Abstract
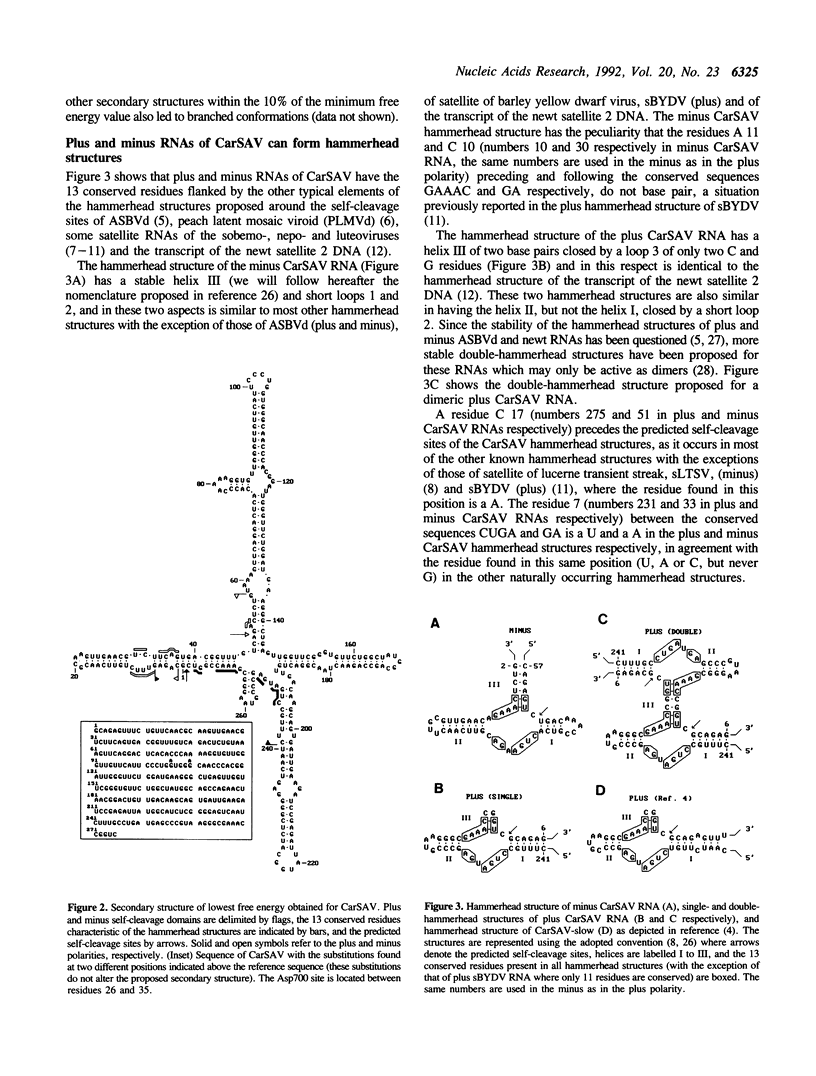
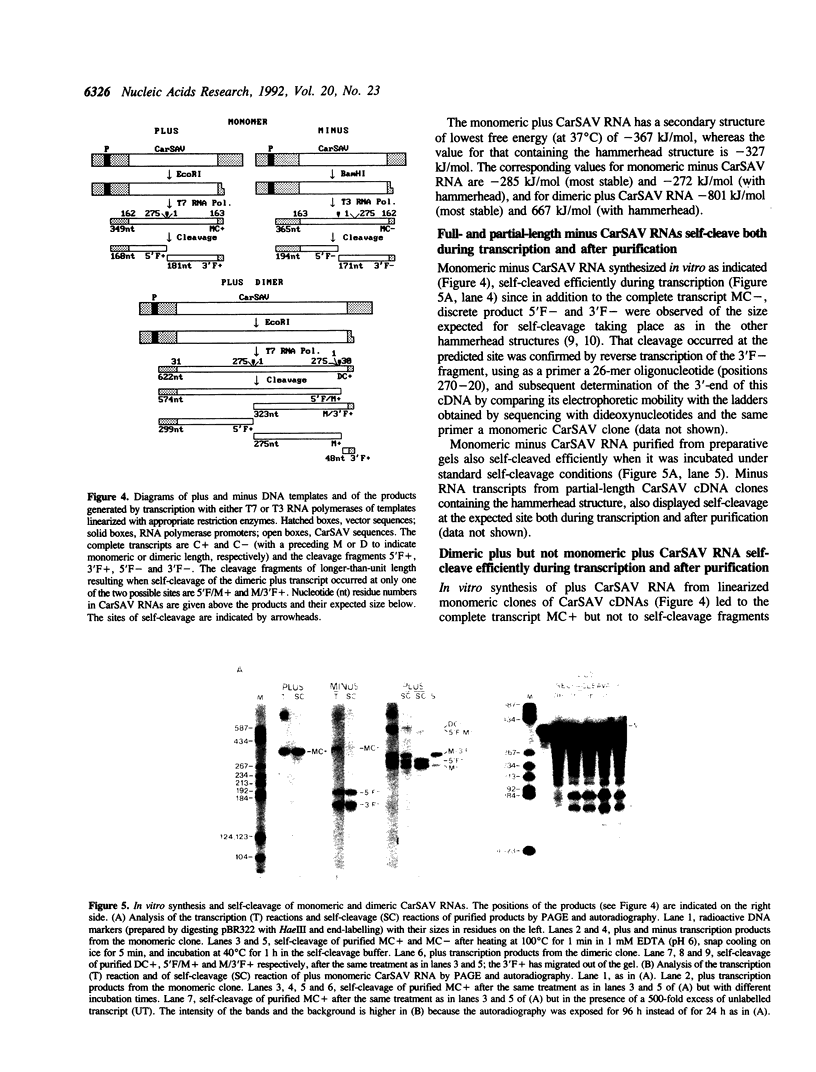
The sequence of a circular RNA from carnation has been determined and found to consist of 275 nucleotide residues adopting a branched secondary structure of minimum free energy. Both plus and minus strands of this RNA can form the hammerhead structures proposed to mediate the in vitro self-cleavage of a number of small infectious plant RNAs and the transcript of satellite 2 DNA from the newt. Minus full- and partial-length transcripts of the carnation circular RNA including the hammerhead structure showed self-cleavage during transcription and after purification, indicating the involvement of a single-hammerhead structure in the self-cleavage reaction. In the case of the plus transcripts only a dimeric RNA, but not a monomeric one, self-cleaved efficiently during transcription and after purification, strongly supporting the implication in this process of a double-hammerhead structure theoretically more stable than the corresponding single cleavage domain. However, a plus monomeric transcript self-cleaved after purification at a slow rate in a concentration-independent reaction which most probably occurs through an intramolecular mechanism. Comparative sequence analysis has revealed that the circular RNA from carnation shares similarities with some representative members of the viroid and viroid-like satellites RNAs from plants, suggesting that it is a new member of either these two groups of small pathogenic RNAs.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruening G. Compilation of self-cleaving sequences from plant virus satellite RNAs and other sources. Methods Enzymol. 1989;180:546–558. doi: 10.1016/0076-6879(89)80123-5. [DOI] [PubMed] [Google Scholar]
- Chaconas G., van de Sande J. H. 5'-32P labeling of RNA and DNA restriction fragments. Methods Enzymol. 1980;65(1):75–85. doi: 10.1016/s0076-6879(80)65012-5. [DOI] [PubMed] [Google Scholar]
- Davies C., Sheldon C. C., Symons R. H. Alternative hammerhead structures in the self-cleavage of avocado sunblotch viroid RNAs. Nucleic Acids Res. 1991 Apr 25;19(8):1893–1898. doi: 10.1093/nar/19.8.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena S. F., Dopazo J., Flores R., Diener T. O., Moya A. Phylogeny of viroids, viroidlike satellite RNAs, and the viroidlike domain of hepatitis delta virus RNA. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5631–5634. doi: 10.1073/pnas.88.13.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein L. M., Gall J. G. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987 Feb 13;48(3):535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- Epstein L. M., Pabón-Peña L. M. Alternative modes of self-cleavage by newt satellite 2 transcripts. Nucleic Acids Res. 1991 Apr 11;19(7):1699–1705. doi: 10.1093/nar/19.7.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Davies C., Hutchins C. J., Symons R. H. Characterization of self-cleavage of viroid and virusoid RNAs. Methods Enzymol. 1990;181:583–607. doi: 10.1016/0076-6879(90)81153-l. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Davies C., Sheldon C. C., Jeffries A. C., Symons R. H. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature. 1988 Jul 21;334(6179):265–267. doi: 10.1038/334265a0. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987 Jul 3;50(1):9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Symons R. H. Chrysanthemum stunt viroid: primary sequence and secondary structure. Nucleic Acids Res. 1981 Jun 25;9(12):2741–2752. doi: 10.1093/nar/9.12.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández C., Flores R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3711–3715. doi: 10.1073/pnas.89.9.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel K. J., Pardi A., Uhlenbeck O. C., Koizumi M., Ohtsuka E., Uesugi S., Cedergren R., Eckstein F., Gerlach W. L., Hodgson R. Numbering system for the hammerhead. Nucleic Acids Res. 1992 Jun 25;20(12):3252–3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins C. J., Rathjen P. D., Forster A. C., Symons R. H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986 May 12;14(9):3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow A. M., Rezaian M. A. Grapevine yellow speckle viroid: structural features of a new viroid group. Nucleic Acids Res. 1988 Feb 11;16(3):849–864. doi: 10.1093/nar/16.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. A., Hercus T., Waterhouse P. M., Gerlach W. L. A satellite RNA of barley yellow dwarf virus contains a novel hammerhead structure in the self-cleavage domain. Virology. 1991 Aug;183(2):711–720. doi: 10.1016/0042-6822(91)91000-7. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Silver S. L. Alternative tertiary structure attenuates self-cleavage of the ribozyme in the satellite RNA of barley yellow dwarf virus. Nucleic Acids Res. 1991 Oct 11;19(19):5313–5320. doi: 10.1093/nar/19.19.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Peña L. M., Zhang Y., Epstein L. M. Newt satellite 2 transcripts self-cleave by using an extended hammerhead structure. Mol Cell Biol. 1991 Dec;11(12):6109–6115. doi: 10.1128/mcb.11.12.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prody G. A., Bakos J. T., Buzayan J. M., Schneider I. R., Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986 Mar 28;231(4745):1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J., Randles J. W., Riesner D. A two-dimensional electrophoretic technique for the detection of circular viroids and virusoids. Anal Biochem. 1983 Dec;135(2):288–295. doi: 10.1016/0003-2697(83)90685-1. [DOI] [PubMed] [Google Scholar]
- Symons R. H. Self-cleavage of RNA in the replication of small pathogens of plants and animals. Trends Biochem Sci. 1989 Nov;14(11):445–450. doi: 10.1016/0968-0004(89)90103-5. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]