Abstract
Misfolded proteins are continuously produced in the cell and present an escalating detriment to cellular physiology if not managed effectively. As such, all organisms have evolved mechanisms to address misfolded proteins. One primary way eukaryotic cells handle the complication of misfolded proteins is by destroying them through the ubiquitin-proteasome system. To do this, eukaryotes possess specialized ubiquitin-protein ligases that have the capacity to recognize misfolded proteins over normally folded proteins. The strategies used by these Protein Quality Control (PQC) ligases to target the wide variety of misfolded proteins in the cell will likely be different than those used by ubiquitin-protein ligases that function in regulated degradation to target normally folded proteins. In this review, we highlight what is known about how misfolded proteins are recognized by PQC ubiquitin-protein ligases.
1. Introduction
To function properly, cells rely on proteins successfully accomplishing specific actions. Fundamental to protein action is the acquisition of a protein’s 3-dimensional structure, and thus the proper folding of a cell’s protein cohort is critical for cells. Due to the central importance of protein folding, cells have evolved a collection of protein quality control (PQC) mechanisms that maintain overall cellular protein homeostasis, or proteostasis [1]. PQC systems can be divided into those that function as either primary or secondary PQC defenses. The cell’s primary PQC defenses are directly involved in repairing or removing misfolded proteins. Repair systems are chiefly composed of protein chaperones, whereas removal systems are principally involved in proteolytic destruction either by the proteasome or via autophagy. In many cases, the PQC repair and removal machinery function together in a triage hierarchy that has the potential to determine if a misfolded protein is salvageable and then direct the PQC action towards either repair or removal [2]. In contrast, the cell’s secondary PQC defenses are adaptive transcriptional responses that balance the primary PQC capacities with the extent of the cellular burden caused by misfolded proteins. They can also reduce global translation as a way to attenuate the production of misfolded proteins. In eukaryotes, PQC systems typically mitigate protein misfolding in a compartment-specific way, with each subcellular compartment housing a distinct set of PQC repair, removal, and adaptive capabilities.
There is now a considerable wealth of information on the different types of cellular PQC, which we cannot cover in its entirety here. We refer the readers to excellent reviews for chaperone-mediated folding and repair and for PQC adaptive stress responses [3–7]. Herein, we will discuss the ubiquitin-dependent PQC removal systems that operate in each eukaryotic cellular compartment (Figure 1).
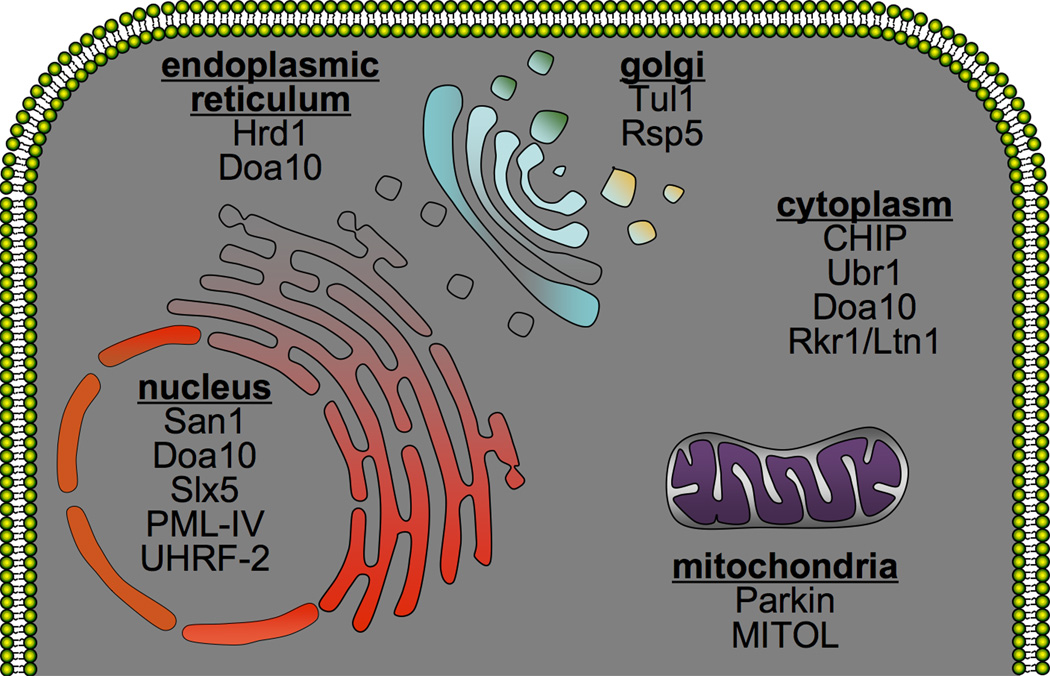
Figure 1. PQC ubiquitin-protein ligases in the cell.
Schematic representation of the cell with PQC ubiquitin-protein ligases listed in the appropriate cellular compartment. Yeast PQC ligases are listed as the main example, except in the cases where there is no yeast homolog.
1.1 Ubiquitin-mediated proteasomal degradation: regulation versus quality control
Protein degradation by the ubiquitin-proteasome system has two primary purposes in the cell: 1) the temporal or spatial regulation of normal proteins, and 2) the removal of misfolded proteins. For each type of degradation, a specific subset of proteins must first be uniquely distinguished from the global pool of cellular proteins and subsequently ubiquitylated. Ubiquitylation is canonically achieved via an enzymatic cascade wherein a ubiquitin-protein ligase (E3) partners with a specific ubiquitin-conjugating enzyme (E2) that has been charged with ubiquitin by a ubiquitin-activating enzyme (E1) [8]. The ubiquitin-protein ligase typically confers substrate specificity within each ubiquitylation cascade, either by possessing intrinsic substrate-binding domains or by recruiting auxiliary proteins that impart substrate specificity.
One critical aspect of ubiquitylation is the ability of a ubiquitin-protein ligase to distinguish its substrates from other proteins. In regulated degradation of normal proteins, ubiquitin-protein ligases typically recognize degrons, which are small specific linear amino acid sequences located within substrates [9]. The degron recognized by the ligase often varies based on the auxiliary proteins that are bound to the ligase. For example, cullin RING ubiquitin-protein ligases (CRLs) are able to recognize many different types of degrons through the directed binding of distinct adapter F-box proteins [10]. The interaction of a particular ubiquitin-protein ligase with different substrate-binding auxiliary proteins allows it to target a larger number of protein substrates. Importantly, these ubiquitin-protein ligases are still only able to target a limited number of substrates due to this sequence specificity requirement.
In contrast to the regulated degradation of normal proteins, substrate recognition in PQC degradation is unlikely to be achieved via the recognition of linear sequence-specific degrons in misfolded substrates for two key reasons. First, any protein has the capacity to misfold and most proteins in each cellular compartment share little, if any sequence homology. Thus, two different misfolded proteins will not likely possess the same sequence-specific degron. Second, a key purpose of PQC degradation is to destroy structurally abnormal proteins that share the same linear sequence with their normal counterparts. Thus, the features of misfolding recognized by PQC ubiquitin-protein ligases must transcend primary structure. One prevalent hypothesis is that PQC ubiquitin-protein ligases distinguish abnormal proteins by recognizing the exposure of hydrophobic residues typically buried in the core of a normal protein. In the subsequent sections, we will introduce the PQC ubiquitin-protein ligases in each cellular compartment, highlighting what is known about substrate recognition mechanisms and the features of structural abnormality recognized within the substrates.
2. Endoplasmic reticulum
The unique properties of the endoplasmic reticulum (ER) present numerous challenges for PQC ubiquitin-protein ligases in substrate recognition. First, de novo protein folding occurs in the ER and ER PQC ubiquitin-protein ligases must be capable of differentiating between nascent polypeptides that are in the process of folding and proteins that have become misfolded. In addition, the ER is a membrane-bound organelle where structural lesions may be present in transmembrane segments or in regions located on the luminal or cytoplasmic side of the membrane. Thus, ER PQC ubiquitin-protein ligases must have location-specific recognition mechanisms that can sense where the lesions are relative to the ER membrane. Another complication in ER PQC substrate recognition is the presence of disulfide bonds and glycosylation moieties in resident proteins. These posttranslational modifications must be queried for defects by PQC machinery. Lastly, ubiquitin and proteasomes are not present in the ER lumen, so ubiquitylation and proteasome degradation can occur only on the cytoplasmic side of the ER membrane. Accordingly, the ER PQC ubiquitin-protein ligases must have a means to recognize substrates on the luminal side of the ER membrane while directing substrate ubiquitylation on the cytoplasmic side of the ER membrane.
2.1 The Hrd1 pathway
The first ubiquitin-protein ligase found to play a role in ER PQC degradation is the yeast protein Hrd1/Der3 [11, 12]. Hrd1 is an integral ER-membrane protein containing a transmembrane domain that traverses the ER membrane six times and a cytoplasmic RING domain that mediates the transfer of ubiquitin to its substrates [12–14]. Several mammalian homologs of Hrd1 have been identified – gp78, hHrd1 (synovilin), and Rfp2 [15–19], with their roles in ER PQC degradation explored to varying degrees. Here, we will focus on yeast Hrd1 as it has been the best characterized.
All Hrd1 substrates require a core group of proteins to mediate substrate ubiquitylation. In addition to Hrd1, the core complex contains Hrd1’s cognate ubiquitin-conjugating enzyme Ubc7 [11, 20–22], which is localized to the Hrd1 core complex by the protein Cue1 [23]. Cue1 possesses a single transmembrane span and a cytoplasmic domain that is important for its interaction with Ubc7. There is no Cue1 mammalian homolog, but gp78 contains a cytoplasmic CUE domain that is similar to Cue1 [24]. Also in the core Hrd1 complex is Hrd3 (SEL1L in mammals [25, 26]), which is predominantly luminal with a single transmembrane span and a small cytoplasmic region [12, 13, 27]. Hrd3 directly binds misfolded proteins in the ER lumen [28]. Additionally, Hrd3 regulates Hrd1 autoubiquitylation and stability [13]. Hrd1 autoubiquitylation, as well as Hrd1 oligomerization, also depends on the protein Usa1 (Herp in mammals [29, 30]), which contains two transmembrane spans and interacts directly with Hrd1 [31–34]. For many Hrd1 substrates, this core complex of Hrd1, Ubc7, Cue1, Hrd3, and Usa1 is sufficient for substrate ubiquitylation [33, 34].
One class of PQC substrates degraded by the core Hrd1 complex comprises proteins with lesions in their ER-membrane spanning segments and are thus referred to as ERAD-M substrates (ER-Associated Degradation – Membrane) [34]. ERAD-M substrate recognition appears to be performed directly by Hrd1 [35]. It has been proposed that the transmembrane domain of Hrd1 recognizes improperly exposed hydrophilic residues in the hydrophobic environment of the ER membrane [35]. Mutational analysis of Hrd1’s transmembrane domain demonstrated that the degradation of Hrd1’s ERAD-M substrates could be disrupted without affecting degradation of other Hrd1 substrates [35], suggesting separate substrate recognition mechanisms.
Other Hrd1 substrates contain lesions in their ER luminal domains (ERAD-L substrates [34]), and their degradation typically requires the core Hrd1 complex and additional ancillary factors. One such factor is Der1 [36], a transmembrane protein localized to the Hrd1 complex via Usa1 [32, 34]. Der1 function in substrate recognition is unclear, though its mammalian homologue Derlin-1 is thought to be involved in postubiquitylation processes that deliver ER luminal proteins to the cytoplasm [37–39]. Another cofactor required in the degradation of ERAD-L substrates is the Hsp70 chaperone, Kar2 (also known as BiP) [40]. Kar2 likely couples to the core Hrd1 complex via interaction with a tetratricopeptide repeat (TPR) domain in Hrd3. Because Kar2 is an Hsp70 chaperone, its involvement in substrate degradation suggests that initial recognition of a misfolded protein could be mediated by the chaperone, which typically involves binding regions of hydrophobicity surrounded by basic residues [41]. However, exposed hydrophobicity in ERAD-L substrates has not yet been demonstrated as the abnormal structural feature recognized by the Hrd1 complex.
The decision to fold or degrade ER proteins can depend on posttranslational modifications. For instance, specific N-glycan moieties on ERAD-L substrates lead to degradation after their recognition by additional Hrd1 complex factors, such as the lectin Yos9 [42–45] (OS-9 in mammals [46]). Yos9 is coupled to the Hrd1 complex via interactions with Hrd3 and Kar2 [46–48]. N-glycan interaction with Yos9 depends on Yos9’s mannose-6-phosphate receptor function and is required for degradation of glycolsylated ERAD-L substrates [42, 49, 50]. The particular location of the glycan modification within the misfolded protein, including the peptide sequences surrounding the glycosylation site, is important for efficient recognition [51–53]. Interestingly, Yos9 is involved in the degradation of certain non-glycosylated substrates [54, 55], suggesting it might have additional substrate recognition roles.
2.2 The Doa10 pathway
Another PQC ubiquitin-protein ligase in the ER is Doa10, which was initially identified as the ubiquitin-protein ligase involved in degradation of the MATα2 transcription factor [56]. Doa10 has been subsequently shown to target misfolded proteins in the ER [57]. Doa10 contains a transmembrane domain that traverses the ER membrane fourteen times and a cytoplasmic RING domain [58]. In mammalian cells, TEB4 is described as a homolog of Doa10 due to its similar membrane topology [58]. TEB4 has recently been shown to be involved in the degradation of the ER-resident type 2 deiodinase [59].
Doa10, like Hrd1, functions as part of a larger protein complex in substrate recognition and ubiquitylation. There are four proteins required for the Doa10 complex to ubiquitylate substrates: Doa10, the ubiquitin-conjugating enzymes Ubc6 and Ubc7, and Cue1 [60]. Select substrates also require the ubiquitin chain elongator Ufd2 for degradation [61]. The role of Ufd2 is likely postubiquitylation and not in substrate recognition. Also required for degradation of certain Doa10 substrates are Hsp70 (of the Ssa class) and Hsp40 chaperones [57].
The Doa10 complex primarily functions in the degradation of ER PQC substrates with lesions on the cytoplasmic side of the ER [56, 57] (ERAD-C substrates [34]). Although it’s not clear how the Doa10 complex recognizes its substrates, the requirement of Hsp70 and Hsp40 chaperones suggests that substrate recognition could be mediated by the chaperones binding to exposed hydrophobicity. Some evidence suggests the Doa10 pathway likely targets hydrophobicity in its substrates. This was revealed from mutational studies of Doa10’s substrate MATα2, which possesses an amphipathic helix in its N-terminus that is both necessary and sufficient for Doa10-dependent degradation [62]. The hydrophobic portion of the amphipathic helix is the critical determinant for Doa10-dependent degradation of MATα2 [62]. Furthermore, studies examining Doa10-dependent degradation of small peptides fused to a reporter protein also revealed a hydrophobic requirement for Doa10-targeting [63–65]. Further work will be required to determine if the Doa10 complex recognizes hydrophobicity in ERAD-C substrates.
2.3 Late secretory PQC degradation pathways
Golgi-localized ubiquitin-protein ligases have also been implicated as having potential roles in PQC degradation. The yeast RING-domain ubiquitin-protein ligase Tul1 was found to be involved in selectively sorting transmembrane proteins with exposed polar residues in their transmembrane spans into multivesicular bodies for delivery to the vacuole for degradation [66]. In addition to Tul1, two studies found that the yeast transmembrane protein Bsd2 recruits the HECT-domain ubiquitin-protein ligase Rsp5 to target membrane proteins with exposed polar residues in their transmembrane spans for delivery to the vacuole [67, 68]. It is important to note that Tul1 and Bsd2-Rsp5 also assist in the correct trafficking of normal proteins to the vacuole [66–68]. Thus, it is unclear if these ubiquitin-protein ligases serve a PQC function, or if the mutant proteins identified mimic physiological substrates.
3. Cytoplasm
The cytoplasmic environment presents challenges to PQC ubiquitin-protein ligases that are both similar to and distinct from the ER. Similar to the ER, de novo protein folding occurs in the cytoplasm and cytoplasmic PQC ubiquitin-protein ligases must have the ability to differentiate between misfolded proteins and nascent unfolded polypeptides in the process of folding. Unlike the ER, cytoplasmic PQC does not have to contend with multiple classes of substrate lesions that present themselves differently in relation to a membrane. It is possible, however, that there might be specific distinct regions within the cytoplasm in which misfolded proteins behave differently [69], and these regions could require different ubiquitin-protein ligases.
3.1 The CHIP pathway
The mammalian protein CHIP is involved in cytoplasmic PQC degradation. CHIP contains a U-box domain necessary for its ubiquitin-protein ligase activity via interaction with the ubiquitin-conjugating enzyme UbcH5 [70–72], and a tetratricopeptide repeat (TPR) domain that is essential for CHIP’s interactions with Hsp70 and Hsp90 chaperones [71, 72]. Through direct interaction with chaperones, CHIP ubiquitylates client proteins that are bound by the chaperones [73]. While CHIP interacts with both Hsp70 and Hsp90 chaperones, data suggests that CHIP has a preference for ubiquitylating Hsp70-bound proteins [74]. Although substrate recognition by CHIP is heavily dependent on chaperone recognition, CHIP itself also appears capable of binding misfolded proteins directly [75].
CHIP’s interactions with chaperones place it at a central decision-making hub that balances productive folding and PQC degradation of a chaperone-bound client protein. However, an open question is how CHIP determines if a chaperone-bound protein should be ubiquitylated and degraded or allowed to continue with productive folding. One possibility is that Hsp70 and Hsp90 accessory proteins mediate this decision. For example, CHIP ubiquitylation of Hsp70 client proteins is influenced by the BAG class of Hsp70 cochaperones, which contain a BCL2-associated athanogene (BAG) domain that mediates the interaction with Hsp70 [76]. BAG cochaperones vary in their CHIP-related function from negatively regulating CHIP-dependent ubiquitylation of Hsp70 client proteins (BAG-2 and BAG-5) [77, 78], to facilitating the interaction of CHIP-chaperone complexes with the proteasome (BAG-1) [79], or in helping recruit the protein p62 to the CHIP-chaperone complex for substrate delivery to the lysosome (BAG-3) [80]. The Hsp70 cochaperone HspBP1 is also known to negatively regulate CHIP’s substrate-ubiquitylating activity [81].
CHIP’s interaction with chaperones suggests that the feature of structural abnormality recognized by CHIP is likely the same feature that the Hsp70 or Hsp90 chaperones recognize. CHIP has also been shown to have a chaperone function itself and is capable of binding thermally denatured proteins in an Hsp70-independent manner to prevent their aggregation [75]. However, the structurally abnormal feature that CHIP directly binds in its thermally denatured substrates is not known.
3.2 The Ubr1 pathway
In S. cerevisiae there is no identified homolog to CHIP. Rather, the RING-domain ubiquitin-protein ligase Ubr1 appears to mediate cytoplasmic PQC degradation [82–85]. Originally, Ubr1 was characterized for its role in N-end rule degradation, in which certain residues at the N-terminus of proteins serve as degrons [86]. Two specific regions in Ubr1 were identified that direct the ubiquitylation of substrates containing N-terminal residues of either type 1 (Arg, Lys, or His) or type 2 (Leu, Ile, Phe, Trp, or Tyr) [86]. It was recently found that Ubr1 also mediates the PQC degradation of misfolded cytoplasmic proteins [82–85], and this is independent of its role in the N-end rule pathway [83, 84]. Additionally, yeast Ubr2 has been shown to mediate cytoplasmic PQC degradation, [83] as well as human Ubr1 [87].
Ubr1-mediated PQC degradation requires the use of the ubiquitin-conjugating enzymes Rad6 and Ubc4 [83, 84, 88], as well as Hsp70 and Hsp110 chaperones [82–84, 88]. Hsp70 functions to keep substrates soluble [88], but it is not yet clear if these chaperones also direct substrates to Ubr1 similar to CHIP or if Ubr1 directly binds misfolded proteins. In support of direct interaction with substrates, Ubr1 is able to ubiquitylate a thermally denatured substrate in vitro without the aid of a chaperone [83]. Addition of Hsp70 increases the Ubr1-dependent ubiquitylation of the denatured substrate and robust ubiquitylation only occurs when Ubr1 is added during thermal denaturation [83]. Additional work will be necessary to clarify the role of chaperones in Ubr1-mediated PQC ubiquitylation.
3.3 The cytoplasmic Doa10 pathway
In addition to its role in ER PQC degradation, Doa10 is also involved in the degradation of cytoplasmic proteins that are misfolded [63] or contain an acetylated N-terminus [89]. Doa10-dependent degradation of cytoplasmic misfolded proteins requires cytoplasmic Hsp70 and Hsp40 chaperones [63]. It is not known if Doa10 recognizes the same features in cytoplasmic misfolded proteins as it does in ER misfolded proteins.
3.4 The Hul5 pathway
Recently, the HECT-domain ubiquitin-protein ligase Hul5 was shown to function in the PQC degradation of misfolded cytoplasmic proteins generated after heat shock [90]. Hul5 was previously found to be a component of the 19S regulatory subunit of the proteasome where it functions to extend polyubiquitin chains on substrates [91], likely to facilitate their processive degradation [91, 92]. It is not clear if Hul5’s function in cytoplasmic PQC degradation is to extend polyubiquitin chains initially added by other cytoplasmic PQC ubiquitin-protein ligases, or if it functions independently as a primary PQC ubiquitin-protein ligase that directly targets misfolded proteins itself. Because of this, how Hul5 targets its substrates is unknown.
3.5 The Rkr1/Ltn1 pathway
Translation of nonstop mRNAs is one way in which abnormal proteins are generated. Nonstop mRNAs can be created through DNA mutation or transcriptional mistakes that alter the stop codon, or by premature polyadenylation due to errors in processing [93]. Nonstop mRNAs can result in an aberrant sequence appended to a normal protein by read-through of the poly(A) tail, which would typically add a poly-Lys residue tract. Polybasic tracts have been shown to cause translational pausing and arrest [94]. RKR1/LTN1/YMR247C was initially identified as a gene that suppressed the phenotype of certain translated nonstop mRNAs [95]. Subsequently, it was demonstrated that Rkr1/Ltn1 is a ubiquitin-protein ligase [96, 97], and is involved in the degradation of abnormal proteins with a polybasic tract of Lys resulting from nonstop translation [97]. Furthermore, it was found that Rkr1/Ltn1 associates with ribosomes and is involved in the degradation of newly synthesized proteins that have stalled on the ribosome by virtue of a nonstop transcript [97]. However, It is not yet clear what Rkr1/Ltn1 recognizes: the polybasic tract of a target substrate in the context of a stalled ribosome or some feature of a stalled ribosome itself.
4. Nucleus
Unlike the cytoplasm, protein biosynthesis does not occur in the nucleus. Instead, nuclear proteins are typically translated and folded in the cytoplasm and imported into the nucleus. Thus, PQC degradation systems designed to detect errors in nascent protein folding will likely be absent in the nucleus. The nucleus does have different subdomains such as chromatin, the nucleoplasm, the nuclear membrane, and the nucleolus. Thus, different PQC ubiquitin-protein ligases might be required to manage misfolded proteins that arise in each of these different subnuclear regions.
4.1 The San1 pathway
In S. cerevisiae, the nuclear RING ubiquitin-protein ligase San1 mediates the PQC degradation of mutant or misfolded nuclear proteins [98–102]. San1 does not target normal versions of the same proteins [99–102], establishing a specific role for San1 in nuclear PQC degradation. While no mammalian homologs of San1 have yet been identified, a S. pombe homolog was recently identified and its function in PQC degradation established [103].
San1-mediated degradation uses the ubiquitin-conjugating enzymes Cdc34 and Ubc1 [99]. Substrate recognition involves San1 directly binding its substrates through N- and C-terminal regions that possess multiple substrate-binding sites embedded within highly disordered sequences [104]. It has been proposed that San1’s unique topology of high intrinsic disorder interspersed with substrate-binding modules allows San1 to use conformational plasticity to accommodate the binding of San1 to the diverse array of misfolded substrate conformations that it is likely to encounter [104].
The abnormal structural feature that San1 recognizes appears to be exposed hydrophobic residues in substrates. Through the use of a two-hybrid assay, it was found that San1 mediates the degradation of reporter proteins fused to hydrophobic peptides [104, 105]. Further exploration revealed that as few as five contiguous hydrophobic residues in the peptides defined the minimal recognition motif for San1-mediated degradation [105]. San1 can also target exposed hydrophobicity in larger misfolded proteins [105].
Surprisingly, it was recently found that some presumably cytoplasmic misfolded proteins become nuclear-localized and are degraded in a San1-dependent manner [82, 84]. It was found that cytoplasmic Hsp70 and Hsp110 chaperones are required for the nuclear localization of these substrates [82, 84], suggesting that chaperones might be involved in misfolded protein trafficking to the nucleus. This could be through a direct action of the chaperones in the nuclear import process, or an indirect involvement of the chaperones in maintaining the solubility of the misfolded proteins prior to nuclear import. Due to their potential role in the nuclear trafficking of substrates and the fact that San1 can directly bind its substrates, the role of these chaperones in substrate recognition in the San1 pathway is not clear.
4.2 The nuclear Doa10 pathway
In addition to its PQC roles in the ER and cytoplasm, Doa10 is also involved in nuclear PQC degradation. The ER membrane and nuclear envelope are contiguous, and a portion of Doa10 localizes to the inner nuclear envelope [106]. Doa10’s nuclear localization is required for the Doa10-dependent regulated degradation of the MATα2 transcription factor [106]. Doa10 also selectively recognizes a temperature-sensitive mutant of the nuclear protein Ndc10 [60]. Mutant Ndc10 is targeted for degradation by exposure of the hydrophobic side of an amphipathic helix and a hydrophobic C-terminal tail [107], indicating that Doa10 recognizes exposed hydrophobicity in misfolded nuclear proteins.
4.3 The Slx5 pathway
Another potential nuclear PQC ubiquitin-protein ligase is the yeast RING-domain protein Slx5, which was previously characterized for its role in ubiquitylating sumoylated proteins [108, 109]. The Slx5 pathway is required for the ubiquitylation and degradation of the SUMOylated transcription factor Mot1 in its normal form, as well as its mutant form Mot1-301 [110]. Because Mot1-301 is degraded more rapidly than normal Mot1, Slx5 was suggested to be involved in nuclear PQC [110]. Enhanced degradation of a mutant protein compared to its normal form is seen in the PQC degradation of certain abnormal proteins. For example, 95% of mutant cystic fibrosis transmembrane conductance regulator (CFTR) is degraded by the ubiquitin-proteasome system, whereas 75% of normal CFTR is degraded due to slow folding kinetics [111, 112]. Additional PQC substrates will need to be identified to solidify Slx5’s role in nuclear PQC degradation.
4.4 Potential mammalian nuclear PQC ubiquitin-protein ligases
Two mammalian ubiquitin-protein ligases that are posited to function in nuclear PQC degradation are UHRF-2 and PML IV [113–115]. UHRF-2 is a RING-domain ubiquitin-protein ligase that has been shown to be involved in the degradation of a truncated form of the huntingtin (htt) protein [113]. Both the non-toxic, normal polyglutamine tract version of truncated htt and a toxic, expanded polyglutamine tract version are degraded in a UHRF-2 dependent manner [113]. Similar to UHRF-2, PML IV has been shown to be involved in the degradation of a nuclear protein with an expanded polyglutamine tract [114]. PML IV also associates with the nuclear aggregates formed by expressing polyglutamine-expanded proteins [114, 115]. It has not yet been shown if UHRF-2 and PML IV can distinguish between normal and abnormal versions of a protein. Thus, additional studies will be needed to clarify their roles in nuclear PQC degradation.
5. Mitochondria
Mitochondria present a specific set of challenges for PQC degradation. One challenge is that mitochondria are bound by both inner and outer membranes that divide the mitochondria into distinct subcompartments. This means that separate PQC degradation pathways will be required to recognize misfolded proteins in each subcompartment. Similar to the ER, ubiquitin and proteasomes are not located inside the mitochondria. Because of this, inner mitochondria PQC degradation appears to be independent of the proteasome, instead relying on AAA-ATPase proteases similar to bacterial systems [116]. Another challenge for PQC degradation in the mitochondria is that there is a continual production of reactive oxygen species (ROS) as a byproduct of ATP production. An environment with a high level of ROS can lead to increased protein oxidation and misfolding, which the mitochondrial PQC degradation machinery will need to mange robustly.
5.1 The Parkin pathway
Parkin is a mammalian PQC ubiquitin-protein ligase associated with the outer mitochondrial membrane. Inactivation of Parkin is a major cause of juvenile Parkinson’s disease [117]. Parkin contains a RING-in between-RING domain [118], which is required for its ubiquitin-protein ligase activity [119, 120]. Parkin also associates with CHIP and Hsp70, and this association leads to increased Parkin-mediated ubiquitylation of unfolded Pael receptor [121]. Association with CHIP and Hsp70 suggests Parkin may have additional roles in cytoplasmic PQC degradation. To date, the abnormal structural feature in the substrates targeted by Parkin is unknown. Parkin mediates the degradation of specific proteins that are associated with Parkinson’s disease [122], but it is not known how or why Parkin targets these substrates.
5.2 The MITOL pathway
Another mammalian ubiquitin-protein ligase associated with the outer mitochondrial membrane is MITOL (mitochondrial ubiquitin-protein ligase) [123]. MITOL contains a multispanning membrane domain that passes through the outer mitochondrial membrane four times [123]. In addition, MITOL possesses a PHD-variant of the RING domain that is responsible for its ligase activity [123]. Certain point mutants of superoxide dismutase 1 (Sod1), but not normal Sod1 are degraded in a MITOL-dependent manner [124]. Components required for MITOL-mediated ubiquitylation are unknown, and MITOL does not contain any obvious chaperone-binding motifs. The lack of multiple substrates and known partners means substrate recognition mechanisms are unclear at this time.
6. Conclusions
In this review, we have attempted to describe the current state of knowledge for how each ubiquitin-protein ligase implicated in PQC degradation operates in substrate recognition. In a few cases, the substrate-targeting mechanisms are becoming better understood. In most cases, however, there is still considerable work that is needed to discover the modes of substrate recognition for each PQC ligase. In particular, we currently have only a rudimentary knowledge about how the individual PQC ubiquitin-protein ligases actually bind their substrates and what they recognize as abnormal within their substrates.
After exploring the literature on the topic of substrate recognition in PQC degradation, we think there are a few main questions that need to be resolved moving forward. First, it is clear that the survey of PQC ubiquitin-protein ligases is incomplete, so what are the other ligases that participate in cellular PQC degradation? Second, what is the purpose for having multiple PQC ubiquitin ligases in a single compartment? In the ER, Hrd1 and Doa10 recognize structural lesions presented in distinct locations in relation to the ER membrane making the utility of two pathways obvious. But why, for example, do San1 and Doa10 both function in nuclear PQC degradation? Do they recognize different abnormal structural features in their substrates? If so, having two separate PQC degradations systems in the nucleus would broaden the cell’s substrate recognition capabilities. Or, do they function in different subcompartments of the nucleus? Consistent with this hypothesis, Doa10 is membrane bound, while San1 is not. Determining the reasons for multiple PQC degradation pathways in a single compartment will require a larger pool of substrates for each pathway and a better understanding of the abnormal structural features recognized by each PQC ligase.
Highlights.
> We review the mechanisms of misfolded protein recognition by ubiquitin-protein ligases that function in eukaryotic protein quality control degradation.
Acknowledgements
This work was supported by NIH/NIGMS training grant 5T32GM007750 (E.K.F), NIH/NIA grant R01AG031136 (R.G.G.), NIH/NCRR grant R21RR025787 (R.G.G), an Ellison Medical Foundation New Scholar Award in Aging (R.G.G), and a Marian E. Smith Junior Faculty Award (R.G.G).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286(5446):1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 3.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11(8):545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 5.Voisine C, Pedersen JS, Morimoto RI. Chaperone networks: tipping the balance in protein folding diseases. Neurobiol Dis. 2010;40(1):12–20. doi: 10.1016/j.nbd.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 7.de la Torre-Ruiz MA, Mozo-Villarias A, Pujol N, Petkova MI. How budding yeast sense and transduce the oxidative stress signal and the impact in cell growth and morphogenesis. Curr Protein Pept Sci. 2010;11(8):669–679. doi: 10.2174/138920310794557628. [DOI] [PubMed] [Google Scholar]
- 8.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 9.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9(9):679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 11.Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9(1):209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7(12):2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151(1):69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deak PM, Wolf DH. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J Biol Chem. 2001;276(14):10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- 15.Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2001;98(25):14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneko M, Ishiguro M, Niinuma Y, Uesugi M, Nomura Y. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 2002;532(1–2):147–152. doi: 10.1016/s0014-5793(02)03660-8. [DOI] [PubMed] [Google Scholar]
- 17.Lerner M, Corcoran M, Cepeda D, Nielsen ML, Zubarev R, Ponten F, et al. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell. 2007;18(5):1670–1682. doi: 10.1091/mbc.E06-03-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadav E, Shmueli A, Barr H, Gonen H, Ciechanover A, Reiss Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem Biophys Res Commun. 2003;303(1):91–97. doi: 10.1016/s0006-291x(03)00279-1. [DOI] [PubMed] [Google Scholar]
- 19.Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, et al. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279(5):3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- 20.Hampton RY, Bhakta H. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci U S A. 1997;94(24):12944–12948. doi: 10.1073/pnas.94.24.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2(7):379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 22.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3(1):24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 23.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278(5344):1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 24.Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci U S A. 2006;103(2):341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilley BN, Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102(40):14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller B, Lilley BN, Ploegh HL. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175(2):261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plemper RK, Bordallo J, Deak PM, Taxis C, Hitt R, Wolf DH. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J Cell Sci. 1999;112(Pt 22):4123–4134. doi: 10.1242/jcs.112.22.4123. [DOI] [PubMed] [Google Scholar]
- 28.Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ERlumenal substrate selection and Cdc48p recruitment. Embo J. 2006;25(9):1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulze A, Standera S, Buerger E, Kikkert M, van Voorden S, Wiertz E, et al. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J Mol Biol. 2005;354(5):1021–1027. doi: 10.1016/j.jmb.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 30.Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28(4):544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim I, Li Y, Muniz P, Rao H. Usa1 protein facilitates substrate ubiquitylation through two separate domains. PLoS One. 2009;4(10):e7604. doi: 10.1371/journal.pone.0007604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horn SC, Hanna J, Hirsch C, Volkwein C, Schutz A, Heinemann U, et al. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell. 2009;36(5):782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Carroll SM, Hampton RY. Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J Biol Chem. 2010;285(8):5146–5156. doi: 10.1074/jbc.M109.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126(2):361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 35.Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34(2):212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. Embo J. 1996;15(4):753–763. [PMC free article] [PubMed] [Google Scholar]
- 37.Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429(6994):834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 38.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429(6994):841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 39.Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol. 2011;18(10):1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388(6645):891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- 41.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. Embo J. 1997;16(7):1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19(6):765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 43.Kim W, Spear ED, Ng DT. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell. 2005;19(6):753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19(6):741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 45.Buschhorn BA, Kostova Z, Medicherla B, Wolf DH. A genome-wide screen identifies Yos9p as essential for ER-associated degradation of glycoproteins. FEBS Lett. 2004;577(3):422–426. doi: 10.1016/j.febslet.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 46.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10(3):272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8(8):849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 48.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126(2):349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 49.Hosokawa N, Kamiya Y, Kamiya D, Kato K, Nagata K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J Biol Chem. 2009;284(25):17061–17068. doi: 10.1074/jbc.M809725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, et al. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32(6):870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie W, Kanehara K, Sayeed A, Ng DT. Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol Biol Cell. 2009;20(14):3317–3329. doi: 10.1091/mbc.E09-03-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kostova Z, Wolf DH. Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J Cell Sci. 2005;118(Pt 7):1485–1492. doi: 10.1242/jcs.01740. [DOI] [PubMed] [Google Scholar]
- 53.Spear ED, Ng DT. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J Cell Biol. 2005;169(1):73–82. doi: 10.1083/jcb.200411136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benitez EM, Stolz A, Wolf DH. Yos9, a control protein for misfolded glycosylated and nonglycosylated proteins in ERAD. FEBS Lett. 2011;585(19):3015–3019. doi: 10.1016/j.febslet.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 55.Jaenicke LA, Brendebach H, Selbach M, Hirsch C. Yos9p assists in the degradation of certain non-glycosylated proteins from the endoplasmic reticulum. Mol Biol Cell. 2011 doi: 10.1091/mbc.E10-10-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15(20):2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, et al. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279(37):38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 58.Kreft SG, Wang L, Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J Biol Chem. 2006;281(8):4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- 59.Zavacki AM, Arrojo EDR, Freitas BC, Chung M, Harney JW, Egri P, et al. The E3 ubiquitin ligase TEB4 mediates degradation of type 2 iodothyronine deiodinase. Mol Cell Biol. 2009;29(19):5339–5347. doi: 10.1128/MCB.01498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25(3):533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu C, van Dyk D, Xu P, Choe V, Pan H, Peng J, et al. Ubiquitin chain elongation enzyme Ufd2 regulates a subset of Doa10 substrates. J Biol Chem. 2010;285(14):10265–10272. doi: 10.1074/jbc.M110.110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94(2):217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- 63.Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem. 2008;283(47):32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilon T, Chomsky O, Kulka RG. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998;17(10):2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilon T, Chomsky O, Kulka RG. Degradation signals recognized by the Ubc6p–Ubc7p ubiquitin-conjugating enzyme pair. Mol Cell Biol. 2000;20(19):7214–7219. doi: 10.1128/mcb.20.19.7214-7219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reggiori F, Pelham HR. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat Cell Biol. 2002;4(2):117–123. doi: 10.1038/ncb743. [DOI] [PubMed] [Google Scholar]
- 67.Hettema EH, Valdez-Taubas J, Pelham HR. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. Embo J. 2004;23(6):1279–1288. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pizzirusso M, Chang A. Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1–7, to the endosomal/vacuolar system in yeast. Mol Biol Cell. 2004;15(5):2401–2409. doi: 10.1091/mbc.E03-10-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454(7208):1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, et al. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276(46):42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 71.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3(1):93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 73.Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2(12):1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stankiewicz M, Nikolay R, Rybin V, Mayer MP. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J. 2010;277(16):3353–3367. doi: 10.1111/j.1742-4658.2010.07737.x. [DOI] [PubMed] [Google Scholar]
- 75.Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM. Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem. 2007;282(31):22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- 76.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274(2):781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 77.Dai Q, Qian SB, Li HH, McDonough H, Borchers C, Huang D, et al. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J Biol Chem. 2005;280(46):38673–38681. doi: 10.1074/jbc.M507986200. [DOI] [PubMed] [Google Scholar]
- 78.Kalia LV, Kalia SK, Chau H, Lozano AM, Hyman BT, McLean PJ. Ubiquitinylation of alpha-synuclein by carboxyl terminus Hsp70-interacting protein (CHIP) is regulated by Bcl-2-associated athanogene 5 (BAG5) PLoS One. 2011;6(2):e14695. doi: 10.1371/journal.pone.0014695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luders J, Demand J, Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem. 2000;275(7):4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- 80.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. Embo J. 2009;28(7):889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alberti S, Bohse K, Arndt V, Schmitz A, Hohfeld J. The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol Biol Cell. 2004;15(9):4003–4010. doi: 10.1091/mbc.E04-04-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21(13):2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, Sultana R, et al. Ubr1 and Ubr2 Function in a Quality Control Pathway for Degradation of Unfolded Cytosolic Proteins. Mol Biol Cell. 2010;21(13):2102–2116. doi: 10.1091/mbc.E10-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A. 2010;107(3):1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eisele F, Wolf DH. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582(30):4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 86.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20(8):1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sultana R, Theodoraki MA, Caplan AJ. UBR1 promotes protein kinase quality control and sensitizes cells to Hsp90 inhibition. Exp Cell Res. 2011;318(1):53–60. doi: 10.1016/j.yexcr.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, et al. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18(1):153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327(5968):973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011;13(11):1344–1352. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10(3):495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 92.Aviram S, Kornitzer D. The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol Cell Biol. 2010;30(4):985–994. doi: 10.1128/MCB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Akimitsu N. Messenger RNA surveillance systems monitoring proper translation termination. J Biochem. 2008;143(1):1–8. doi: 10.1093/jb/mvm204. [DOI] [PubMed] [Google Scholar]
- 94.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284(16):10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilson MA, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007;177(2):773–784. doi: 10.1534/genetics.107.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Braun MA, Costa PJ, Crisucci EM, Arndt KM. Identification of Rkr1, a nuclear RING domain protein with functional connections to chromatin modification in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27(8):2800–2811. doi: 10.1128/MCB.01947-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467(7314):470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dasgupta A, Ramsey KL, Smith JS, Auble DT. Sir antagonist 1 (San1) is a ubiquitin ligase. J Biol Chem. 2004;279(26):26830–26838. doi: 10.1074/jbc.M400894200. [DOI] [PubMed] [Google Scholar]
- 99.Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120(6):803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 100.Evans DR, Brewster NK, Xu Q, Rowley A, Altheim BA, Johnston GC, et al. The yeast protein complex containing cdc68 and pob3 mediates core-promoter repression through the cdc68 N-terminal domain. Genetics. 1998;150(4):1393–1405. doi: 10.1093/genetics/150.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Estruch F, Peiro-Chova L, Gomez-Navarro N, Durban J, Hodge C, Del Olmo M, et al. A genetic screen in Saccharomyces cerevisiae identifies new genes that interact with mex67-5, a temperature-sensitive allele of the gene encoding the mRNA export receptor. Mol Genet Genomics. 2009;281(1):125–134. doi: 10.1007/s00438-008-0402-x. [DOI] [PubMed] [Google Scholar]
- 102.Lewis MJ, Pelham HR. Inefficient quality control of thermosensitive proteins on the plasma membrane. PLoS One. 2009;4(4):e5038. doi: 10.1371/journal.pone.0005038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matsuo Y, Kishimoto H, Tanae K, Kitamura K, Katayama S, Kawamukai M. Nuclear protein quality is regulated by the ubiquitin-proteasome system through the activity of Ubc4 and San1 in fission yeast. J Biol Chem. 2011 doi: 10.1074/jbc.M110.169953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41(1):93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol Biol Cell. 2011;22(13):2384–2395. doi: 10.1091/mbc.E11-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443(7113):827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- 107.Furth N, Gertman O, Shiber A, Alfassy OS, Cohen I, Rosenberg M, et al. Exposure of Bipartite Hydrophobic Signal Triggers Nuclear Quality Control of Ndc10 at the Endoplasmic Reticulum/Nuclear Envelope. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-05-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mullen JR, Brill SJ. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J Biol Chem. 2008;283(29):19912–19921. doi: 10.1074/jbc.M802690200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem. 2007;282(47):34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- 110.Wang Z, Prelich G. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol. 2009;29(7):1694–1706. doi: 10.1128/MCB.01470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ward CL, Kopito RR. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem. 1994;269(41):25710–25718. [PubMed] [Google Scholar]
- 112.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83(1):121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 113.Iwata A, Nagashima Y, Matsumoto L, Suzuki T, Yamanaka T, Date H, et al. Intra-nuclear degradation of polyglutamine aggregates by the ubiquitin proteasome system. J Biol Chem. 2009;284(15):9796–9803. doi: 10.1074/jbc.M809739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Janer A, Martin E, Muriel MP, Latouche M, Fujigasaki H, Ruberg M, et al. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J Cell Biol. 2006;174(1):65–76. doi: 10.1083/jcb.200511045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fu L, Gao YS, Tousson A, Shah A, Chen TL, Vertel BM, et al. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol Biol Cell. 2005;16(10):4905–4917. doi: 10.1091/mbc.E05-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kaser M, Langer T. Protein degradation in mitochondria. Semin Cell Dev Biol. 2000;11(3):181–190. doi: 10.1006/scdb.2000.0166. [DOI] [PubMed] [Google Scholar]
- 117.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 118.Morett E, Bork P. A novel transactivation domain in parkin. Trends Biochem Sci. 1999;24(6):229–231. doi: 10.1016/s0968-0004(99)01381-x. [DOI] [PubMed] [Google Scholar]
- 119.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25(3):302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A. 2000;97(24):13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, et al. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10(1):55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- 122.Tanaka K, Suzuki T, Hattori N, Mizuno Y. Ubiquitin, proteasome and parkin. Biochim Biophys Acta. 2004;1695(1–3):235–247. doi: 10.1016/j.bbamcr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 123.Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. Embo J. 2006;25(15):3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yonashiro R, Sugiura A, Miyachi M, Fukuda T, Matsushita N, Inatome R, et al. Mitochondrial ubiquitin ligase MITOL ubiquitinates mutant SOD1 and attenuates mutant SOD1-induced reactive oxygen species generation. Mol Biol Cell. 2009;20(21):4524–4530. doi: 10.1091/mbc.E09-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]