. Therefore, an mRNP diffusing through the nucleoplasm and bouncing into the nuclear envelope will encounter a nuclear pore in only ~10% of the time. Indeed, single molecule studies in mammalian cells have shown that interactions of mRNPs with the nuclear periphery lead only infrequently to a stable interaction with an NPC, and that mRNPs are frequently released back into the nucleoplasm [1]. The large size of nuclei in higher eukaryotes might therefore further decrease the rate of mRNA export compared to cells with smaller nuclei. The probability of an mRNP to find its way back to the periphery after bounding off the nuclear envelope will be significantly lower in cells with a large nucleus as compared to a yeast cell, where nuclei are small (2µm in diameter) and an mRNA can move across it in a few seconds. However, mRNA diffusion behavior in yeast has not yet been measured.
If mRNAs really find their way to the periphery simply by diffusion, the mechanisms in place to ensure rapid export appear hardly very sophisticated. Cells, however, have many options to increase efficiency, even if mRNPs move by diffusion. The organization of higher order chromatin is probably the most obvious. More than 25 years ago, Günther Blobel already suggested a connection between gene expression processes in his gene gating hypothesis he proposed that active genes bind to nuclear pores and facilitate the processing and export of mRNAs by creating a “circumscribed space subjacent to the nuclear pore complex and extending into the interior of the nucleus in the form of channels” (quoted from [61]). Furthermore, he suggested that this could be “envisioned to serve as the locale where transcription and much of the co- and posttranscriptional processing would occur” (quoted from [61]). Studies over the last two decades showed that although this concept is not entirely true, it is still partially valid (reviewed in [62]). Actively transcribing genes, in particular in mammalian cells, are usually located in the nuclear interior from where mRNPs are released into the nucleoplasm to find their way to a nuclear pore. In higher eukaryotes, regions surrounding nuclear pores are usually devoid of heterochromatin, whereas more condensed chromatin is found in regions at the nuclear periphery lacking NPCs [60]. While mRNPs diffuse easily into euchromatin, dense heterochromatin, however, is less accessible [50]. This simple organization per se might create ‘tracks’ that guide mRNAs towards the NPC. Ensuring pore accessibility might be an actively regulated process mediated by the NPC itself. This idea was first supported by studies that demonstrated chromatin boundary activity of the NPC. In yeast the nucleoporin Nup2p was shown to prevent the spreading of heterochromatin when a reporter gene was tethered to the NPC, thereby keeping dense heterochromatin away from the NPC [63]. In accordance with this, the Drosophila nucleoporin TPR (yeast Mlp) was shown to be required for the formation and maintenance of heterochromatin exclusion zones surrounding the NPC [64].
Another possibility to increase the efficiency of mRNA export kinetics is by ensuring that mRNPs stay at the periphery once they arrive there. Grünwald and Singer showed that often beta-actin mRNPs are not released back into the nucleoplasm even if they did not encounter and/or interact with a pore. Instead, they were shown to slide along the periphery, appearing to scan the region for the presence of nuclear pores [1]. It is not know if this is a general mechanism as Mor and colleagues, using a similar approach (although using much slower image acquisition compared to the Grünwald study), did not observe complexes scanning at the periphery, nor is it known which proteins could be implicated in such process [54]. However, a scanning mechanism, if it exists, is likely to significantly increase export kinetic.
Gene gating: Bringing genes to the pore
One way of efficiently bringing newly transcribed mRNPs into close proximity of an NPC was uncovered in yeast. Using nucleoporins and transport factors as baits in genome-wide chromatin immuno-precipitation experiments, the Silver laboratory discovered that many active genes associate directly with nucleoporins (nups) [65]. Most of the nups found interacting with active genes are part of the nuclear pore basket (see below) and thus nups that can be easily accessed by chromatin and would tether a gene closer to the export channel. Following these first observations, a number of studies showed that many inducible genes are targeted to the nuclear periphery upon activation, including INO1, GAL1, HXK1 and HSP104 [66–71]. The mechanism, by which this occurs, however, is less clear and multiple models have been proposed. It is likely that a number of different events participate in tethering genes to the pore and that these may vary for different genes or classes of genes. In yeast, chromatin is very mobile and most loci diffuse within a volume of around 0.5µm in diameter [72–74]. This suggests that almost any gene can encounter the nuclear periphery passively once in a while; to stay in close proximity to a nuclear pore, it simply would have to be tethered there by a factor that mediates the interaction between specific chromatin-associated factors and NPC components. Many genes found at the periphery are highly transcribed, suggesting that strong, active transcription might be a pre-requisite to tethering genes to nuclear pores. One possible mediator for this is the export receptor Mex67. Mex67 was shown to be recruited co-transcriptional to a number of these genes (GAL2, GAL10, HSP104) and might simply ‘pull’ genes to the periphery by its ability to directly interact with nucleoporins [30].
The transcriptional co-activator complex SAGA and the nuclear pore associated complex TREX-2 have also been implicated in the recruitment of transcribing genes to the NPC [66,75,76]. Both complexes share the component Sus1, and a crystal structure of the TREX-2 complex suggests that its component Sac3 acts as a scaffold to assemble TREX-2 at the pore and so assist in recruiting SAGA-associated genes to the NPC [77]. SAGA-regulated genes, however, represent only about 10% of genes in yeast, many of them regulated or induced by different stresses or changes in environmental conditions, suggesting that this mechanism might affect only a small subset of genes [78]. Consistent with a role for promoter complexes in perinuclear recruitment, it was shown for galactose regulated genes that these associate with the periphery prior to transcription activation [67]. A further mechanism to target genes to periphery was recently identified by the Brickner lab [71]. They found that DNA itself has the intrinsic ability to interact with the nuclear pore and keeps the INO1 gene located at the periphery, even after transcription has shut down. This is mediated by a “zip-code” sequence within the promoter region that is evolutionally conserved and has also been identified in S. pombe. It remains to be seen whether such sequences are a common way to tether genes to the periphery and close to nuclear pores, and which proteins are implicated in this mechanism.
Whether gene gating through pore-tethering is wide spread in higher eukaryotes is not clear. TREX-2 is conserved in higher eukaryotes and was shown to be required for mRNA export and NPC anchoring for a subset of genes in Drosophila [79,80]. However, most genes in higher eukaryotes are not transcribed at the nuclear periphery, and were instead shown to move to the nuclear interior upon activation [81,82]. Interestingly, a number of nucleoporins in higher eukaryotes were shown to have a nucleoplasmic phase and to be located not at the NPC but at promoter regions in the nuclear interior, where they stimulate transcription [83,84]. Future studies will help to further elucidate the role of NPC targeting in higher eukaryotes.
So what could be the actual benefit of NPC-gene interactions? The most obvious advantage for tethering specific genes close or directly to the nuclear pore would be to facilitate rapid export of their mRNAs. While tethering genes to nuclear pores has mainly been observed in yeast, an organism with very small nuclei, is it necessary to ask if tethering would lead to an actual kinetic advantage, especially in yeast. In a yeast nucleus, diffusion of an mRNA from any location within the nucleoplasm to a pore or the nuclear periphery is likely to be a very efficient way to quickly encounter an NPC (as discussed above). Therefore, the difference in delay between an mRNA being released into the nucleoplasm, instead of having been delivered directly to the NPC prior to that event by tethering its gene, can only be in the range of a couple of seconds. Compared to the time it takes for an mRNA to be transcribed or to be translated (which is both in the order of minutes), this seems to be a very minor delay. It is therefore possible that gene tethering serves alternative functions, which are as yet unknown. However, and very interestingly, many of the genes that were shown to have a strong localization at the periphery during their expression are inducible genes, such as heat-shock or the galactose regulated genes, which are expressed in response to environmental changes or stress. Thus we could speculate that in these cases the ability to express these genes close to a nuclear pore for subsequent rapid mRNA export and translation in the cytoplasm would be advantageous for the cell. However, the direct export of mRNAs expressed by periphery-tethered genes through adjacent pores has not yet been demonstrated [3,70,71,85,86].
5.Almost there: Recruitment to the nuclear pore
The nuclear pore complex
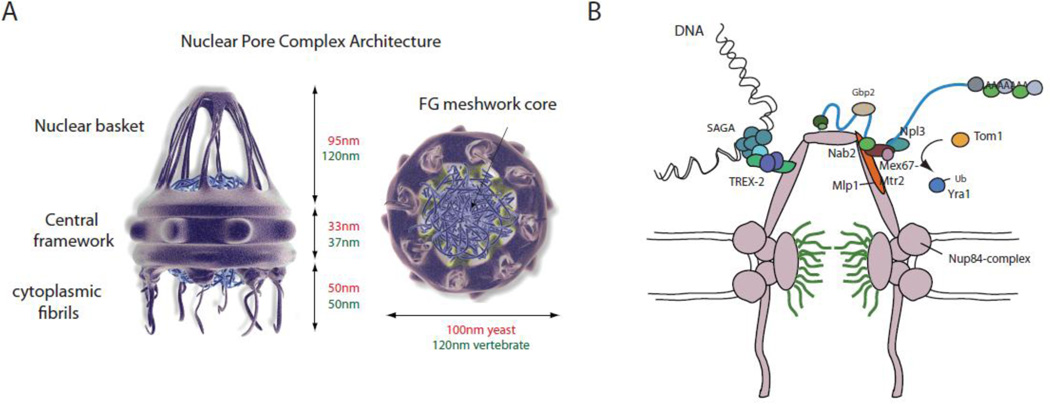
The primary function of the NPC is to mediate selective bidirectional transport between the nucleus and the cytoplasm [3]. To that effect it composes a large protein complex of ~60MDa embedded in the nuclear envelope (NE) that is evolutionarily conserved [87–90]. A detailed architectural map of the yeast NPC was recently determined using a computational approach that combined proteomic, biophysical and imaging data, which allowed each nuclear pore protein or nucleoporin (nup) to be assigned to particular substructures within the NPC [88,91]. The nuclear pore complex is a cylindrical structure comprised of eight spokes surrounding a central tube and can be divided into three sections: the nuclear face, the central channel and the cytoplasmic face (Figure 3A). The NPC is composed of ~30 different nups that can be placed in four classes: transmembrane, core scaffold (inner ring and outer ring), linker and Phe-Gly (FG) nups. In both yeast and vertebrates, three transmembrane nups span the pore membrane and constitute an outer transmembrane ring that anchors the NPC to the nuclear envelope. The outer and inner rings are formed by a dozen core scaffold nups, which together comprise the core scaffold of the NPC [92–94]. This scaffold encloses the central transport tube which is about ~35nm in diameter [95]. Anchored to the core scaffold are the largely unfolded FG nups. FG nups are characterized by regions of multiple Phe-Gly repeats and can be classified into two groups: symmetric (on both sides of the NPC) and asymmetric (either located closer to the nuclear or cytoplasmic side) [88–90]. They line the surface of the central tube from the nuclear to the cytoplasmic face and have a pivotal role in determining the mechanism of nuclear transport as they contain the docking sites for most cargo complexes and mediate nuclear trafficking [96–98]. In yeast, it has been shown that around 160 individual FG nups line the walls of the transport channel in each nuclear pore [88,89]. FG nups are anchored to the core scaffold by the linker nups.
Figure 3. Recruitment of mRNPs to the nuclear pore.
A. Schematic representation of the nuclear pore complex. The nuclear pore complex is a cylindrical structure comprised of eight spokes surrounding a central tube, a basket extending into the nucleus and cytoplasmic fibrils. The central transport tube (or channel) is filled with FG nups that interact with transport receptors and mediate the translocation trough the channel (see text for details). Dimensions of yeast (green) and vertebrate (red) NPC are shown. Illustrations reprint with permission from S. Patel (http://sspatel.googlepages.com/nuclearporecomplex) B. Cartoon showing NPC interactions relevant for mRNA export. Different genes in yeast were shown to be tethered to the NPC by the TREX-2 complex. Interaction of mRNPs with the NPC are mediated by the interaction of the basket protein Mlp1 with the RNA-binding protein Nab2. Before entering the pore, modification of Yra1 by the ubiquitin modifying enzyme Tom1 leads to the dissociation of Yra1 from the mRNP, one possible trigger to allow the mRNP access to the nuclear pore. An alternative access might be mediated by the Nup84 complex, forming the outer ring of the central structure of the NPC and directly interacting with the export receptor Mex67 (see text for details).
The NPC-associated peripheral structures consist of cytoplasmic filaments, the basket and a distal ring. Two FG nups are found on the cytoplasmic face of the NPC in yeast, Nup42 and Nup159 (three in metazoans, NLP1, NuP214 and NuP358) [3]. From these nups, eight filaments project into the cytoplasm to interface with the protein synthesis machinery and the cytoskeleton. These cytoplasmic filaments are thought to be formed from extended domains of Nup42 and Nup159, possible through their interaction with dynein light chain (Dyn2), as was recently shown [99]. Both Nup42 and Nup159 filaments contain numerous binding sites to interact with the Gle1–DEAD box protein 5 (Dbp5) RNA helicase complex during the final phases of mRNP export and the initiation of mRNA translation on ribosomes [100].
The structure of the nuclear basket
Due to the limited understanding of the basket’s composition it has been a matter of debate for some time. It was previously suggested that it might be composed of FG nups (NuP153 in humans, and Nup60 and Nup1 in S. cerevisiae), however, recent immuno-EM experiments have shown that its main molecular component is the highly conserved protein Tpr in human cells, myosin-like protein 1 (Mlp1) and Mlp2 in yeast and megator in Drosophila, all of which form long, filamentous, coiled-coil dimers [3,101–104]. The current view of the basket is that of eight protein filaments protruding ~60–80nm from the nuclear face of the NPC into the nucleoplasm and converging in a distal ring structure. The basket is anchored to the nuclear pore by the nucleoporins Nup60 and it is believed to have an important role as recruitment and docking site of the NPC [3].
The basket: docking or quality control platform? Or both?
Over the past years evidence has accumulated that places the basket at the center of post-transcriptional processes, in particular mRNA surveillance, which prevents defective mRNAs, such as unspliced or partially spliced polyadenylated RNAs, from reaching the cytoplasm [105] (Figure 3B). Although the composition of an mRNP arriving at the NPC is not completely clear, in yeast it is believed that it contains at least the RNA binding proteins Nab2, Npl3, Yra1, Gbp2, Hrb1, the cap binding proteins Cbc20 and Cbc80, the poly(A) binding protein Pab1, the ATP-dependent RNA helicases Sub2 and Dbp5, and the mRNA export receptor Mex67 with its binding partner Mtr2 [2]. While transcript specificity has been previously suggested for some of these factors (e.g. Npl3, Nab2) recent data showed that at least Nab2 is bound to most, if not all, mRNAs in the nucleus [106–108]. Whether this is true for all of these proteins still remains to be clarified. Further transcript-specific proteins might be bound as well at this stage, but are not discussed here. Mex67’s interaction with the mRNA is mediated by its binding to Yra1, Nab2 and Npl3 [106,109–112]. However, the first interaction of the mRNP with the nuclear pore is thought not to be mediated by the export receptor, but by Nab2 through binding directly to the C-terminal region of Mlp1, which is believed to act as the docking site of the mRNPs at the basket [113]. Surprisingly, Nab2 but not Mlp1 is essential for mRNA export, as deletion of MLP1 does not result in an mRNA export phenotype. However, over-expression of the C-terminal region of Mlp1, which is required for interaction with Nab2 and hence the mRNP, leads to the accumulation of poly(A)RNA in the nucleus [113]. This C-terminal fragment does not localize to the nuclear periphery but is distributed throughout the nucleoplasm, suggesting that it might cause an mRNA export defect by sequestering mRNPs away from the pore, consistent with a role for Mlp1 in recruiting mRNPs to the pore.
While not affecting export of mRNPs, deletion of Mlp1 surprisingly results in the leakage of unspliced mRNAs and other aberrant mRNAs to the cytoplasm [36,114]. This observation led to the suggestion that the role of Mlps, and in particular Mpl1, might not just be to provide access to the pore but rather to act as quality control retention filter, a ‘gatekeeper’, ensuring that only fully mature, ‘quality-controlled’ mRNPs can enter the nuclear pore. So what could be the final quality control step before an mRNP can pass the ‘gatekeeper’ and proceed on into the nuclear pore? One possible scenario could be a structural remodeling step that facilitates the addition or removal of specific proteins, or interactions. Supporting the existence of such an event, Stutz and coworkers recently showed that ubiquitination of Yra1 by the E3 ligase Tom1 at the pore promotes its dissociation from mRNPs prior to export [112]. Deletion of TOM1 was previously shown to result in the nuclear retention of poly(A) RNAs and their localization within nuclear foci together with Nab2 [106]. Taken together this suggests that the removal of Yra1 might be an important event in defining an export-competent mRNP
Thus, Mlp proteins in their function as ‘gatekeepers’ might introduce a rate-limiting step into the export process that could interfere with rapid export of mRNAs. Interestingly, under heat-shock conditions, where rapid production of heat-shock proteins is required, Mlps were shown to accumulate in nucleoplasmic foci away from the pore, together with Yra1 and Nab2 [115]. Under these conditions, heat-shock, but not other mRNAs, are exported to the cytoplasm. Simultaneously, upon heat–shock, Nab2 is phosphorylated by the mitogen-activated protein (MAP) kinase Slt2/Mpk1 [115]. This modification was shown to increase the amount of Nab2 in complex with Mlp1 while at the same time reducing the amount of Nab2 associated with the NPC, suggesting that under these conditions mRNP complexes together with Mlps get sequestered into the nuclear foci. These observations highlight that the cell has the ability to regulate mRNA export by allowing only a specific set of mRNAs to be exported. However, to achieve this ‘export specificity’ it might sacrifice a quality control step usually occurring at the basket. As heat-shock is a harsh environmental stress situation for the cell, loosing a quality control step might be an acceptable tradeoff in favor of assuring fast export and the subsequent production of heat-shock proteins. However, without the Mlps as nuclear pore docking sites, alternative recruitment or entry sites have to exist at the NPC.
One possibility is that mRNPs could find their way to the central channel simply by diffusion where they would then bind to FG nucleoporins through the export receptor Mex67 and its direct interaction with FG nups [116]. Alternatively, Mex67 has also been shown to bind to Nup85, a component of the Nup84 complex that forms the outer ring of the NPC and disruption of the interaction resulted in an mRNA export defect [117]. Therefore, it is possible that the Nup84 complex serves as an alternate docking site for mRNPs. Consistent with this idea, EM probing of poly(A) RNA showed that mRNAs are found surrounding the NPC, not just associated with the distal site of the basket [118]. The distance between the basket spokes is approximately 20–25nm. EM data for Nab2-purified mRNPs indicated an average length of 20–30nm and a width of 5–7nm, suggesting that, in principle, mRNPs would be able to pass between the basket spokes, whereas larger mRNAs such as the Balbiani ring (BR) mRNP, with a diameter of around 50nm, would not [108]. Such a scenario, and whether this would or could occur only under certain circumstances, remains to be explored. It is interesting to speculate, however, whether mRNPs entering the pore through an alternative docking site would still undergo Mlp1-dependent – or any other kind - of quality control.
6. Out of the nucleoplasm into the pore: Translocation of mRNPs
Translocation kinetics: the principle
The actual transport through the pore is likely one of the shortest episodes in the lifetime of an mRNA. This makes studying this process a rather difficult venture as catching single translocation events is difficult to accomplish, in particularly in an in vivo environment. Nevertheless, in principle, the nuclear pore is likely to be a crowded environment. In mammalian cells, it has been estimated that 500–1000 molecules cross each nuclear pore per second, therefore the translocation of molecules through the pore has to be fast [119,120]. Single molecule measurements of import kinetics of protein reporters indeed showed that individual proteins cross the NPC within a few milliseconds [121]. However, proteins are much smaller than mRNPs, which, together with pre-ribosomal particles, are by far the largest molecules to cross this channel. The central channel’s maximum diameter is ~35 nm, with the disordered FG nups lining the channel reducing that size to only around 10nm [88]. Considering the typical size of an mRNP of ~5nm in diameter and 20–30nm in length, such a particle could easily fill up a significant space within the central channel, suggesting that slow transport would strongly reduce the ability of a pore to transport other molecules. It is therefore reasonable to assume that translocation of mRNPs through the pore is also a fast process.
On the other hand, mRNP transport rates for an individual pore might actually not be that high. The number of mRNAs crossing each nuclear pore in yeast can be calculated relatively easily by taking into account i) the total mRNA levels, ii) their half-lives, and iii) the number of pores in a cell. Different studies have estimated the number of mRNAs in an exponentially growing yeast cell to be between 15,000–40,000mRNAs/cell [78,122]. Using the higher estimate of 40,000mRNAs/cell and considering an average half-life of ~20min per RNA, this would suggest that yeast synthesizes about 1000 mRNAs per minute. With approximately 90 pores/nucleus, every pore has to transport just about 10 mRNAs per minute; a small number compared to the total number of proteins that cross the NPC. However, we also have to take into consideration that in yeast the pores adjacent to the nucleolus are most likely not used for mRNA export, which would increase the number of mRNPs to be exported per pore to ~16, if we assume that approximately one third of nuclear pores are 'nucleolar'. Pre-ribosomal subunits, the second large complexes are a slightly higher burden for the NPC to transport; it has been estimated that ~25 pre-ribosomes cross each NPC per minute [123]. Taken together, this suggests that just to allow all mRNAs to be exported, the transport would not necessarily have to be very fast, even if only a single mRNA can pass a single NPC at the time. These numbers are only estimates and might vary, as it is not known if mRNAs and pre-ribosomes travers the same pores. Specialization of pores has so far only been suggested in HL-60 cells, where, using immunogold labeling, NFT2 and poly(A) mRNAs were observed to use different sets of pores [124]. However, since NFT2 was shown to label all pores in HeLa nuclei, it remains unclear if this phenomenon is cell-type specific or if it exists in other organisms.
Translocation of mRNPs: the practice
The first images of mRNPs crossing the pore were static and came from electron microscopy studies. Although earlier EM studies in the 1960s and 70s observing electron dense material within nuclear pores suggested to show translocating mRNPs, the most convincing data came from studies of the large Balbiani ring (BR) mRNPs [5,125,126]. These large, 35–40kB RNAs fold into 50nm particles, easily visible by EM. Snapshots of BR mRNPs at different steps of their translocation through an NPC showed that these mRNPs are being rearranged after arriving at the basket, changing from a globular to a ribbon-like structure and entering the NPC 5’ end first. The mRNA was then shown to translocate as a linear molecule and to directly associate with ribosomes once it had reached the cytoplasm [126].
Using transmission electron microscopy (TEM) Kiseleva and coworkers obtained a higher resolution image and could visualize an mRNP entering the NPC by first binding to the distal portion of the basket (outer/terminal ring). The outer ring was shown to expand upon binding allowing access to the NPC and resulting in translocation into and through the pore [127]. After the mRNP had passed, the terminal ring closed. The flexible nature of the ring and the basket makes it a prime candidate for having a ‘gatekeeper’ function that regulates entry to the nuclear pore. As discussed above, the C-terminal region of Mlp/TRP has been localized to this region, which binds directly to most mRNPs and is believed to act as a site of first contact and final quality control (see above).
Overall these data suggest that the BR mRNPs undergo significant rearrangements that facilitate their entry into the pore, as their huge size does not allow them to enter otherwise. However, we have to consider that BR mRNPs are much larger than most common mRNPs; the average length of a yeast mRNA is only about 2kb, and nuclear mRNPs purified from yeast cells show a rod like shape with a length of 20–30nm and a width of 5–7nm [108]. Although mRNPs used in recent single molecule studies were much smaller than the BR RNPs, Mor et al nevertheless, showed that these mRNPs changed their structure at the NPC prior to export [54]. mRNPs displayed a spherical structure before export, whereas after export, on the cytoplasmic side, they presented a disorganized open structure. This was accompanied by a decrease in fluorescence signal on the mRNP, suggestive of restructuring and unfolding during export, which possibly included some of the MS2 stem loop structures. Soon after the translocation, mRNPs regained their rounded structure. Although it is not clear whether such rearrangements occur on, or are a requirement, for all mRNPs to fit through the pore, it is known that mRNA binding proteins are stripped off the mRNP before it translocates to the cytoplasm. It is therefore likely that all mRNPs are restructured in some way before they get exported. However, it will be interesting too see if this is a general phenomenon for all mRNPs that cross the NPC.
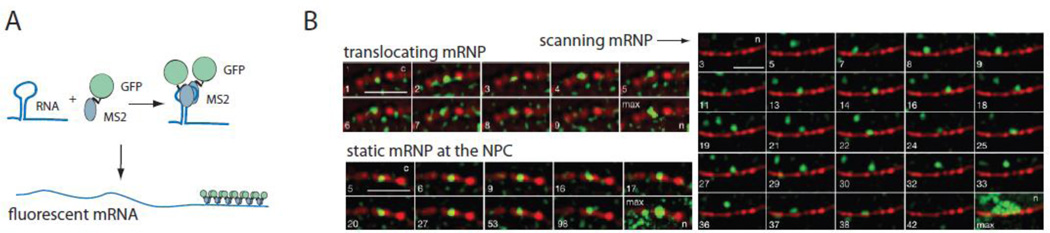
Only very recently, single molecule studies finally allowed the observation of actual translocation events in real time [1,54]. By creating fluorescent RNAs in vivo using the MS2 bacteriophage system, individual mRNAs were followed in real time using high-resolution fluorescent microscopy (Figure 4). These studies revealed that mRNPs are indeed exported rather quickly, albeit still slower than proteins, and tracking different sized mRNAs Mor and colleagues obtained an average translocation time of about 0.5 seconds [54]. Using a more sophisticated imaging setup that allowed faster acquisition rates as well as the tracking of mRNPs localized at the different regions of the NPC (basket-central, channel and the cytoplasmic fibrils), Grünwald and Singer showed that the mouse beta-actin mRNA requires on average only 180ms from docking to the basket to its release into the cytoplasm. Furthermore, they showed that export happens in three steps which occurred at different timescales; docking of the mRNA which lasts ~80 ms, transport through the central channel (5–20 ms), and release of the mRNA which takes another 80 ms. This observation mirrors previous studies monitoring protein transport rates that also showed that docking and release, but not translocation are the rate limiting steps during transport. Interestingly, both studies also observed mRNPs that seem to be ‘stuck’ at pores, sometimes for quite a considerable amount of time. This makes it is interesting to speculate whether these mRNPs might be held at the pore by a quality control machinery, which is preventing their access.
Figure 4. Real-time observation of scanning, docking and translocation of an individual mRNP.
A. Description of the MS2 system, the most commonly used method for single molecule mRNA detection for live cell imaging. The system uses a bacteriophage RNA binding protein fused to GFP that binds a specific RNA stem loop structure with high affinity. Adding multiple binding sites for to an mRNA sequence allows single mRNA detection. B. Observation of mRNA export in real time. Images show single beta-actin mRNAs at different stages of mRNA export (from Grünwald and Singer, 2010 [1]). Single mRNAs are observed scanning the nuclear periphery, static at the NPC or translocating to the cytoplasm. Images were acquired at frame rates of 20ms per frame. Frames are shown in the upper left corner of each image. ‘Max’ shows the sum of all previous frames. Nuclear pore complexes are labeled in red, beta-actin mRNAs in green. N=nucleus, c=cytoplasm. Reprint with permission of Nature Publishing Group.
Once in the central channel, mRNPs do not translocate unidirectional to the other site of the NPC in a single step, but move bi-directionally within the channel [1]. This is consistent with current models that the central channel does not have any directionality, but that directionality is obtained by binding events on either site of it [128]. The current view of the role of the central channel is to form a dynamic network of filaments that block translocation of inert molecules above a certain size threshold. This barrier is overcome by the binding of transport receptors, which in turn bind to the FG nups located within the channel (Mex67/NFX1 for mRNA export) and so allow movement of cargos into and through it.
7. On the other side: Release of mRNPs into the cytoplasm
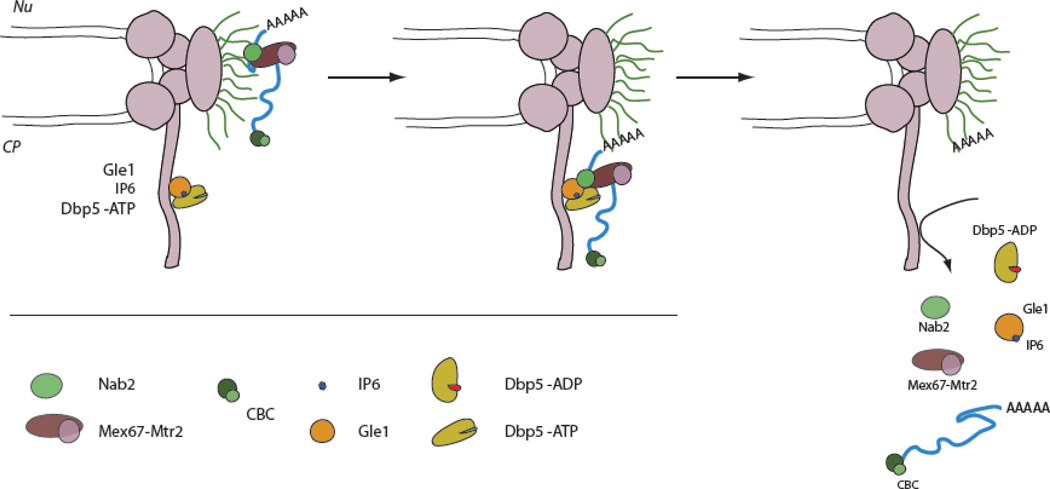
As the central channel itself does not provide directionality, as far as release of mRNPs after export, directionality is achieved by factors located at the cytoplasmic site of the NPC, whose role is to ‘trap’ the mRNP at the cytoplasmic side, rearrange it and release the mRNP into the cytoplasm (e.i. by stripping of mRNA export factors). Different NPC components and NPC-associated factors located at the cytoplasmic side of the NPC were already know to be required for mRNA export, but only recently has a detailed mechanism emerged with the shuttling RNA-dependent ATPase Dbp5 playing a crucial role in this process [100,129–134]. Studies from different groups showed that when Dbp5-bound mRNPs reach the cytoplasmic site of the pore, binding of Dbp5 to the nuclear pore protein Gle1 and its cofactor IP6 activates the ATPase function of Dbp5 and stimulates a conformational change in the protein (Figure 5). This activity is able to induce mRNP remodeling, resulting in the dissociation of the export receptor heterodimer Mex67-Mtr2 and the RNA binding protein Nab2 from the mRNA, and the release of the mRNA into the cytoplasm (Figure 5). Dbp5 has ATP-dependent helicase activity in vitro and it is possible that Dbp5 utilizes ATP binding or hydrolysis to facilitate duplex unwinding, what in turn could participate in the release of proteins from the mRNA. Consistent with this idea, in vitro experiments showed that Dbp5 does not interact directly with Nab2 or displaces it from poly(A)RNA, but most likely inhibits its re-association by restructuring the free mRNA in such a way that it is incompatible with Nab2 binding [135]. However, the exact mechanism of how Mex67–Mtr2 and Nab2 are released is not yet understood.
Figure 5. mRNP release.
The ATP dependent RNA helicase/ATPase Dbp5 plays a crucial role in the removal of Nab2 and Mex67-Mtr2 from the poly(A)mRNAs and the release of mRNAs into the cytoplasm. When the mRNP reaches the cytoplasmic site of the nuclear pore, the formation of a Gle1-IP6-Dbp5-RNA complex induces the ATPase activity of Dbp5. This leads to a conformational change in Dbp5 and ATP hydrolysis, inducing the dissociation of Mex67 and Nab2 from the mRNP and subsequent release of the mRNA into the cytoplasm (see text for details). Nu=nucleus, CP= cytoplasm.
To be ready for another cycle of ATP hydrolysis and mRNP remodeling, Dbp5p has to release ADP and rebind ATP. ADP-release is triggered by binding to the nucleoporin Nup159, which is located at the cytoplasmic side of the NPC and acts as an ADP release factor. Binding of Nup159 probably occurs after Dbp5 has released the remodeled mRNP, as binding of Dbp5 to RNA and Nup159 are mutually exclusive due to them occupying the same binding site on Dbp5 [132,136,137]. Although data suggests that binding of Dbp5 to mRNAs occurs already in the nucleus, it is also possible that Dbp5 can remodel mRNPs at the cytoplasmic face of the NPC, independently of being loaded onto the mRNP in the nucleus, by binding to mRNPs/mRNAs when they exit the central channel. Fluorescent-recovery-after-photobleaching (FRAP) experiments showed that signal of fluorescently labeled Dbp5 at the NPC recovers very fast (~1 second) [134]. Such fast rebinding of a large pool of Dbp5 to the pore is unlikely to be mediated by a flow of new nuclear mRNPs trough the NPC, as only about 10–15 mRNAs pass through a pore every second (see above). Therefore it is likely that at least a fraction of Dbp5 ‘sits’ at the exit of the pore/central channel, ready to rearrange mRNPs as they come through.
Besides Dbp5, two other proteins are suggested to participate in the release of mRNPs at the cytoplasmic site of the nuclear pore, whose function, however, is not yet understood. One is the nucleoporin Nup42, which interacts directly with Gle1 and may act as an additional anchor for Gle1 at the pore [138]. Interestingly, Nup42 is specifically required for the export of heat-shock mRNAs and might therefore have a specific role in mRNA release under these conditions [139]. The second protein is Gfd1, which is also found in a complex with Gle1 and, in addition, interacts directly with Nab2 [138,140]. Neither Nup42 nor Gfd1 are essential for viability or mRNA export and their role in mRNP remodeling and release still has to be determined.
One interesting question remaining is how precisely Dbp5 remodels an mRNP and how its activity is regulated such that only nuclear factors but not RNA binding proteins required in the cytoplasm are striped off the mRNA. Little is known about where mRNA binding proteins bind on the mRNA, not for Mex67p-Mtr2p and not for the more general RNA binding proteins such as Nab2p or Npl3p. Defining the composition of mRNPs and the binding sites of these proteins on the mRNA will be essential for our understanding of how the rearrangements occurring on mRNPs along the gene expression pathway affect and allow for coordinated and efficient export of these large macromolecules from their site of synthesis to the cytoplasm.
8. Conclusions and outlook
In the recent years, we have obtained a much clearer picture on how cells achieve the export of mRNA. It has become evident that at least three critical steps define the kinetic of the export process, two of them directed and tightly regulated, with the third being much more passive and ‘random’. The regulated steps are comprised of i) the co-transcriptional events which are required for linking transcription, 3’ processing and formation of an export competent mRNP, and ii) the mRNP binding and gaining access to a nuclear pore; both these steps are monitored by distinct quality control machineries. Whatever happens in between, however, seems to occur in a rather random manner, as mRNPs simply diffuse throughout the nucleoplasm undirected, to find a nuclear pore. The randomness of this event might be surprising considering how tightly controlled the processing and assembly of export-competent mRNPs is otherwise. Nevertheless, the occurrence of random movement in cells is quite common and often considered effective and energy-efficient. Still, it will be interesting to see whether the tethering of specific genes in yeast is a mechanism developed by the cell to ensure faster export kinetics in ‘times of need’ and whether a similar mechanism also exists in higher eukaryotes.
Carrying out regulatory and quality control steps on fixed cellular structures is likely to be more efficient than when components freely diffuse within the nucleoplasm. In cellular terms, transcription is a very slow process and the nascent mRNA has plenty of time to correctly fold and assemble into an mRNP, the Pol II transcription machinery thereby acting as a scaffold for mRNP formation and quality control. The Mlps at the nuclear basket might similarly act as a scaffold where mRNPs are checked and rearranged to allow them entry into the pore. Both these events might not only be required to allow for efficient export of mRNAs but also to ensure that mRNPs, which are not supposed to leave the nucleus, are retained. RNA Pol II transcribes many different kinds of RNAs besides mRNAs, and at least some of them are capped and polyadenylated, including some long non-coding RNAs (lncRNAs) [141]. Some of these lncRNAs are retained in the nucleus, despite undergoing normal 3’ end processing, and are likely to have a very similar composition than an export-competent mRNP. Understanding how the assembly of different Pol II-transcribed mRNPs is achieved will help to understand the mechanistic details of mRNA export as well as shed light onto the biogenesis of other classes of RNPs. Determining the composition of mRNPs will therefore be instrumental in understanding the export process. Very little is know about the composition of mRNPs; we know the identity of many proteins bound to mRNPs, however, for most of the mRNA binding proteins implicated in mRNA export, we do not know how many copies of them are present on the RNA nor do we know where on the mRNA they are bound. Along the same lines, we do not understand how mRNAs are folded and if RNA structure is important in obtaining export competence and/or access to the NPC. Recent technical developments in using cross-linking approaches to map protein binding sites on RNAs as well as high throughput methods to determine RNA structure will be instrumental in understanding what mRNPs actually look like and how changes in their composition and structure may be instrumental in regulating mRNA export [142–145].
Finally, single molecule microscopy approaches are likely to play an important role in determining how mRNA export is executed within a cell, being the only tool that allows following individual mRNAs in real time. Currently, it is still a challenge to track mRNAs through the entire nuclear space but fast developments of imaging technologies are likely to overcome these limitations at some point. In addition, super-resolution imaging approaches will allow an even more detailed description of mRNPs docking to the NPC and during translocation, enabling the dissection of the mechanistic steps occurring during these processes [1,146,147]. Having identified most factors involved in mRNA export, using this knowledge to combine protein knockdowns in higher eukaryotes or mutant strains in yeast with single molecule imaging will open up an interesting new chapter in studying this rather short time span in the live of an mRNA.
Highlights.
-
>
The co-transcriptional recruitment of mRNA maturation factors is linked to export.
-
>
mRNPs have to undergo surveillance after transcription and before entry to the pore.
-
>
mRNA transport rates per pore in yeast are about 16mRNPs per minute
-
>
Docking and release are the rate-limiting steps of mRNA export through the pore
-
>
Translocation trough the nuclear pore occurs in tens of milliseconds
Acknowledgments
M.O. holds a CIHR New Investigator Award and a FRSQ Chercheur Boursier Junior I. M. O. is supported by funding from the CIHR, NSERC, FRSQ, NHI (U54 022220) and CFI. D.Z. holds a FRSQ Chercheur Boursier Junior I. D. Z. is supported by funding from the CIHR, NSERC and CFI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grünwald D, Singer RH. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tutucci E, Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nat Rev Mol Cell Biol. 2011;12:377–384. doi: 10.1038/nrm3119. [DOI] [PubMed] [Google Scholar]
- 3.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 4.Stewart M. Nuclear export of mRNA. Trends Biochem Sci. 2010;35:609–617. doi: 10.1016/j.tibs.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Stevens BJ, Swift H. RNA transport from nucleus to cytoplasm in Chironomus salivary glands. J Cell Biol. 1966;31:55–77. doi: 10.1083/jcb.31.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mor A, Shav-Tal Y. Dynamics and kinetics of nucleo-cytoplasmic mRNA export. WIREs RNA. 2010;1:388–401. doi: 10.1002/wrna.41. [DOI] [PubMed] [Google Scholar]
- 7.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid M, Jensen TH. Nuclear quality control of RNA polymerase II transcripts. WIREs RNA. 2010 doi: 10.1002/wrna.24. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 9.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends in Genetics. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Buratowski S. Progression through the RNA Polymerase II CTD Cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson SA, Cubberley G, Bentley DL. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3' end processing factor Pcf11. Mol Cell. 2009;33:215–226. doi: 10.1016/j.molcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson SA, Kim H, Erickson B, Bentley DL. The export factor Yra1 modulates mRNA 3′ end processing. Nat Struct Mol Biol. 2011;18:1164–1171. doi: 10.1038/nsmb.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenklusen D, Vinciguerra P, Wyss J-C, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodríguez-Navarro S, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 15.Chavez S, Beilharz T, Rondon A, Erdjument-Bromage H, Tempst P, Svejstrup J, et al. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. Embo J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rondón AG, Jimeno S, García-Rubio M, Aguilera A. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J Biol Chem. 2003;278:39037–39043. doi: 10.1074/jbc.M305718200. [DOI] [PubMed] [Google Scholar]
- 17.Mason P, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 20.Gómez-González B, García-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marín A, et al. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. Embo J. 2011;30:3106–3119. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. Embo J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopytova DV, Orlova AV, Krasnov AN, Gurskiy DY, Nikolenko JV, Nabirochkina EN, et al. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev. 2010;24:86–96. doi: 10.1101/gad.550010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, et al. THO/Sub2p functions to coordinate 3'-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Saguez C, Schmid M, Olesen JR, Ghazy MAE-H, Qu X, Poulsen MB, et al. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol Cell. 2008;31:91–103. doi: 10.1016/j.molcel.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 25.Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3' end formation by RNA polymerase II. Embo J. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bucheli ME, He X, Kaplan CD, Moore CL, Buratowski S. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. Rna. 2007;13:1756–1764. doi: 10.1261/rna.607207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- 28.Siebel CW, Guthrie C. The essential yeast RNA binding protein Np13p is methylated. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13641–13646. doi: 10.1073/pnas.93.24.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu MC. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 2004;18:2024–2035. doi: 10.1101/gad.1223204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dieppois G, Iglesias N, Stutz F. Cotranscriptional Recruitment to the mRNA Export Receptor Mex67p Contributes to Nuclear Pore Anchoring of Activated Genes. Mol Cell Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hobeika M, Brockmann C, Iglesias N, Gwizdek C, Neuhaus D, Stutz F, et al. Coordination of Hpr1 and ubiquitin binding by the UBA domain of the mRNA export factor Mex67. Molecular Biology of the Cell. 2007;18:2561–2568. doi: 10.1091/mbc.E07-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, et al. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16376–16381. doi: 10.1073/pnas.0607941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmid M, Jensen TH. Quality control of mRNP in the nucleus. Chromosoma. 2008;117:419–429. doi: 10.1007/s00412-008-0166-4. [DOI] [PubMed] [Google Scholar]
- 34.Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, et al. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. Embo J. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3'-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 36.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- 37.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 39.Hessle V, Björk P, Sokolowski M, de Valdivia EG, Silverstein R, Artemenko K, et al. The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol Biol Cell. 2009;20:3459–3470. doi: 10.1091/mbc.E09-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu X, Lykke-Andersen S, Nasser T, Saguez C, Bertrand E, Jensen TH. Assembly of an export-competent mRNP is needed for efficient release of the 3'-end processing complex after polyadenylation. Mol Cell Biol. 2009;29:5327–5338. doi: 10.1128/MCB.00468-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng H, Dufu K, Lee C-S, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5' end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 42.Dias AP, Dufu K, Lei H, Reed R. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Comms. 2010;1:97. doi: 10.1038/ncomms1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masuda S. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proceedings of the National Academy of Sciences. 2008;105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Politz JC, Tuft RA, Pederson T, Singer RH. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr Biol. 1999;9:285–291. doi: 10.1016/s0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- 46.Politz JCR, Tuft RA, Prasanth KV, Baudendistel N, Fogarty KE, Lifshitz LM, et al. Rapid, diffusional shuttling of poly(A) RNA between nuclear speckles and the nucleoplasm. Molecular Biology of the Cell. 2006;17:1239–1249. doi: 10.1091/mbc.E05-10-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh OP, Bjorkroth B, Masich S, Wieslander L, Daneholt B. The intranuclear movement of Balbiani ring premessenger ribonucleoprotein particles. Exp Cell Res. 1999;251:135–146. doi: 10.1006/excr.1999.4490. [DOI] [PubMed] [Google Scholar]
- 48.Calapez A, Pereira HM, Calado A, Braga J, Rino J, Carvalho C, et al. The intranuclear mobility of messenger RNA binding proteins is ATP dependent and temperature sensitive. J Cell Biol. 2002;159:795–805. doi: 10.1083/jcb.200203046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molenaar C, Abdulle A, Gena A, Tanke HJ, Dirks RW. Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J Cell Biol. 2004;165:191–202. doi: 10.1083/jcb.200310139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vargas DY, Raj A, Marras SAE, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shav-Tal Y. Dynamics of Single mRNPs in Nuclei of Living Cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veith R, Sorkalla T, Baumgart E, Anzt J, Häberlein H, Tyagi S, et al. Balbiani Ring mRNPs Diffuse through and Bind to Clusters of Large Intranuclear Molecular Structures. Biophys J. 2010;99:2676–2685. doi: 10.1016/j.bpj.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siebrasse JP, Veith R, Dobay A, Leonhardt H, Daneholt B, Kubitscheck U. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proceedings of the National Academy of Sciences. 2008;105:20291–20296. doi: 10.1073/pnas.0810692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mor A, Mor A, Suliman S, Suliman S, Ben-Yishay R, Ben-Yishay R, et al. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 55.Braga J, Rino J, Carmo-Fonseca M. Photobleaching microscopy reveals the dynamics of mRNA-binding proteins inside live cell nuclei. Prog. Mol. Subcell. Biol. 2004;35:119–134. doi: 10.1007/978-3-540-74266-1_6. [DOI] [PubMed] [Google Scholar]
- 56.Janicki S, Tsukamoto T, Salghetti S, Tansey W, Sachidanandam R, Prasanth K, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winey M, Yarar D, Giddings T, Mastronarde D. Nuclear Pore Complex Number and Distribution throughout the Saccharomyces cerevisiae Cell Cycle by Three-Dimensional Reconstruction from Electron Micrographs of Nuclear Envelopes. Mol Biol Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maul GG, Price JW, Lieberman MW. Formation and distribution of nuclear pore complexes in interphase. J Cell Biol. 1971;51:405–418. doi: 10.1083/jcb.51.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blobel G. Gene gating: a hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arib G, Akhtar A. Multiple facets of nuclear periphery in gene expression control. Curr Opin Cell Biol. 2011;23:346–353. doi: 10.1016/j.ceb.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 63.Ishii K, Arib G, Lin C, van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 64.Krull S, Ouml Rries JD, Boysen BOR, Reidenbach S, Magnius L, Norder H, et al. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. The EMBO Journal. 2010;29:1659. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Casolari J, Brown C, Komili S, West J, Hieronymus H, Silver P. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 66.Cabal G, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 67.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 68.Taddei A, van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 69.Brickner J, Walter P. Gene Recruitment of the Activated INO1 Locus to the Nuclear Membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, et al. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- 73.Berger A, Cabal G, Fabre E, Duong T, Buc H, Nehrbass U, et al. High-resolution statistical mapping reveals gene territories in live yeast. Nat Meth. 2008;5:1031–1037. doi: 10.1038/nmeth.1266. [DOI] [PubMed] [Google Scholar]
- 74.Taddei A, Schober H, Gasser SM. The budding yeast nucleus. Cold Spring Harb Perspect Biol. 2010;2:a000612. doi: 10.1101/cshperspect.a000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodriguez-Navarro S, Fischer T, Luo M, Antunez O, Brettschneider S, Lechner J, et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 76.Rodríguez-Navarro S, Rodríguez-Navarro S, Hurt E, Hurt E. Linking gene regulation to mRNA production and export. Curr Opin Cell Biol. 2011;23:302–309. doi: 10.1016/j.ceb.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 77.Jani D, Lutz S, Marshall NJ, Fischer T, Köhler A, Ellisdon AM, et al. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol Cell. 2009;33:727–737. doi: 10.1016/j.molcel.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 79.Kurshakova M, Krasnov A, Kopytova D, Shidlovskii Y, Nikolenko J, Nabirochkina E, et al. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC FREE. The EMBO Journal. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wickramasinghe VO, McMurtrie PIA, Mills AD, Takei Y, Penrhyn-Lowe S, Amagase Y, et al. mRNA export from mammalian cell nuclei is dependent on GANP. Curr Biol. 2010;20:25–31. doi: 10.1016/j.cub.2009.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nature Reviews Genetics. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 82.Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- 83.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 84.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brickner D, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee P-C, Widom J, et al. H2A.Z-Mediated Localization of Genes at the Nuclear Periphery Confers Epigenetic Memory of Previous Transcriptional State. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, et al. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 87.DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, et al. Evidence for a Shared Nuclear Pore Complex Architecture That Is Conserved from the Last Common Eukaryotic Ancestor. Mol Cell Proteomics. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 89.Rout M, Aitchison J, Suprapto A, Hjertaas K, Zhao Y, Chait B. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 92.Lutzmann M. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. The EMBO Journal. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.D'Angelo MA. Nuclear Pores Form de Novo from Both Sides of the Nuclear Envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- 94.Makio T, Stanton LH, Lin C-C, Goldfarb DS, Weis K, Wozniak RW. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J Cell Biol. 2009;185:459–473. doi: 10.1083/jcb.200810029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Elad N, Maimon T, Frenkiel-Krispin D, Lim RYH, Medalia O. Structural analysis of the nuclear pore complex by integrated approaches. Curr Opin Struct Biol. 2009;19:226–232. doi: 10.1016/j.sbi.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 96.Terry LJ, Wente SR. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J Cell Biol. 2007;178:1121–1132. doi: 10.1083/jcb.200704174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 98.Bayliss R, Littlewood T, Stewart M. Structural Basis for the Interaction between FxFG Nucleoporin Repeats and Importin-β in Nuclear Trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 99.Stelter P, Kunze R, Flemming D, Höpfner D, Diepholz M, Philippsen P, et al. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat Cell Biol. 2007;9:788–796. doi: 10.1038/ncb1604. [DOI] [PubMed] [Google Scholar]
- 100.Ledoux S, Guthrie C. Regulation of the Dbp5 ATPase cycle in mRNP remodeling at the nuclear pore: a lively new paradigm for DEAD-box proteins. Genes Dev. 2011;25:1109–1114. doi: 10.1101/gad.2062611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qi H, Rath U, Wang D, Xu Y-Z, Ding Y, Zhang W, et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Molecular Biology of the Cell. 2004;15:4854–4865. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Krull S, Thyberg J, Björkroth B, Rackwitz H-R, Cordes VC. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Molecular Biology of the Cell. 2004;15:4261–4277. doi: 10.1091/mbc.E04-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fasken MB, Corbett AH. Mechanisms of nuclear mRNA quality control. RNA Biol. 2009;6:237–241. doi: 10.4161/rna.6.3.8330. [DOI] [PubMed] [Google Scholar]
- 106.Duncan K, Umen J. ScienceDirect - Current Biology : A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Current Biology. 2000 doi: 10.1016/s0960-9822(00)00527-3. [DOI] [PubMed] [Google Scholar]
- 107.Kim Guisbert K, Duncan K, Li H, Guthrie C. Functional specificity of shuttling hnRNPs revealed by genome-wide analysis of their RNA binding profiles. Rna. 2005;11:383–393. doi: 10.1261/rna.7234205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Batisse J, Batisse C, Budd A, Böttcher B, Hurt E. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J Biol Chem. 2009;284:34911–34917. doi: 10.1074/jbc.M109.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zenklusen D, Vinciguerra P, Strahm Y, Stutz F. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol. 2001;21:4219–4232. doi: 10.1128/MCB.21.13.4219-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stutz F, Bachi A, Doerks T, Braun I, Seraphin B, Wilm M, et al. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. Rna. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. Embo J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, et al. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010;24:1927–1938. doi: 10.1101/gad.583310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Green DM, Johnson CP, Hagan H, Corbett AH. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1010–1015. doi: 10.1073/pnas.0336594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. Embo J. 2005;24:813–823. doi: 10.1038/sj.emboj.7600527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carmody SR, Tran EJ, Apponi LH, Corbett AH, Wente SR. The Mitogen-Activated Protein Kinase Slt2 Regulates Nuclear Retention of Non-Heat Shock mRNAs during Heat Shock-Induced Stress. Mol Cell Biol. 2010;30:5168–5179. doi: 10.1128/MCB.00735-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Strasser K, Bassler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yao W, Lutzmann M, Hurt E. A versatile interaction platform on the Mex67-Mtr2 receptor creates an overlap between mRNA and ribosome export. Embo J. 2008;27:6–16. doi: 10.1038/sj.emboj.7601947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang S, Deerinck TJ, Ellisman MH, Spector DL. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ribbeck K, Görlich D. Kinetic analysis of translocation through nuclear pore complexes. Embo J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Smith AE. Systems Analysis of Ran Transport. Science. 2002;295:488–491. doi: 10.1126/science.1064732. [DOI] [PubMed] [Google Scholar]
- 121.Mincer JS, Simon SM. Simulations of nuclear pore transport yield mechanistic insights and quantitative predictions. Proceedings of the National Academy of Sciences. 2011;108:E351–E358. doi: 10.1073/pnas.1104521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Warner J. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 124.Iborra FJ, Jackson DA, Cook PR. The path of RNA through nuclear pores: apparent entry from the sides into specialized pores. J Cell Sci. 2000;113(Pt 2):291–302. doi: 10.1242/jcs.113.2.291. [DOI] [PubMed] [Google Scholar]
- 125.Mehlin H, Daneholt B, Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 126.Daneholt B. Assembly and transport of a premessenger RNP particle. Proceedings of the National Academy of Sciences. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kiseleva E, Goldberg MW, Allen TD, Akey CW. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J Cell Sci. 1998;111(Pt 2):223–236. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- 128.Grünwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475:333–341. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhao J, Jin S-B, Björkroth B, Wieslander L, Daneholt B. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. Embo J. 2002;21:1177–1187. doi: 10.1093/emboj/21.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Estruch F, Cole CN. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Molecular Biology of the Cell. 2003;14:1664–1676. doi: 10.1091/mbc.E02-09-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 132.Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Noble KN, Tran EJ, Alcazar-Roman AR, Hodge CA, Cole CN, Wente SR. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011;25:1065–1077. doi: 10.1101/gad.2040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, et al. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 2011;25:1052–1064. doi: 10.1101/gad.2041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 136.Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proceedings of the National Academy of Sciences. 2009;106:3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 138.Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbach M. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. Embo J. 1999;18:5761–5777. doi: 10.1093/emboj/18.20.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saavedra CA, Hammell CM, Heath CV, Cole CN. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 1997;11:2845–2856. doi: 10.1101/gad.11.21.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zheng C, Fasken MB, Marshall NJ, Brockmann C, Rubinson ME, Wente SR, et al. Structural basis for the function of the Saccharomyces cerevisiae Gfd1 protein in mRNA nuclear export. J Biol Chem. 2010;285:20704–20715. doi: 10.1074/jbc.M110.107276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 142.Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nature Reviews Genetics. 2011;12:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Solmaz SR, Chauhan R, Blobel G, Melčák I. Molecular Architecture of the Transport Channel of the Nuclear Pore Complex. Cell. 2011;147:590–602. doi: 10.1016/j.cell.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proceedings of the National Academy of Sciences. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Klein T, Löschberger A, Proppert S, Wolter S, van de Linde S, Sauer M. Live-cell dSTORM with SNAP-tag fusion proteins. Nat Methods. 2011;8:7–9. doi: 10.1038/nmeth0111-7b. [DOI] [PubMed] [Google Scholar]