Abstract
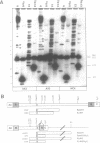
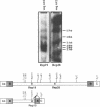
Approximately 2% of the Dictyostelium discoideum genome consists of multiple copies of a retrotransposable element termed DRE (Dictyostelium Repetitive Element). These elements have always been found integrated in a position and orientation-specific manner 50 +/- 4 nucleotides upstream of the coding region of tRNA genes (tDNAs). An intact DRE is 5.7 kb long. It carries an extensive coding region flanked by non-identical long terminal repeats (LTRs), composed of three distinct modules A, B and C. The left LTR proximal to the tRNA gene contains one or several A-modules followed by a single B-module (AnB). By contrast, the right LTR is composed of a B-module followed by a C-module (BC). Approximately 50% of the DRE elements in NC4 derivatives of D. discoideum are structurally different from the 5.7 kb DRE described above. They carry the following alterations: a) a 3.1 kb deletion in the coding region; b) two small deletions of 8 and 29 nucleotides in the B-module of the right LTR; c) a 72 bp deletion in the B-C junction; and d) three distinct point mutations within the A-module of the left LTR. The deletion in the open reading frame encompasses the putative coding regions for reverse transcriptase adn integrase. At least 60 copies of this smaller 2.4 kb DRE subtype are found in the genome of D. discoideum NC4 strains associated with tRNA genes. Thus, inspite of their lack in reverse transcriptase and integrase those 2.4 kb elements are presumably transposable and at least all isolated copies are found exclusively in the proximity of tRNA gene loci. The enzymes needed for their replication and transposition are likely to be provided by the intact 5.7 kb DREs.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aksoy S., Williams S., Chang S., Richards F. F. SLACS retrotransposon from Trypanosoma brucei gambiense is similar to mammalian LINEs. Nucleic Acids Res. 1990 Feb 25;18(4):785–792. doi: 10.1093/nar/18.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990 Jul 27;62(2):339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello J., Handelsman K., Lodish H. F. Sequence of Dictyostelium DIRS-1: an apparent retrotransposon with inverted terminal repeats and an internal circle junction sequence. Cell. 1985 Nov;43(1):105–115. doi: 10.1016/0092-8674(85)90016-9. [DOI] [PubMed] [Google Scholar]
- Chalker D. L., Sandmeyer S. B. Transfer RNA genes are genomic targets for de Novo transposition of the yeast retrotransposon Ty3. Genetics. 1990 Dec;126(4):837–850. doi: 10.1093/genetics/126.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. J., Bilanchone V. W., Haywood L. J., Dildine S. L., Sandmeyer S. B. A yeast sigma composite element, TY3, has properties of a retrotransposon. J Biol Chem. 1988 Jan 25;263(3):1413–1423. [PubMed] [Google Scholar]
- Dingermann T., Amon E., Williams K. L., Welker D. L. Chromosomal mapping of tRNA genes from Dictyostelium discoideum. Mol Gen Genet. 1987 Apr;207(1):176–187. doi: 10.1007/BF00331507. [DOI] [PubMed] [Google Scholar]
- Hansen L. J., Chalker D. L., Sandmeyer S. B. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol Cell Biol. 1988 Dec;8(12):5245–5256. doi: 10.1128/mcb.8.12.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D., Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990 May;15(5):186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J., Schumann G., Borschet G., Gösseringer R., Bach M., Bertling W. M., Marschalek R., Dingermann T. Transfer RNA genes from Dictyostelium discoideum are frequently associated with repetitive elements and contain consensus boxes in their 5' and 3'-flanking regions. J Mol Biol. 1991 Dec 5;222(3):537–552. doi: 10.1016/0022-2836(91)90495-r. [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H. E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985 Dec 13;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Kimmel B. E., ole-MoiYoi O. K., Young J. R. Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol Cell Biol. 1987 Apr;7(4):1465–1475. doi: 10.1128/mcb.7.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R., Brechner T., Amon-Böhm E., Dingermann T. Transfer RNA genes: landmarks for integration of mobile genetic elements in Dictyostelium discoideum. Science. 1989 Jun 23;244(4911):1493–1496. doi: 10.1126/science.2567533. [DOI] [PubMed] [Google Scholar]
- Marschalek R., Hofmann J., Schumann G., Gösseringer R., Dingermann T. Structure of DRE, a retrotransposable element which integrates with position specificity upstream of Dictyostelium discoideum tRNA genes. Mol Cell Biol. 1992 Jan;12(1):229–239. doi: 10.1128/mcb.12.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Fulton S. M., Dobson M. J., Wilson W., Kingsman S. M., Kingsman A. J. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature. 1985 Jan 17;313(5999):243–246. doi: 10.1038/313243a0. [DOI] [PubMed] [Google Scholar]
- Sandmeyer S. B., Bilanchone V. W., Clark D. J., Morcos P., Carle G. F., Brodeur G. M. Sigma elements are position-specific for many different yeast tRNA genes. Nucleic Acids Res. 1988 Feb 25;16(4):1499–1515. doi: 10.1093/nar/16.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer S. B., Olson M. V. Insertion of a repetitive element at the same position in the 5'-flanking regions of two dissimilar yeast tRNA genes. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7674–7678. doi: 10.1073/pnas.79.24.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts D. J., Ashworth J. M. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem J. 1970 Sep;119(2):171–174. doi: 10.1042/bj1190171. [DOI] [PMC free article] [PubMed] [Google Scholar]