Abstract
Molecular mechanics has been used to predict the structure of the Y+.R-.R(+)-type DNA triple helix, in which a second polypurine strand binds antiparallel to the homopurine strand of a homopurine/homopyrimidine stretch of duplex DNA. From calculations on the sequence d(C)10.d(G)10.d(G)10, two likely structures emerge. One has the glycosidic torsions of the third strand bases in the anti-conformation and Hoogsteen hydrogen-bonds to the purine strand of the duplex, the other has the third strand purines in the syn orientation and uses a reverse-Hoogsteen hydrogen-bonding pattern. Despite the large structural differences between these two types of triplex, calculations performed in vacuo with a distance-dependent dielectric constant to mimic the shielding effect of solvent show them to be energetically very similar, with the latter (syn) slightly preferred. However, if explicit solvent molecules are included in the calculation, the anti conformation is found to be much preferred. This difference in the results seems to stem from an underestimation of short-range electrostatic interactions in the in vacuo simulations. When TAA or TAT base triples are substituted for the sixth CGG triple in the sequence, it is found that, for the solvated model, the third strand base of the TAA triple prefers the syn orientation while that in the TAT triple retains a preference, though reduced, for the anti conformation.
Full text
PDF
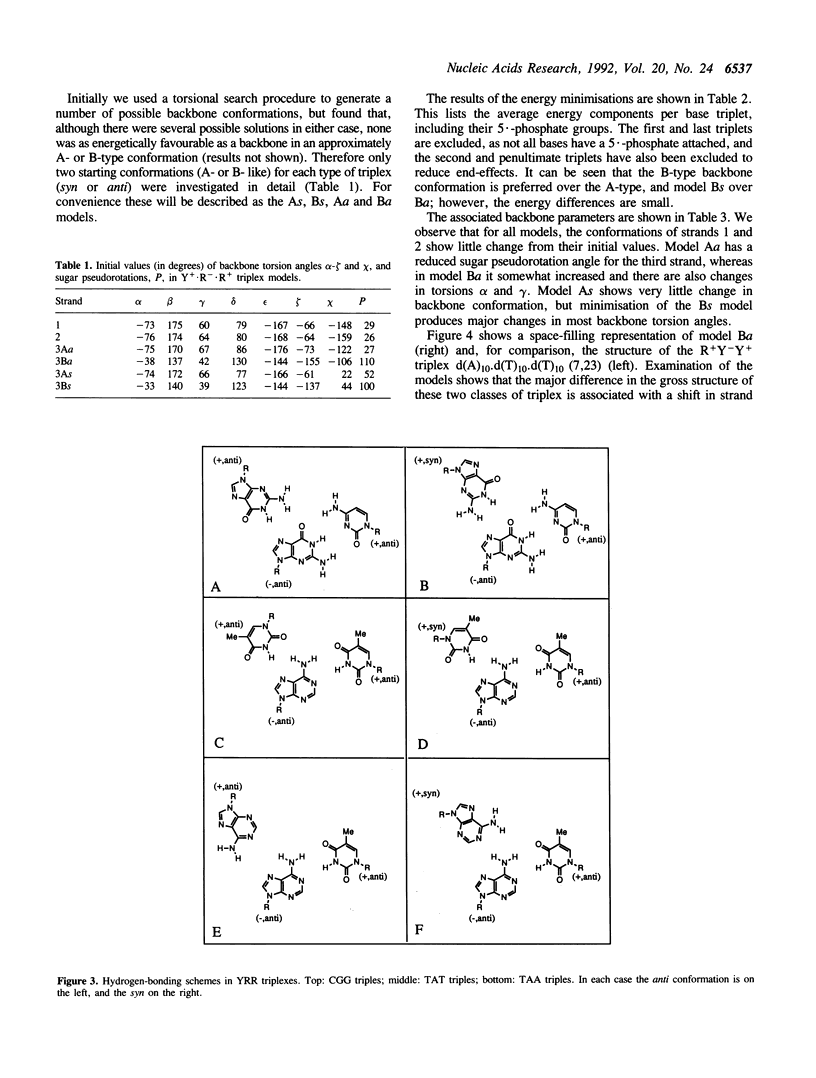
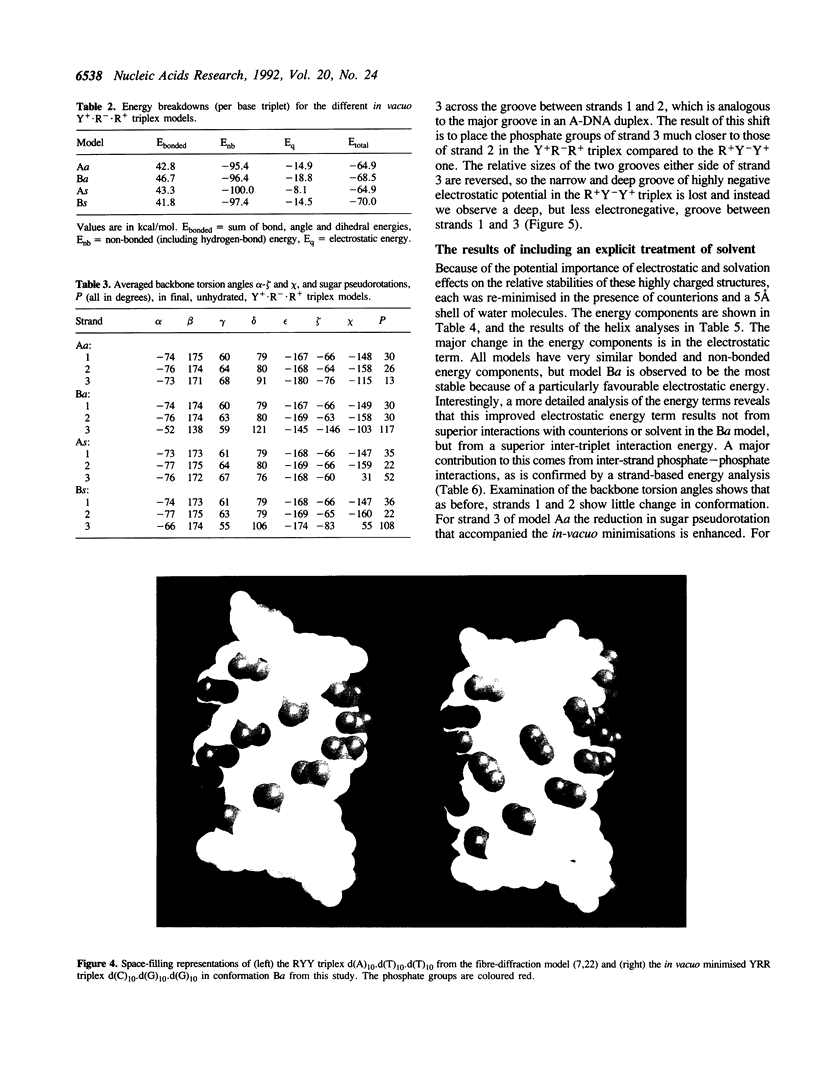
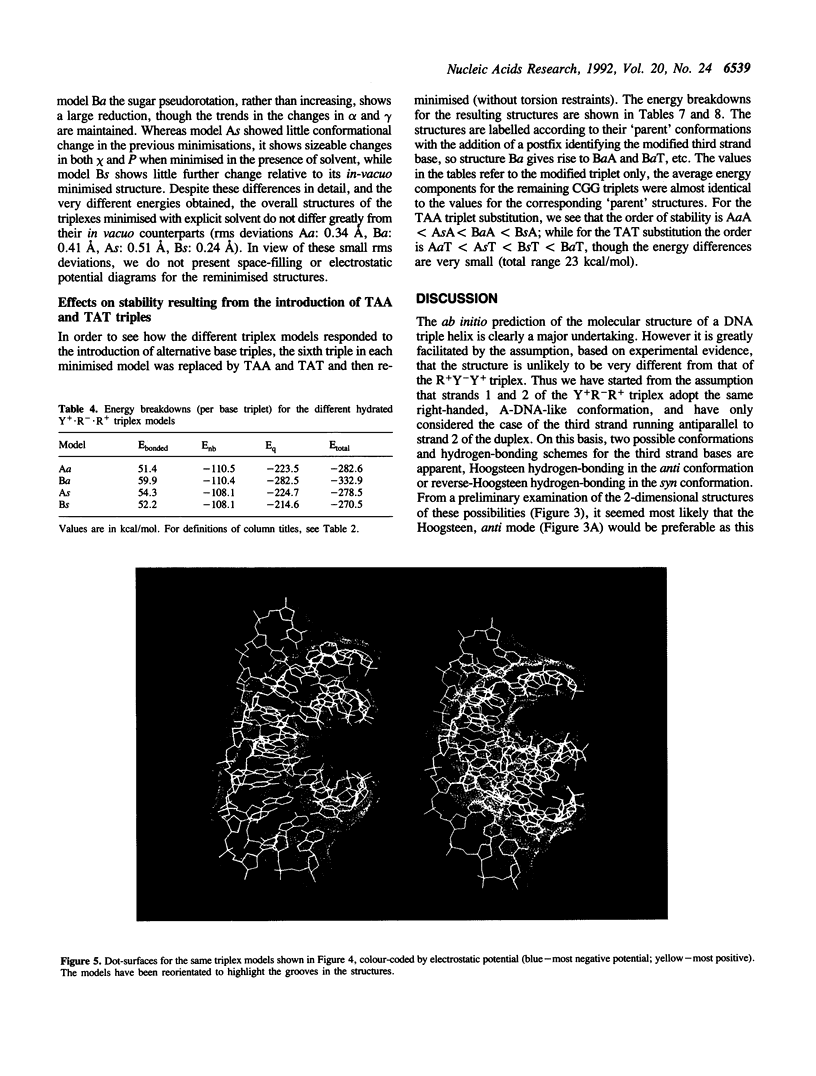
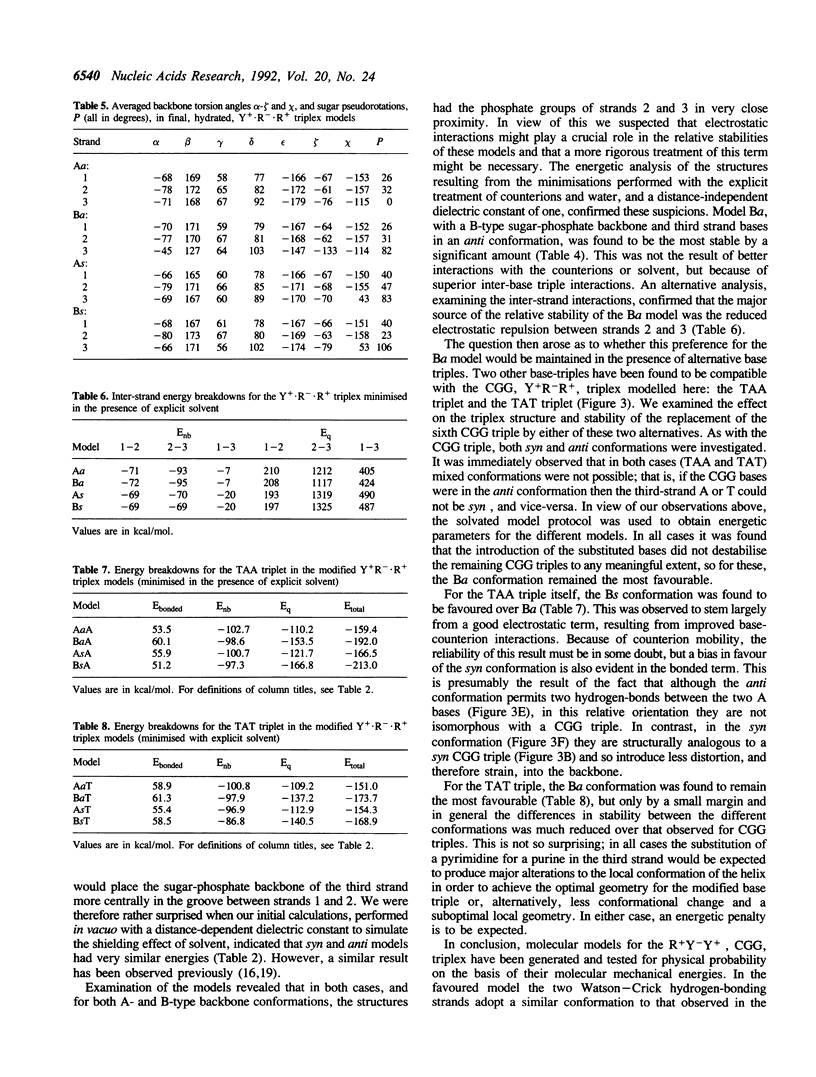

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Broitman S. L., Im D. D., Fresco J. R. Formation of the triple-stranded polynucleotide helix, poly(A.A.U). Proc Natl Acad Sci U S A. 1987 Aug;84(15):5120–5124. doi: 10.1073/pnas.84.15.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durland R. H., Kessler D. J., Gunnell S., Duvic M., Pettitt B. M., Hogan M. E. Binding of triple helix forming oligonucleotides to sites in gene promoters. Biochemistry. 1991 Sep 24;30(38):9246–9255. doi: 10.1021/bi00102a017. [DOI] [PubMed] [Google Scholar]
- Grigoriev M., Praseuth D., Robin P., Hemar A., Saison-Behmoaras T., Dautry-Varsat A., Thuong N. T., Hélène C., Harel-Bellan A. A triple helix-forming oligonucleotide-intercalator conjugate acts as a transcriptional repressor via inhibition of NF kappa B binding to interleukin-2 receptor alpha-regulatory sequence. J Biol Chem. 1992 Feb 15;267(5):3389–3395. [PubMed] [Google Scholar]
- Jayasena S. D., Johnston B. H. Intramolecular triple-helix formation at (PunPyn).(PunPyn) tracts: recognition of alternate strands via Pu.PuPy and Py.PuPy base triplets. Biochemistry. 1992 Jan 21;31(2):320–327. doi: 10.1021/bi00117a002. [DOI] [PubMed] [Google Scholar]
- Johnson K. H., Durland R. H., Hogan M. E. The vacuum UV CD spectra of G.G.C triplexes. Nucleic Acids Res. 1992 Aug 11;20(15):3859–3864. doi: 10.1093/nar/20.15.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi Y., Kohwi-Shigematsu T. Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3781–3785. doi: 10.1073/pnas.85.11.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughton C. A., Neidle S. Molecular dynamics simulation of the DNA triplex d(TC)5.d(GA)5.d(C+T)5. J Mol Biol. 1992 Jan 20;223(2):519–529. doi: 10.1016/0022-2836(92)90667-9. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Orozco M., Laughton C. A., Herzyk P., Neidle S. Molecular-mechanics modelling of drug-DNA structures; the effects of differing dielectric treatment on helix parameters and comparison with a fully solvated structural model. J Biomol Struct Dyn. 1990 Oct;8(2):359–373. doi: 10.1080/07391102.1990.10507810. [DOI] [PubMed] [Google Scholar]
- Pilch D. S., Levenson C., Shafer R. H. Structural analysis of the (dA)10.2(dT)10 triple helix. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1942–1946. doi: 10.1073/pnas.87.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch D. S., Levenson C., Shafer R. H. Structure, stability, and thermodynamics of a short intermolecular purine-purine-pyrimidine triple helix. Biochemistry. 1991 Jun 25;30(25):6081–6088. doi: 10.1021/bi00239a001. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Gao X., de los Santos C., Live D., Patel D. J. NMR structural studies of intramolecular (Y+)n.(R+)n(Y-)nDNA triplexes in solution: imino and amino proton and nitrogen markers of G.TA base triple formation. Biochemistry. 1991 Sep 17;30(37):9022–9030. doi: 10.1021/bi00101a016. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., de los Santos C., Patel D. J. Nuclear magnetic resonance structural studies of intramolecular purine.purine.pyrimidine DNA triplexes in solution. Base triple pairing alignments and strand direction. J Mol Biol. 1991 Oct 20;221(4):1403–1418. [PubMed] [Google Scholar]
- Rajagopal P., Feigon J. NMR studies of triple-strand formation from the homopurine-homopyrimidine deoxyribonucleotides d(GA)4 and d(TC)4. Biochemistry. 1989 Sep 19;28(19):7859–7870. doi: 10.1021/bi00445a048. [DOI] [PubMed] [Google Scholar]
- Van Vlijmen H. W., Ramé G. L., Pettitt B. M. A study of model energetics and conformational properties of polynucleotide triplexes. Biopolymers. 1990;30(5-6):517–532. doi: 10.1002/bip.360300505. [DOI] [PubMed] [Google Scholar]
- de los Santos C., Rosen M., Patel D. NMR studies of DNA (R+)n.(Y-)n.(Y+)n triple helices in solution: imino and amino proton markers of T.A.T and C.G.C+ base-triple formation. Biochemistry. 1989 Sep 5;28(18):7282–7289. doi: 10.1021/bi00444a021. [DOI] [PubMed] [Google Scholar]