Abstract
Supramolecular nanoassemblies that respond to the presence of proteins are of great interest, as aberrations in protein concentrations represent the primary imbalances found in a diseased state. We present here a molecular design, syntheses, and study of facially amphiphilic dendrimers that respond to the presence of the protein, immunoglobulin G. It is of particular interest that the ligand functionality, utilized for causing the binding-induced disassembly, be lipophilic. Demonstration of binding with lipophilic ligands greatly expands the repertoire of binding-induced disassembly, since this covers a rather large class of ligand moieties designed for proteins and these provide specific insights into the mechanistic pathways that are available for the binding-induced disassembly process. Here, we describe the details of the binding induced disassembly, including the change in size of the assembly in response to proteins, concurrent release of noncovalently encapsulated guest molecules, and the specificity of the disassembly process.
Keywords: dendrimers, disassembly, micelles, proteins, supramolecules
Introduction
Stimuli-responsive supramolecular systems have gained significant attention in recent years, as these systems have implications in a variety of areas, especially in drug delivery.[1] Similarly, amphiphilic assemblies have been attractive because of the potential to encapsulate water-insoluble drug molecules within their interior, which can then be released at a specific location in response to a trigger.[2] In this regard, amphiphilic systems that respond to physical or chemical internal or external stimuli have been widely explored. Systems that respond to change in pH,[3] temperature,[4] redox environment,[5] and light[6] are popular in this area. Location-dependent variations in these factors could be considered to be secondary imbalances in biology. The primary imbalances in biology that result in a diseased state involve variations in protein concentrations. Therefore, it is interesting to design supramolecular assemblies that respond to proteins, an area that is relatively under-explored.[7]
Among responsive supramolecular assemblies, nanosized structures have been of great interest, since these have the propensity to accumulate in the inflamed tissues of certain diseases, notably cancer, due to the enhanced permeability and retention (EPR) effect.[8] This has spurred interest in molecular assemblies, based on surfactants,[9] lipids,[10] polymers,[2,11] and dendrimers.[12] While small molecule surfactant based amphiphilic assemblies exhibit the ability to sequester lipophilic molecules in aqueous media, these suffer from low stability and high critical aggregation concentrations (CACs). On the other hand, phospholipid based liposomes exhibit enhanced stability and low CACs.[10] However, their application is largely limited to the delivery of hydrophilic molecules or lipophilic molecules that can be rendered water soluble.[10,13] Considering the fact that most drug molecules are lipophilic, it is desirable that systems possessing lipophilic microenvironments, to accommodate guest molecules, are used. In this context, amphiphilic polymers have been widely studied for drug delivery applications due to their ability to form micelles and encapsulate lipophilic guest molecules. These efforts have resulted in some excellent contributions to the field.[14] However, dendrimers provide a distinct advantage in fundamentally understanding the structural factors that control amphiphilic supramolecular assemblies and stimuli-responsive disassemblies. This is mainly due to the excellent control over their size and, thus, the perfectly monodisperse nature of these macromolecules.[12,15]
With all these scaffolds, a limited number of reports exist on protein-sensitive assemblies.[7,16] These reports, however, are mainly based on the enzymatic activity of the protein that causes the disassembly rather than a specific ligand–protein interaction. Designing systems that respond to a protein-binding event will bring the large class of nonenzymatic proteins into the realm of stimuli-responsive supramolecular nanoassemblies and, thus, greatly expand the repertoire of these systems for applications, such as drug delivery and sensing.
Results and Discussion
Molecular design and synthesis
Recently, we introduced a new class of facially amphiphilic biaryl dendrimers in which every repeating unit in the dendritic backbone contains both lipophilic and hydrophilic functionalities.[4a,12e] As a result of the orthogonal placement of these mutually incompatible units, these molecules exhibit a unique assembly switch, depending on their solvent environment. In other words, these dendrimers are able to form micelle-type assemblies in an aqueous milieu and inverted micelle-type assemblies in apolar solvents, such as toluene.[12e,17] In contrast to classical amphiphilic dendrimers,[12a,d] the micellar assemblies from our dendrimers are formed through aggregation of several dendrimer molecules. This feature presents a unique opportunity for stimuli-induced disassembly in these dendrimers through disaggregation.
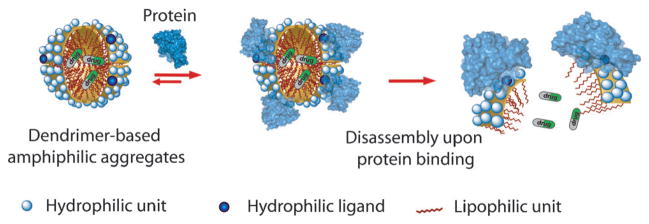
Certain hydrophilic–lipophilic balance (HLB) is required for these facially amphiphilic dendrimers to self-assemble in the aqueous phase. Therefore, we envisaged the possibility of disturbing the HLB of a dendrimer, through interaction with a protein, to cause disassembly and afford a protein-sensitive amphiphilic nanoassembly. Our basic premise for the hypothesis is that a typical ligand is a relatively small moiety that can be incorporated on to a dendrimer side chain. This functionality makes a certain contribution towards the overall HLB of the dendron and thus the stability of the assembly. When a protein binds to this ligand moiety, the ligand is masked and a rather large protein is presented to the solvent surface. Since water-soluble globular proteins exhibit a hydrophilic surface, the small ligand functionality is essentially changed to a large hydrophilic moiety (Figure 1). We hypothesized that such an alteration should result in a drastic change of HLB and cause disassembly. Indeed, we have recently shown that disassembly of these amphiphilic aggregates can be achieved with a specific ligand–protein interaction, in which the ligand is hydrophilic and thus is displayed on the surface of the amphiphilic aggregate.[7c] This ligand display provides a convenient access for protein to bind to the ligand and thus cause the binding-induced disassembly (Figure 1).
Figure 1.
Binding-induced disassembly with hydrophilic ligand containing dendrons.
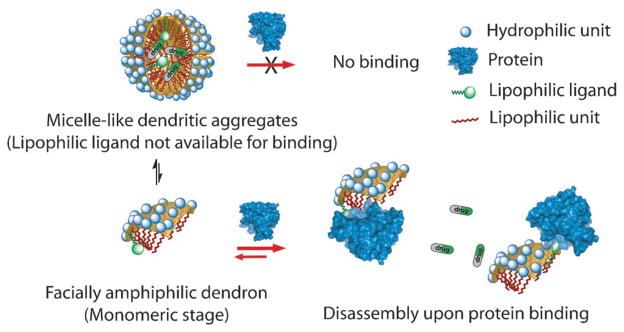
A potential limitation of the above finding is that the presumed mechanism (Figure 1), allows for the binding-induced disassembly to be executed with hydrophilic ligands. However, most of the custom-designed ligands target the hydrophobic binding pockets of proteins; thus, most promising ligands are lipophilic in nature. When these lipophilic ligands are incorporated into our proteins, these functionalities will be buried within the interior of the assembly and thus might not be available for binding to complementary proteins (Figure 2, top).
Figure 2.
Binding-induced disassembly with lipophilic ligand containing dendrons.
We hypothesized that an alternate mechanistic possibility in these types of supramolecular assemblies might allow for binding-induced disassembly with lipophilic ligands. Here, we describe and test whether this alternate pathway is available for binding-induced disassembly. Note that noncovalent amphiphilic assemblies are in equilibrium with their corresponding monomers (illustrated by the equilibrium in Figure 2, left). If binding between the ligand and the protein is favored in the monomeric dendron, then binding can preferentially occur at the monomeric stage. In this case, the equilibrium would ultimately cause disassembly due to the Le Chatlier-type effect (Figure 2, bottom). If this pathway is available for binding-induced disassembly, then the versatility of this approach will greatly increase, as this approach would then not be limited to just hydrophilic ligands.
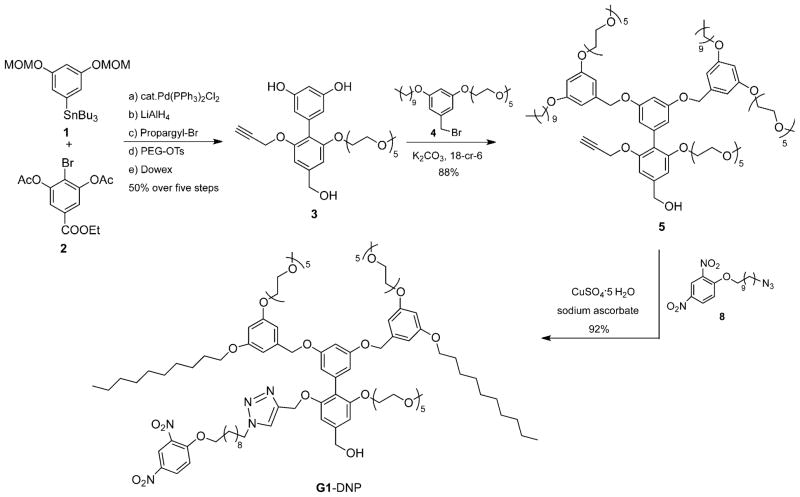
To test our hypothesis, we chose dinitrophenyl (DNP) moiety as the ligand functionality, because: 1) it is lipophilic and will be buried within the interior of the amphiphilic aggregate and thus the only reasonable pathway available for binding involves the equilibrium-driven disassembly; 2) DNP has been shown to bind to rat anti-DNP immunoglobulin G antibody (IgG) with subnanomolar binding affinity,[18] and 3) synthetic modification of the 2,4-DNP group is relatively straightforward and allows for the proof-of-concept to be achieved with relative ease. Structures of the targeted dendrons are shown as G1–DNP and G2–DNP. In these dendrons, the decyl chain acts as the lipophilic unit, and pentaethylene glycol (PEG) as the hydrophilic unit. PEG was chosen as a charge-neutral hydrophilic functionality to reduce nonspecific interactions between dendrimer and protein. The target dendrons are designed in such a way that these dendrimers are uniformly amphiphilic over the entire dendritic structure. To achieve this, the dendrons were constructed from a biaryl monomer, represented by structure 3 in Scheme 1. The biaryl monomer has several unique features: 1) it has the AB2 functional groups required for dendritic growth; 2) dendrimers constructed from this biaryl building block are fully amphiphilic because of the ability of the biaryl monomer to carry both lipophilic and hydrophilic functional groups. Thus, once the dendron is constructed every repeating layer in the dendritic backbone contains amphiphilic functionalities; 3) hydrophilic ligands can be placed in the dendrons by replacing the PEG unit with the ligand functionality, while the lipophilic ligand is placed by replacing the decyl moiety within a monomeric repeat unit.
Scheme 1.
Synthesis scheme for G1–DNP dendron.
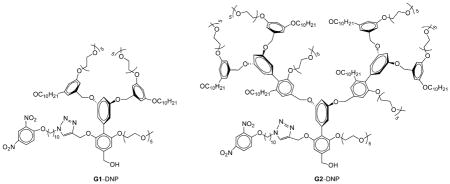
We were interested in having the syntheses of the dendrons modular, as this will allow for the incorporation of other lipophilic ligands on to the dendritic building block. We have previously utilized the 1,3-dipolar Huisgen cycloaddition reaction, the so-called click reaction, as the last step in our syntheses to install the ligand functionalities. Thus, the target building block unit for our dendron syntheses involves the presence of acetylene functionality at the lipophilic side of the dendron (Scheme 1, 3). The key step in the synthesis of 3 involves the formation of the biaryl bond. Thus, synthesis of biaryl compound 3 was achieved by using Stille coupling as the key step, from the aryl stannane 1 and bromoaryl ester 2 (Scheme 1). Reaction between the reported peripheral amphiphilic unit 4[4a] and biaryl building block 3[16b] in the presence of potassium carbonate afforded the G1 dendron, 5, in 88% yield. Similarly, the corresponding G2 dendron was synthesized from 3 and the brominated version of the G1 dendron 5.[4a] Once the first and second generation dendrons were assembled from the biaryl monomer 3, the 2,4-DNP ligand moiety was attached to these dendrons through the Huisgen reaction with 8 to obtain the targeted G1–DNP and G2–DNP dendrons. This reaction was chosen to install the DNP ligand to the dendrons, as it allows for easier future ligand variations. All dendrons were characterized by 1H and 13C NMR spectroscopy, and MALDI-ToF mass spectrometry, and details of the syntheses and characterizations are outlined in the Supporting Information.
Self-assembling properties of dendrimers
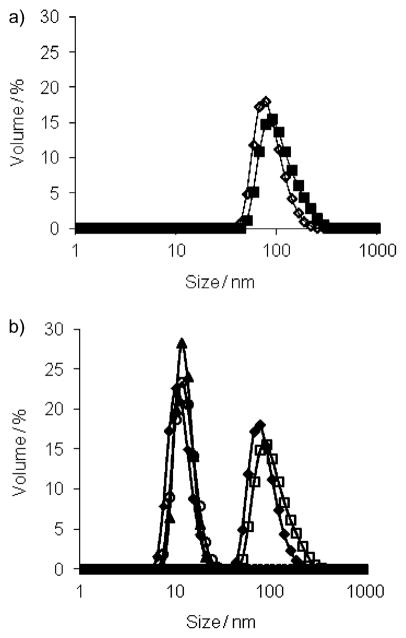
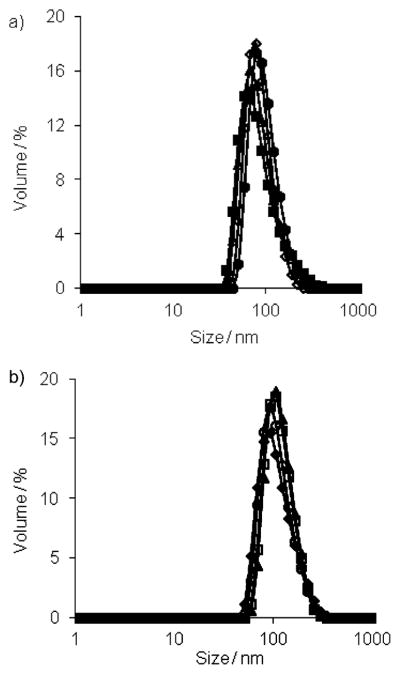
We first studied the assembly properties of the dendrons G1–DNP and G2–DNP using Nile red as a fluorescent probe. Nile red is a lipophilic dye and is not soluble in water unless it is accommodated in a hydrophobic pocket of a micelle-like assembly. The emission spectra of Nile red, at different G1–DNP and G2–DNP dendron concentrations in water, indicate that these dendrons are capable of providing an apolar microenvironment that sequesters the lipophilic dye molecule (Figure S1 in the Supporting Information). The emission spectra at different dendron concentrations were used to calculate the critical aggregation concentrations (CACs) of the dendrons. Plotting the emission intensity of Nile red as a function of dendron concentration afforded an inflection point, which was taken to be the dendron’s CAC (see the Supporting Information). Using this method, CACs for G1–DNP and G2–DNP dendrons were found to be 0.035 (18 μM) and 0.020 mgmL−1 (5 μM), respectively. Formation of the micelle-like assembly was further verified with dynamic light scattering (DLS) experiments. The dendron solutions (26 μM, above the CACs) were prepared in water and DLS results showed that 105 and 124 nm sized aggregates are formed for G1–DNP and G2–DNP dendrons, respectively (Figure 3a). This suggests that these dendrons are indeed aggregated to form micelle-like nanoassemblies in water, which are responsible for sequestering Nile red as the lipophilic guest.
Figure 3.
a) Sizes of G1–DNP (◇) and G2–DNP (■) dendrons; b) size changes in G1–DNP and G2–DNP dendron based amphiphilic assemblies due to the presence of anti-DNP IgG; ◆, G1–DNP; □, G2–DNP;▲, G1–DNP+IgG; ○, G2–DNP+IgG; ■, IgG.
Dendrimer–protein interactions
Since the DNP ligand in the dendrons is known to bind anti-2,4-DNP IgG, we were interested in testing the effect of the presence of this protein on the self-assembled structures. Our hypothesis is that the binding interaction between the DNP ligand in the dendrons and the IgG protein will afford a dendron–protein complex, the HLB of which would be drastically different from that of the dendron itself. We anticipated that this change would result in disruption of the micelle-type assembly. In order to examine if these dendritic micellar aggregates are indeed responsive to IgG, we monitored the size of the assembly using DLS. Upon addition of anti-DNP IgG (7.5 μM) to the solution of G1–DNP (26 μM) a remarkable decrease in the size of the assembly was observed (Figure 3b). A similar change in the assembly size was also observed with G2–DNP. The sizes of the G1–DNP and G2–DNP dendrons decreased to approximately 10 nm; this corresponds to the size of the protein itself, and indicates that the dendritic assembly is disaggregated due to ligand–protein binding.
Although the disassembly of the micellar aggregates provides good evidence for the anti-DNP IgG sensitivity of the dendrons, it is necessary to determine whether this disassembly indeed takes place due to a specific ligand–protein interaction. Therefore, we examined the effect of the presence of noncomplementary proteins, pepsin (pI = 1.0), cytochrome c (CytC, pI=10.2), and α-chymotrypsin (ChT, pI = 8.8). These proteins were chosen for their diversity in pI values, since this is often the source of nonspecific interactions. Thus, solutions of G1–DNP and G2–DNP dendrons were exposed to these three proteins (Figure 4). We were gratified to find that the size of the dendritic assemblies were unchanged in the presence of these proteins. These results support our hypothesis that IgG binding indeed caused micellar disassembly.
Figure 4.
Size changes: a) G1–DNP (◇), G1–DNP+pepsin (■), G1–DNP+α-chymotrypsin (△), G1–DNP+cytochrome c (●); b) G2–DNP (◆), G2–DNP+pepsin (□), G2–DNP+α-chymotrypsin (▲), G2–DNP+ cytochrome c (○).
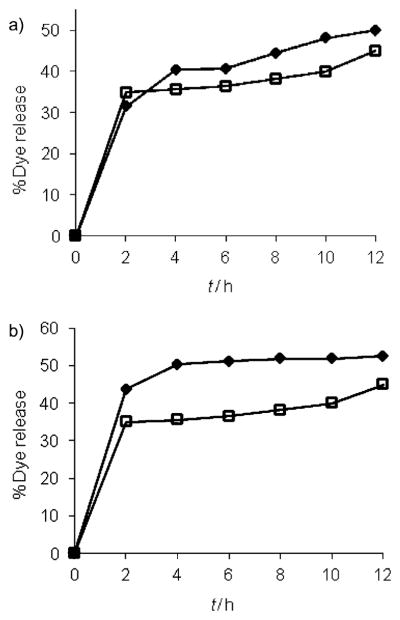
Next, we investigated the possibility of binding-induced guest release from the micellar interiors. If the size change observed in DLS was indeed due to the disassembly of the micellar aggregate, then it is reasonable to anticipate that the disassembly event should cause a concomitant release of guest molecules noncovalently encapsulated within the dendritic interiors. For this purpose, Nile red encapsulated solutions of G1–DNP and G2–DNP dendrons were exposed to the anti-DNP IgG. Indeed, we observed Nile red release due to protein binding, as evident from the decrease in emission intensity of the solution (Figure 5). Interestingly, the guest release exhibited a time-dependent behavior, which is most likely due to the limited accessibility of the lipophilic ligand functionalities that are buried inside the cores of the micellar assemblies. Also, note that we observed only 50 and 45% of the dye molecules to be released within the total time frame, for G1–DNP and G2–DNP dendrimers, respectively. The lack of 100% release could be attributed to the fact that the dendron–protein complex still presents hydrophobic functionalities, and provides an opportunity for lipophilic guest molecules to be bound to the complex. Also, we were interested in finding if the guest release would be enhanced if the percentage of free dendrons were decreased by increasing the relative ratio of the protein to the dendron. The commercial form of the protein imposes an upper limit on its concentration. Fortunately, however, the lower CAC of G2–DNP dendron enables us to work with lower dendron concentration. Thus, when 13 μM G2–DNP dendron was incubated with the IgG (7.5 μM) for 12 h, we observed 52% dye release, as compared to 45% release with 26 μM of G2–DNP dendron (Figure 5b).
Figure 5.
a) Guest molecule release from 26 μM solutions of G1–DNP (◆) and G2–DNP (□) dendrons; b) release from lower G2–DNP concentrations at 13 (◆) and 26 μM (□).
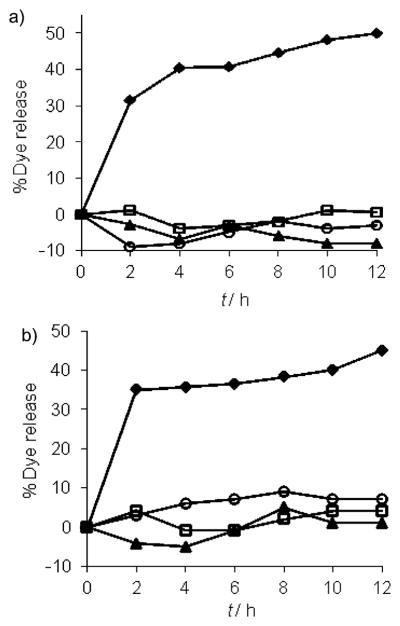
To determine whether dye release is specific only to IgG, we exposed G1–DNP and G2–DNP dendrons to pepsin, ChT, and CytC. We indeed found that exposure of G1–DNP and G2–DNP dendrons to these proteins did not cause any significant change in the fluorescence of Nile red; this indicates that there is no dye released from dendritic assembly (Figure 6). It should also be noted that CytC is a metallo-protein and we have previously shown that addition of positively charged CytC to negatively charged carboxylate polymers and dendrimers results in quenching of fluorescence of the noncovalently encapsulated fluorophores; this indicates the nonspecific binding of CytC to these assemblies.[19] Here, the lack of fluorescence change of Nile red in the presence of CytC provides additional support for the lack of nonspecific interactions between the dendritic assemblies and proteins.
Figure 6.
Guest molecule release from: a) G1–DNP, and b) G2–DNP dendrons, in the presence of IgG (◆), pepsin (□), cytochrome c (▲) and α-chymotrypsin (○).
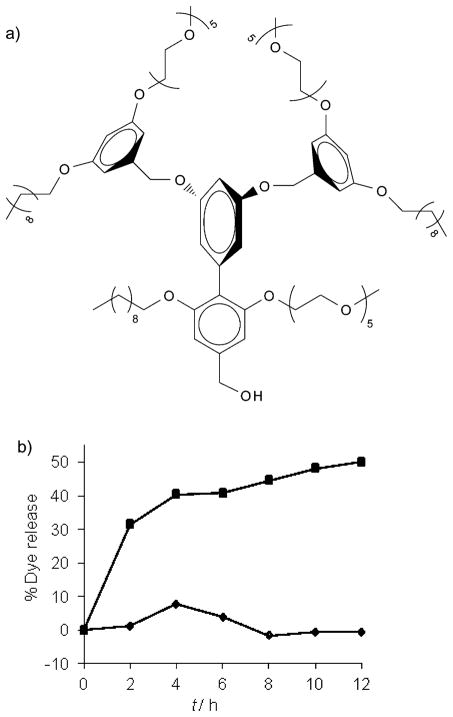
To further examine whether the binding events and the concurrent dye release are specifically due to the DNP–IgG interactions, we also synthesized a G1-control[4a] dendron (Figure 7a). This dendron is structurally similar to G1–DNP, except that it lacks the DNP ligand functionality. When the IgG was added to the solution of Nile red encapsulated G1-control dendron (26 μM) we observed no guest release from this assembly (Figure 7b); this indicates that the release obtained with DNP-containing dendrons was indeed due to the specific ligand–protein interactions.
Figure 7.
a) The structure of G1-control dendron; b) guest release from G1-control (◆) versus G1–DNP (■) after binding IgG.
Conclusion
We have designed and synthesized facially amphiphilic biaryl dendrimers that contain lipophilic ligand functionalities that are complementary to anti-DNP immunoglobulin G (IgG). These dendrimers were shown to self-assemble into micelle-like aggregates in aqueous solution and are capable of encapsulating lipophilic guest molecules, such as Nile red. We have shown that these aggregates disassemble in response to the ligand–protein interaction. We attribute the disassembly event to the significant HLB change caused by the ligand–protein interaction. This disassembly process clearly suggests that the binding event between a protein and the ligand-containing dendron does not have to occur in the self-assembled aggregate stage. In this scenario, it is likely that the equilibrium between the monomeric state of the dendron and the micellar aggregate provides a viable pathway for the binding-induced disassembly process. Alternately, it is also possible that there is conformational flexibility within the building block units in the assembly that allows for transient exposure of the lipophilic ligand on the surface of the dendron and provides the opportunity for binding with the protein while remaining in the assembly. We do not have a way of discounting this possibility at this time. Nonetheless, it is clear that the binding-induced disassembly strategy is not limited to hydrophilic ligands. Demonstration of the protein-sensitive disassembly by using lipophilic ligands opens up the possibility of utilizing such molecular design for a variety of applications, particularly in drug delivery and sensing.
Supplementary Material
Acknowledgments
We thank the NIGMS of the NIH (GM-065255) for support.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/chem.201102727.
References
- 1.a) Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]; b) Davis ME, Chen Z, Shin DM. Nat Rev Drug Discovery. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]; c) Ganta S, Devalapally H, Shahiwala A, Amiji M. J Controlled Release. 2008;126:187–204. doi: 10.1016/j.jconrel.2007.12.017. [DOI] [PubMed] [Google Scholar]; d) Rijcken CJF, Soga O, Hennink WE, van Nostrum CF. J Controlled Release. 2007;120:131–148. doi: 10.1016/j.jconrel.2007.03.023. [DOI] [PubMed] [Google Scholar]; e) Rapoport N. Prog Polym Sci. 2007;32:962–990. [Google Scholar]; f) Kojima C. Expert Opin Drug Delivery. 2010;7:307–319. doi: 10.1517/17425240903530651. [DOI] [PubMed] [Google Scholar]
- 2.a) Jeong B, Bae YH, Lee DS, Kim SW. Nature. 1997;388:860–862. doi: 10.1038/42218. [DOI] [PubMed] [Google Scholar]; b) Bellomo EG, Wyrsta MD, Pakstis L, Pochan DJ, Deming TJ. Nat Mater. 2004;3:244–248. doi: 10.1038/nmat1093. [DOI] [PubMed] [Google Scholar]; c) Kale TS, Klaikherd A, Popere B, Thayumanavan S. Langmuir. 2009;25:9660–9670. doi: 10.1021/la900734d. [DOI] [PubMed] [Google Scholar]; d) Haag R. Angew Chem. 2004;116:280–284. [Google Scholar]; Angew Chem Int Ed. 2004;43:278–282. [Google Scholar]
- 3.a) Giacomelli C, Men LL, Borsali R, Lai-Kee-Him J, Brisson A, Armes SP, Lewis AL. Biomacromolecules. 2006;7:817–828. doi: 10.1021/bm0508921. [DOI] [PubMed] [Google Scholar]; b) Gillies ER, Jonsson TB, Fréchet JMJ. J Am Chem Soc. 2004;126:11936–11943. doi: 10.1021/ja0463738. [DOI] [PubMed] [Google Scholar]; c) Jones MC, Ranger M, Leroux JC. Bioconjugate Chem. 2003;14:774–781. doi: 10.1021/bc020041f. [DOI] [PubMed] [Google Scholar]; d) Sawant RM, Hurley JP, Salmaso S, Kale A, Tolcheva E, Levchenko TS, Torchilin VP. Bioconjugate Chem. 2006;17:943–949. doi: 10.1021/bc060080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Aathimanikandan SV, Savariar EN, Thayumanavan S. J Am Chem Soc. 2005;127:14922–14929. doi: 10.1021/ja054542y. [DOI] [PubMed] [Google Scholar]; b) Soppimath KS, Liu LH, Seow WY, Liu SQ, Powell R, Chan P, Yang YY. Adv Funct Mater. 2007;17:355–362. [Google Scholar]; c) Klaikherd A, Nagamani C, Thayumanavan S. J Am Chem Soc. 2009;131:4830–4838. doi: 10.1021/ja809475a. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Morishima Y. Angew Chem. 2007;119:1392–1394. [Google Scholar]; Angew Chem Int Ed. 2007;46:1370–1372. doi: 10.1002/anie.200603405. [DOI] [PubMed] [Google Scholar]
- 5.a) Ryu J, Roy R, Ventura J, Thayumanavan S. Langmuir. 2010;26:7086–7092. doi: 10.1021/la904437u. [DOI] [PubMed] [Google Scholar]; b) Zhang L, Liu W, Lin L, Chen D, Stenzel MH. Biomacromolecules. 2008;9:3321–3331. doi: 10.1021/bm800867n. [DOI] [PubMed] [Google Scholar]; c) Li Y, Lokitz BS, Armes SP, McCormick CL. Macromolecules. 2006;39:2726–2728. [Google Scholar]; d) Bae KH, Mok H, Park TG. Biomaterials. 2008;29:3376–3383. doi: 10.1016/j.biomaterials.2008.04.035. [DOI] [PubMed] [Google Scholar]; e) Li YL, Zhu L, Liu Z, Cheng R, Meng F, Cui JH, Ji SJ, Zhong Z. Angew Chem. 2009;121:10098–10102. doi: 10.1002/anie.200904260. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:9914–9918. doi: 10.1002/anie.200904260. [DOI] [PubMed] [Google Scholar]
- 6.a) Goodwin AP, Mynar JL, Yingzhong M, Fleming GR, Fréchet JMJ. J Am Chem Soc. 2005;127:9952–9953. doi: 10.1021/ja0523035. [DOI] [PubMed] [Google Scholar]; b) Jiang J, Tong X, Morris D, Zhao Y. Macromolecules. 2006;39:4633–4640. [Google Scholar]; c) Jiang J, Tong X, Zhao Y. J Am Chem Soc. 2005;127:8290–8291. doi: 10.1021/ja0521019. [DOI] [PubMed] [Google Scholar]
- 7.a) Savariar EN, Ghosh S, Gonzalez DC, Thayumanavan S. J Am Chem Soc. 2008;130:5416–5417. doi: 10.1021/ja800164z. [DOI] [PubMed] [Google Scholar]; b) Takaoka Y, Sakamoto T, Tsukiji S, Narazaki M, Matsuda T, Tochio H, Shirakawa M, Hamachi I. Nat Chem. 2009;1:557–561. doi: 10.1038/nchem.365. [DOI] [PubMed] [Google Scholar]; c) Azagarsamy MA, Yesilyurt V, Thayumanavan S. J Am Chem Soc. 2010;132:4550–4551. doi: 10.1021/ja100746d. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Mizusawa K, Ishida Y, Takaoka Y, Masayoshi M, Tsukiji S, Hamachi I. J Am Chem Soc. 2010;132:7291–7293. doi: 10.1021/ja101879g. [DOI] [PubMed] [Google Scholar]
- 8.a) Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. J Controlled Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]; Baban DF, Seymour LW. Adv Drug Delivery Rev. 1998;34:109–119. doi: 10.1016/s0169-409x(98)00003-9. [DOI] [PubMed] [Google Scholar]; c) Gillies ER, Fréchet JMJ. Drug Discovery Today. 2005;10:35–43. doi: 10.1016/S1359-6446(04)03276-3. [DOI] [PubMed] [Google Scholar]
- 9.a) Minkenberg CB, Florusse L, Eelkema R, Koper GJM, van Esch JH. J Am Chem Soc. 2009;131:11274–11275. doi: 10.1021/ja902808q. [DOI] [PubMed] [Google Scholar]; b) Wang Y, Ma N, Wang Z, Zhang X. Angew Chem. 2007;119:2881–2884. [Google Scholar]; Angew Chem Int Ed. 2007;46:2823–2826. doi: 10.1002/anie.200604982. [DOI] [PubMed] [Google Scholar]; c) Ghosh S, Irvin K, Thayumanavan S. Langmuir. 2007;23:7916–7919. doi: 10.1021/la700981z. [DOI] [PubMed] [Google Scholar]; d) Orihara Y, Matsumura A, Saito Y, Ogawa N, Saji T, Yamaguchi A, Sakai H, Abe M. Langmuir. 2001;17:6072–6076. [Google Scholar]
- 10.a) Torchilin VP. Nat Rev Drug Discovery. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]; b) Andresen TL, Jensen SS, Kent J. Prog Lipid Res. 2005;44:68–97. doi: 10.1016/j.plipres.2004.12.001. [DOI] [PubMed] [Google Scholar]; c) Eliaz RE, Nir S, Marty C, Szoka FC. Cancer Res. 2004;64:711–718. doi: 10.1158/0008-5472.can-03-0654. [DOI] [PubMed] [Google Scholar]
- 11.a) Duncan R. Nat Rev Drug Discovery. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]; b) Savic R, Laibin L, Eisenberg A, Maysinger D. Science. 2003;300:615–618. doi: 10.1126/science.1078192. [DOI] [PubMed] [Google Scholar]; c) Discher DE, Eisenberg A. Science. 2002;297:967–973. doi: 10.1126/science.1074972. [DOI] [PubMed] [Google Scholar]; d) Basu S, Vutukuri DR, Shyamroy S, Sandanaraj BS, Thayumanavan S. J Am Chem Soc. 2004;126:9890–9891. doi: 10.1021/ja047816a. [DOI] [PubMed] [Google Scholar]
- 12.For some examples of amphiphilic dendrimers, see: Newkome GR, Moorefield CN, Baker GR, Johnson AL, Behera RK. Angew Chem. 1991;103:1205–1207.Angew Chem Int Ed Engl. 1991;30:1176–1178.Percec V, Wilson DA, Leowanawat P, Wilson CJ, Hughes AD, Kaucher MS, Hammer DA, Levine DH, Kim AJ, Bates FS, Davis KP, Lodge TP, Klein ML, DeVane RH, Aqad E, Rosen BM, Argintaru AO, Sienkowska MJ, Rissanen K, Nummelin S, Ropponen J. Science. 2010;328:1009–1014. doi: 10.1126/science.1185547.Rosen BM, Wilson CJ, Wilson DA, Peterca M, Imam MR, Percec V. Chem Rev. 2009;109:6275–6540. doi: 10.1021/cr900157q.Hawker CJ, Wooley KL, Fréchet JMJ. J Chem Soc Perkin Trans 1. 1993:1287.Vutukuri DR, Basu S, Thayumanavan S. J Am Chem Soc. 2004;126:15636–15637. doi: 10.1021/ja0449628.Joester D, Losson M, Pugin R, Heinzelmann H, Walter E, Merkle HP, Diederich F. Angew Chem. 2003;115:1524–1528. doi: 10.1002/anie.200250284.Angew Chem Int Ed. 2003;42:1486–1490. doi: 10.1002/anie.200250284.Ambade AV, Savariar EN, Thayumanavan S. Mol Pharm. 2005;2:264–272. doi: 10.1021/mp050020d.
- 13.a) Elegbede AI, Banerjee J, Hanson AJ, Tobwala S, Ganguli B, Wang R, Lu X, Srivastava DK, Mallik S. J Am Chem Soc. 2008;130:10633–10642. doi: 10.1021/ja801548g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Boyer C, Zasadzinski JA. ACS Nano. 2007;1:176–182. doi: 10.1021/nn7002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Yoo HS, Park TG. J Controlled Release. 2004;96:273–283. doi: 10.1016/j.jconrel.2004.02.003. [DOI] [PubMed] [Google Scholar]; b) Lavasanifar A, Samuel J, Kwon GS. Adv Drug Delivery Rev. 2002;54:169–190. doi: 10.1016/s0169-409x(02)00015-7. [DOI] [PubMed] [Google Scholar]; c) Gaucher G, Dufresne MH, Sant VP, Kang N, Maysinger D, Leroux JC. J Controlled Release. 2005;109:169–188. doi: 10.1016/j.jconrel.2005.09.034. [DOI] [PubMed] [Google Scholar]; d) Kataoka K, Harada A, Nagasaki Y. Adv Drug Delivery Rev. 2001;47:113–131. doi: 10.1016/s0169-409x(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 15.a) Jansen JFGA, de Brabandervan den Berg EMM, Meijer EW. Science. 1994;266:1226–1229. doi: 10.1126/science.266.5188.1226. [DOI] [PubMed] [Google Scholar]; b) Astruc D, Boisselier E, Ornelas C. Chem Rev. 2010;110:1857–1959. doi: 10.1021/cr900327d. [DOI] [PubMed] [Google Scholar]; c) Grayson SM, Fréchet JMJ. Chem Rev. 2001;101:3819–3867. doi: 10.1021/cr990116h. [DOI] [PubMed] [Google Scholar]; d) Menjoge AR, Kannan RM, Tomalia DA. Drug Discovery Today. 2010;15:171–185. doi: 10.1016/j.drudis.2010.01.009. [DOI] [PubMed] [Google Scholar]; e) Kose MM, Yesilbag G, Sanyal A. Org Lett. 2008;10:2353–2356. doi: 10.1021/ol800553t. [DOI] [PubMed] [Google Scholar]; f) Avital-Shmilovici M, Shabat D. Soft Matter. 2010;6:1073–1080. [Google Scholar]
- 16.a) Shamis M, Lode HN, Shabat D. J Am Chem Soc. 2004;126:1726–1731. doi: 10.1021/ja039052p. [DOI] [PubMed] [Google Scholar]; b) Azagarsamy MA, Sokkalingam P, Thayumanavan S. J Am Chem Soc. 2009;131:14184–14185. doi: 10.1021/ja906162u. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Amir RJ, Zhong S, Pochan DJ, Hawker CJ. J Am Chem Soc. 2009;131:13949–13951. doi: 10.1021/ja9060917. [DOI] [PubMed] [Google Scholar]; d) Sarkar N, Banerjee J, Hanson AJ, Elegbede AI, Rosendahl T, Krueger AB, Banerjee AL, Tobwala S, Wang R, Lu X, Mallik S, Srivastava DK. Bioconjugate Chem. 2008;19:57–64. doi: 10.1021/bc070081p. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Escudero A, Azagarsamy MA, Theddu N, Vachet RW, Thayumanavan S. J Am Chem Soc. 2008;130:11156–11163. doi: 10.1021/ja803082v. [DOI] [PubMed] [Google Scholar]
- 18.a) Bilgiçer B, Moustakas DT, Whitesides GM. J Am Chem Soc. 2007;129:3722–3728. doi: 10.1021/ja067159h. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bilgiçer B, Thomas SW, Shaw BF, Kaufman GK, Krishnamurthy VM, Estroff LA, Yang J, Whitesides GM. J Am Chem Soc. 2009;131:9361–9367. doi: 10.1021/ja9023836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Sandanaraj BS, Demont R, Aathimanikandan SV, Savariar EN, Thayumanavan S. J Am Chem Soc. 2006;128:10686–10687. doi: 10.1021/ja063544v. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sandanaraj BS, Demont R, Thayumanavan S. J Am Chem Soc. 2007;129:3506–3507. doi: 10.1021/ja070229f. [DOI] [PubMed] [Google Scholar]; c) González DC, Savariar EN, Thayumanavan S. J Am Chem Soc. 2009;131:7708–7716. doi: 10.1021/ja900579g. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Savariar EN, Koppelman J, Thayumanavan S. Isr J Chem. 2009;49:41–47. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.