Abstract
BIMD of Aspergillus nidulans belongs to a highly conserved protein family implicated, in filamentous fungi, in sister-chromatid cohesion and DNA repair. We show here that BIMD is chromosome associated at all stages, except from late prophase through anaphase, during mitosis and meiosis, and is involved in several aspects of both programs. First, bimD+ function must be executed during S through M. Second, in bimD6 germlings, mitotic nuclear divisions and overall cellular program occur more rapidly than in wild type. Thus, BIMD, an abundant chromosomal protein, is a negative regulator of normal cell cycle progression. Third, bimD6 reduces the level of mitotic interhomolog recombination but does not alter the ratio between crossover and noncrossover outcomes. Moreover, bimD6 is normal for intrachromosomal recombination. Therefore, BIMD is probably not involved in the enzymology of recombinational repair per se. Finally, during meiosis, staining of the Sordaria ortholog Spo76p delineates robust chromosomal axes, whereas BIMD stains all chromatin. SPO76 and bimD are functional homologs with respect to their roles in mitotic chromosome metabolism but not in meiosis. We propose that BIMD exerts its diverse influences on cell cycle progression as well as chromosome morphogenesis and recombination by modulating chromosome structure.
Cell division results from an integrated sequence of events, occurring in an orderly way, to ensure faithful transmission of the genetic information to the next generation. As part of this process, chromosome morphogenesis and progression of other cellular events occur coordinately. The exact relationship between the chromosomal cycle and cell cycle regulation remains poorly understood.
It is clear that cell cycle molecules influence chromosome behavior (review in ref. 1). The opposite is likely also true, not only during “checkpoint” responses but also in a normal unperturbed mitotic cell (see review, ref. 2). Notably, several key chromosomal proteins have been implicated in regulation of cell cycle progression. The human homologs of the Pds1/Cut2p securin and of the Spo76/BIMD/AS3 family of chromosomal proteins have been identified as possible tumor suppressors (3, 4). Correspondingly, androgen-induced G1 arrest in human is mediated by up-regulation of AS3 (5), and overexpression of bimD confers arrest in G1 (6). Also, overexpression of the mouse homolog of the Mcd1/Scc1/Rad21p cohesin leads to inhibition of fibroblast proliferation (7).
The evolutionarily conserved Spo76 family includes Spo76p of Sordaria macrospora (4, 8), BIMD of Aspergillus nidulans (6), Pds5p of budding yeast (9, 10), as well as AS3 and hPDS5 of human (5, 11). Spo76p is essential for mitotic and meiotic chromosome morphogenesis and is associated with the chromosomes in both programs (4). All three fungal proteins are implicated in maintenance of sister-chromatid cohesion (4, 6, 8–10). Moreover, the mitotic catastrophe defect of bimD6 is suppressed by mutations in the Aspergillus homolog of the cohesin Smc3 (12).
To gain further insight into the function and behavior of Spo76 family proteins with respect to both chromosome morphogenesis and regulation of cell cycle progression, we have used the filamentous fungus A. nidulans, which is particularly well suited for such analyses (13, 14). We have followed the chromosomal localization of BIMD in mitosis and meiosis. In the mitotic program, we have analyzed execution points and examined effects of bimD6 on cell cycle progression and spontaneous recombination. Finally, we tested bimD and SPO76 for heterologous complementation of mutant defects.
Materials and Methods
Strains.
Because all laboratory strains of the homothallic A. nidulans are derived from a single nucleus, they are essentially isogenic (15). All bimD6 strains used were progeny of strain D6.9 riboA1; sC12; bimD6 pyroA4 (6) and are available (as A1063) from the Fungal Genetics Stock Center (University of Kansas Medical Center, Kansas City, KS). A cross of A1064 (F1 of A1063) to FGSC strain A733, pyrG89; wA3; pyroA4 produced six strains: pyroA4, wA3; pyroA4, pyrG89; pyroA4, and three corresponding bimD6 strains. Quantitative assessment of complementation by SPO76 was performed in two additional pyrG89 bimD6 strains (A1061 and A1062). The recipient strain for transformation of S. macrospora was spo76–1 (4). For bimD6 strains, permissive temperature was 25 or 30°C, and restrictive temperature was 42°C.
Mitotic allelic recombination in adE8/adE20 (16) was examined in isogenic strains homozygous for bimD+ or bimD6 (for genotypes, see Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org). For intrachromosomal recombination, suitable pyrG89; bimD6 and pyrG89; uvsC114 benA duplication strains were obtained by crosses to strain IS88 (17).
Media and Genetic Procedures.
Standard Aspergillus media and genetic techniques (15), modifications of the media, methods of mitotic mapping, congenic strain construction (18), and procedures for genetic analysis of DNA repair mutants and of their effects on mitotic recombination in diploids were all as described previously (16). For tests of intrachromosomal recombination, the benA interrupted duplication system (17) was adapted for visual assessment of recombination frequencies.
Transformation.
The Aspergillus protocols for protoplasting and transformation by using pyrG89 as the selective marker were modifications of refs. 19 and 20. Recipient pyrG89 strains were grown in liquid YG medium (0.5% yeast extract, 2% glucose, and trace elements) supplemented with vitamins, 10 mM uracil, and 5 mM uridine (UU). Putative pyr+ transformants were selected on solid YG medium (with 2% agar in bottom, 1% in overlays, 1 M sucrose, without UU), either at 30°C or by selection for pyr+ and ts+ at 42°C. Transformation procedures for S. macrospora were as in ref. 4.
Plasmids.
We used: pANscos1 (HygR; ref. 21); pPL6 (=pPyrG; ref. 22); pRG3 (pyr-4; ref. 23); pBimD1 (bimD; ref. 6); pDH1 (SPO76) and pDH13 (SPO76-GFP; ref. 4). For the construction of pGW1454, a 6.7-kb NcoI-SmaI fragment from pBimD1 was cloned into pBR328 EcoRV-NcoI; for pGW1460, enhanced green fluorescent protein (EGFP) was amplified from pEGFP-1 (CLONTECH) and inserted into the XhoI site of pGW1454; for pGW1463, a 1.4-kb 3′-BglII/XhoI bimD cDNA fragment was cloned into the expression vector pQE32 (Qiagen, Chatsworth, CA) BamHI-SalI. The Escherichia coli strain DH5α was used as a host in plasmid propagation (24).
Sequencing.
A 1.2-kb fragment spanning the bimD6 mutation was amplified by PCR by using gene-specific primers on genomic DNA from strains WG542 and WG540. Genomic DNA was isolated as described (25). PCR products were sequenced directly with gene-specific primers (4). For DNA sequencing, we used the Dyedeoxy Terminator, Cycle Sequencing kit (Applied Biosystems) and a 373 DNASequencer (Applied Biosystems).
Cytogenetic Methods.
For mitotic and cell cycle analyses, conidiospores were either grown in a drop of supplemental minimal medium on a slide or inoculated (5 × 106 per ml) in SM with 50 mM hydroxy urea (HU) and incubated for 13 h at 25°C. After HU treatment, cells were washed twice with SM, and 10 μl samples were taken. C source was glucose (1%) or lactose (3%); N source was 50 mM urea or ammonium. Nuclei were stained with the DNA-specific dye 4,6-diamidino-2-phenylindole (DAPI). For meiotic analyses, fruiting bodies were mechanically squashed with a blunted metal needle.
Cells were processed for immunofluorescence as described for Sordaria (26). For BIMD antiserum, E. coli SG13009 cells (Qiagen) were transformed with plasmid pGW1463. After induction with isopropyl-β-d-thiogalactopyranoside, the fusion protein was purified from the bacterial cell lysate by affinity chromatography on a nickel column (Qiagen) and dialyzed against PBS (140 mM NaCl/10 mM sodium phosphate, pH 7.3). Antiserum 560 was raised against the fusion protein in a rabbit, affinity-purified as described (27), and used in a dilution corresponding to 1:200 diluted serum. Secondary antibody was CyTM3 anti-rabbit (Jackson ImmunoResearch), diluted 1:4,000. Cells incubated with primary or secondary antibodies alone gave no signal. Three BIMD-GFP transformants (of WG546 with pGW1460) were analyzed. Cells were observed on a Zeiss Axioplan microscope with images captured by a charge-coupled device Princeton camera.
Western Blot Analysis.
For crude protein extracts of A. nidulans, mycelium grown at permissive temperature was powdered in liquid nitrogen in the presence of the complete miniprotease inhibitor mixture (Boehringer, no. 1836153), boiled in electrophoresis sample buffer, and centrifuged. Proteins extracted from 7 mg of mycelium were loaded per 3.5-cm-wide slot of a 10% polyacrylamide gel and electrophoresed in parallel with molecular weight markers (Bio-Rad). Of each lane, 0.5 cm was stained with Coomassie blue, and the remainder was transferred to nitrocellulose. After immunoblotting, markers and mycelial proteins were detected with Ponceau S. Binding of BIMD antibodies to 0.3-cm-wide strips was detected by goat–anti-rabbit secondary antibodies conjugated to alkaline phosphatase and incubation in nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate, as described (27).
Results
BIMD Localizes to Chromosomes During Mitosis and Meiosis.
We determined the localization of BIMD by using both affinity-purified antibodies and a fully functional BIMD-GFP-tagged derivative. The antibody preparation recognizes a single prominent protein species of ≈170 kDa in wild-type extracts, corresponding to a predicted BIMD molecular mass of 166 kDa. The corresponding signal is absent in bimD6 (Fig. 1A); thus, the antibodies are specific for BIMD.
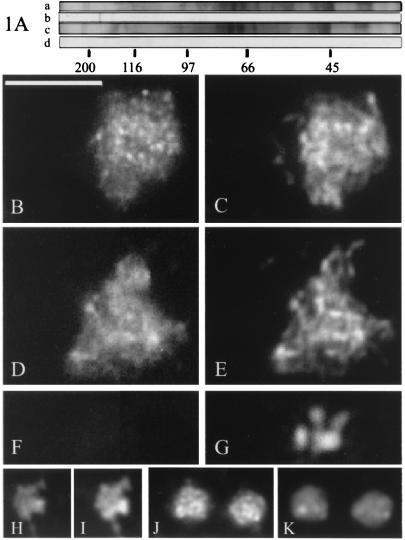
Figure 1.
Localization of BIMD. (A) Immunoblot analysis of proteins extracted from wild-type (lanes a and b) and bimD6 (lanes c and d) mycelium, by using antibodies against the C terminus of BIMD. Lanes a and c, Coomassie blue-stained gels; lanes b and d, corresponding immunoblots probed with affinity-purified anti-BIMD antibodies from serum 560. Molecular size standards are indicated below lane d in kilodaltons. (B–E) Wild-type early meiotic prophases (leptotene/zygotene): (B) stained with affinity-purified anti-BIMD antibodies from serum 560; (C) corresponding DAPI; (D) stained with GFP; and (E) corresponding DAPI. (F and G) Metaphase I nucleus stained with (F) GFP and (G) DAPI. (H–K) Mitotic prophase nuclei in wild-type germlings stained with (H) GFP and (J) anti-BIMD antibodies. I and K are the corresponding DAPI. (Bars = 5 μm.)
By both approaches, BIMD was observed exclusively in nuclei and, within those nuclei, specifically on chromatin. During meiosis, BIMD is chromosome-associated during prophase I (e.g., Fig. 1 B–E) but disappears from the chromosomes as they emerge from the diffuse stage into diplotene (e.g., at metaphase I, Fig. 1 F and G). It reappears on the chromosomes at telophase(s). Likewise, during mitosis, BIMD dissociates from chromosomes in prometaphase and reassociates at telophase. BIMD was present on the chromatin in resting conidia (G0 or G1), in HU arrested cultures (S phase) and in mitotically cycling nuclei (500 germlings observed) (Fig. 1 H–K). However, slight differences in staining intensity were discernible at different stages (e.g., brighter in S than in G1/G0). In dividing nuclei of mitosis and meiosis, antibody staining detects numerous tiny foci, whereas GFP staining seems more continuous (Fig. 1, compare B and H with D and J). These minor differences might reflect different sensitivities of the two techniques. No BIMD-antibody staining was detected in bimD6 resting or dividing nuclei at either temperature.
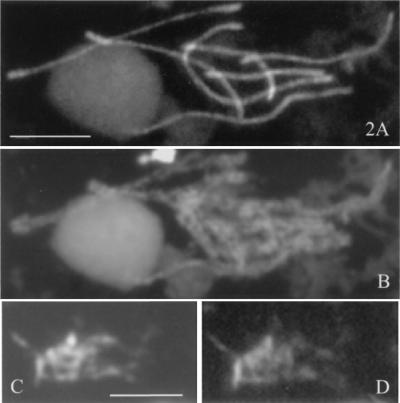
The temporal pattern of BIMD localization to chromosomes in A. nidulans is identical to that found for Spo76p in S. macrospora (4). However, two interesting qualitative differences in staining patterns are observed, both during meiotic prophase. First, in Sordaria, prophase nuclei are more brightly stained than any other nuclei of either vegetative or sexual cycles (4), which is not the case in Aspergillus (Fig. 1, compare D and J). Second, Spo76p is preferentially localized along the chromosome axes at prophase (Fig. 2 A and B), whereas BIMD staining does not reveal any axes but instead stains all chromatin (Fig. 2 C and D).
Figure 2.
(A) Spread pachytene nucleus of S. macrospora: Spo76-GFPp decorates the axes of the synapsed homologs (gray ball, nucleolus). (B) Corresponding DAPI. (C) DAPI of a spread A. nidulans pachytene. (D) Corresponding BIMD-GFP stains the entire chromatin mass. (Bars = 5 μm.)
Unexpected Nature of the bimD6 Mutation.
bimD is an essential gene, and bimD6 was mapped to its 5′ region (6). Sequence analysis of this region in bimD6 and wild type revealed a single sequence difference: a T to A mutation at position 420 in the ORF, which changes the codon TAT (Tyr) to TAA (Stop). The most straightforward consequence would be the production of a short truncated protein (≈15 kDa) containing the first 140 N-terminal amino acids. Translational readthrough or reinitiation could give a longer polypeptide (reviewed by ref. 28); however, antibodies against (approximately) the C-terminal third of the BIMD protein do not detect any prominent new polypeptide in extracts of bimD6 (above). Thus, residual function of the BIMD6 protein might reside in the short N-terminal region or be provided by a rather low level of an (undetected) longer protein. Alternatively, Aspergillus BIMD may be essential only at high temperature (analogous to pds1; ref. 29).
Meiotic Defects of bimD6.
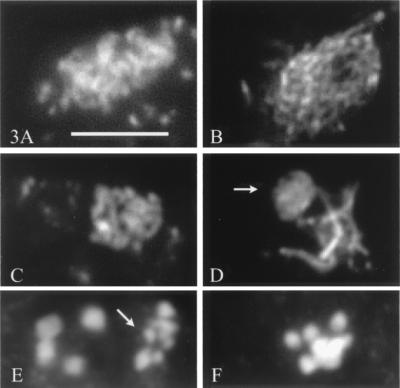
At permissive temperature, bimD6 fruiting bodies are comparable to those of wild type in number and morphology, but they remain barren. Wild-type fruiting bodies contain hundreds of asci, within which meiosis occurs. Mutant fruiting bodies contain only few asci (average 10), which indicates that bimD function is required during premeiosis. Moreover, bimD6 asci are abnormal in several respects. (i) During prophase I, the chromatin appears always more diffuse in bimD6 than in wild-type nuclei (Fig. 3, compare A and B), and a typical pachytene stage with orderly condensed chromosomes is not observed (Fig. 3, compare C and D). (ii) Thirty-three percent of all observed asci (65/200) were in metaphase I (a stage rarely seen in wild type, probably because of its short duration), and only 3% progressed beyond metaphase I, even in older fruiting bodies. The remaining 64% asci were at pachytene or diffuse stage. (iii) Metaphase I nuclei exhibit higher numbers of chromatin units than the eight bivalents seen in wild type (Fig. 3, compare E and F). These units should correspond either to prematurely separated homologs and/or to precociously separated sister chromatids. Because of their small sizes (arrow in Fig. 3E), the latter is more likely. However, all analyzed metaphase I nuclei also contained some regularly sized bivalents (Fig. 3E Left).
Figure 3.
Progression of meiosis in bimD6 and wild-type asci. All nuclei are stained with DAPI. (A) Early bimD6 prophase. (B) Early wild-type prophase. (C) bimD6 pachytene. (D) Wild-type pachytene; arrow indicates the nucleolar organizer bivalent. (E) Spread metaphase I nucleus of bimD6. Note the small size of some of the chromatin units (arrow) when compared with normal bivalents (four seen, Left). (F) Wild-type metaphase I (four clearly separated and four overlapping bivalents). (Bar = 5 μm.)
BimD6+ Function Is Required During Early S Through M and Not in G1 Phases of the Mitotic Cycle.
Execution points for bimD6+ function were evaluated by germination tests of haploid conidia (in G1/G0) by using HU arrest in combination with temperature shifts.
HU arrest per se has no effect on the mutant phenotype at either temperature; the same is true for wild-type conidia. In bimD6 conidia maintained in HU at permissive temperature for 13 and 19 h and then released at permissive temperature, all ensuing mitoses were normal (500 analyzed). When conidia were maintained in HU at restrictive temperature throughout (13 h) and then released at restrictive temperature, they exhibited the same catastrophic mitosis observed in the absence of HU treatment (300 analyzed). For HU arrest in combination with temperature shifts, four different conditions were tested. (i) In conidia arrested with HU at permissive temperature and then shifted to restrictive temperature on release of arrest, all mitoses (n > 400) were catastrophic in bimD6 but not in wild type, indicating the existence of an essential function(s) for bimD during early S through M. (ii) When bimD6 conidia were arrested at restrictive temperature with HU (13 h), then shifted to permissive temperature still in HU (6 h), and then released at permissive temperature, almost all (over 95%) ensuing mitoses were normal (200 analyzed). Thus BIMD does not play an essential role before the HU-arrest point. (iii) When bimD6 conidia were arrested at restrictive temperature with HU for 13 h and then released at permissive temperature, almost all (over 95%) ensuing mitoses were catastrophic (200 analyzed). These two latter experiments together indicate that BIMD function is irreversibly inactivated by high temperature. (iv) When arrested at restrictive temperature with HU for a longer time (19 h) and then released at permissive temperature, almost all conidia were blocked earlier, before the onset of mitosis (300 analyzed): this experiment indicates that BIMD may play a role during S phase or HU arrest.
BIMD Is a Negative Regulator of Cell Cycle Progression.
We also asked whether BIMD might have a general role in cell cycle regulation. Aspergillus germination tubes are well suited for analysis of cell cycle kinetics (14). Each tube is a single cell that develops from a uninucleate haploid conidium (see Fig. 5A, which is published as supplemental data on the PNAS web site, www.pnas.org) by the progressive occurrence of sequential mitotic divisions (Fig. 5B). The nuclei within each tube divide synchronously; concomitantly, the tube elongates, and its 2, 4, 8, or more nuclei migrate to spatially specific positions after each division (Fig. 5 C–E). The cell cycle history of each germling can thus be deduced from the size of the tube and the number and position of its nuclei.
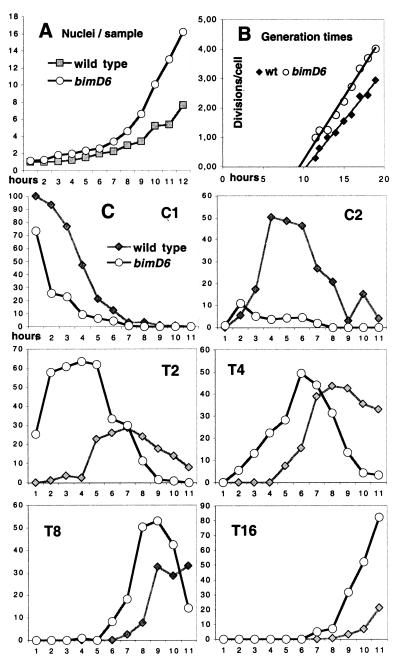
Wild-type and bimD6 germlings were compared at permissive temperature, where normal mitoses occur in both strains, and were examined for four mitotic cycles. For each strain, at each time point analyzed, 200–400 germlings were scored. Three protocols were examined. First, conidia were germinated after cold storage, which provides some degree of synchrony, in SM + glucose and urea. Compared with wild type, bimD6 cycles faster: its first mitosis occurs earlier, and the number of nuclei per sample increases faster (Fig. 4A). By linear regression analysis, calculated generation times (Gts) were 2 h 21 and 2 h 59 for bimD6 and wild type, respectively. The difference between these values is statistically significant (P < 0.05; Fig. 4B). Second, an even greater difference was observed when cold-stored conidia were germinated in SM with a less favorable combination of C- and N-sources (lactose + urea): Gts were 6 h 28 for bimD6 and 9 h 14 for wild type (P < 0.1). Third, conidia were synchronized with HU, and only one division was followed: Gts calculated for this division were of 7 h 49 for bimD6 and 13 h 46 for wild type (P < 0.05). Thus, bimD6 nuclei consistently divided earlier and cycled faster than wild type; furthermore, the extent of the difference increases in parallel with the length of the cycle.
Figure 4.
Rate of mitoses and germling phenotypes in bimD6 and wild-type conidia germinated in SM (glucose + urea) during 20 h. (A) Number of nuclei per sample. Samples were analyzed every hour, and hour 1 in graph corresponds to the start of divisions, namely at 8 h after inoculation of the conidia. (B) For each time point (X), the average number of divisions per cell (Y) was calculated by logarithm (average number of nuclei per cell)/logarithm 2. After linear regression (Y = bX), Gt was determined by using the equation Gt = 1/b; 300–2,000 nuclei were analyzed for each time point. (C) Wild-type phenotypes are indicated by gray squares and lines, whereas bimD6s (m) are given as black curves with plain circles; C1 and C2 represent, respectively, the percentage of uninucleated and binucleated conidia, and T2–T16 correspond to the percentage of germ tubes with 2, 4, 8, or 16 nuclei.
Importantly, however, whereas bimD6 exhibits faster nuclear divisions, it maintains normal coordination between nuclear divisions and other aspects of germination, such that the entire cellular program proceeds faster than normal (Fig. 4C). Each step of germination is faster in bimD6 than in wild type (Fig. 4 C1–T16). In particular, initiation of tube formation after the first division is faster in bimD6 (as seen Fig. 4C by the percentage of conidia with two nuclei in C2 compared with binucleate tubes in T2). At later stages, tube elongation occurs with an essentially normal delay after completion of the corresponding nuclear divisions. Wild-type and mutant tube lengths were, respectively, 19 ± 4 and 17 ± 5 μm when tubes contained two well separated nuclei (as in Fig. 5D) and 25 ± 6 and 23 ± 7 μm when they contained four nuclei (Fig. 5E; 100 analyzed). The nuclear migration process, however, is not perfectly normal in bimD6. For example, after the second mitosis, the four nuclei of wild-type germlings are usually distributed uniformly throughout the tube (Fig. 5E); in four nucleated bimD6 germlings, in contrast, 30% had not yet started tube formation (Fig. 5F) or contained randomly distributed nuclei (Fig. 5G). Also, the average distance between nuclei, although variable in both strains, was smaller in bimD6 than in wild type. Thus, coupling of nuclear migration to nuclear division appears less strict in bimD6.
bimD6 Is Defective for Interhomolog, but Not Intrachromosomal, Recombination.
bimD6 mutants show increased sensitivities to methyl methanesulfonate and UV only during germination and not in quiescent conidia (6). Germinating conidia of bimD6 also showed x-ray sensitivity (2- to 2.5-fold; see Fig. 6, which is published as supplemental data on the PNAS web site, www.pnas.org). To further probe the role of BIMD for repair, we examined bimD6 for spontaneous mitotic recombination, which is thought to occur in response to endogenous DNA damage. We analyzed in parallel the Aspergillus mutant uvsC114, defective for a recA/RAD51 homolog (25); uvsC114 is also sensitive to radiation and methyl methanesulfonate only while dividing (30).
Well-developed genetic systems for assessment of mitotic recombination are available in Aspergillus (30). We selected for rare ad+ recombinants arising in diploids heterozygous for the two distinguishable alleles adE8 and adE20 (ref. 16; Table 1). Because of the unusually close linkage of outside markers to adE, the fraction of ad+ recombinants that have undergone crossing over of flanking markers can be determined; more specifically, genetic analysis of haploid segregants reveals the arrangement of markers on the two homologs. Both bimD6 and uvsC114 reduce the frequency of ad+ recombinants in adE8/adE20 diploids about 9-fold (Table 1 and Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org). However, among these recombinants, the relative frequencies of crossover and noncrossover types resemble those of wild type (Table 1). This result contrasts with findings for two other Aspergillus DNA repair mutants, musN227 (hyperrec) and musL222 (hyporec), both of which exhibit an increased ratio of crossover to noncrossover recombinants (16).
Table 1.
Allelic recombination in adE8/adE20 diploids: Absolute frequencies of recombinants and relative frequencies of conversion vs. crossing over
| Control (+) | bimD6 | uvsC114 | |
|---|---|---|---|
| Absolute frequencies of ad+ × 10−6 | 11.43 ± 1.33 | 1.22 ± 0.21 | 1.20 ± 0.35 |
| Class* Convertants/Crossovers | Relative frequencies of recombination types | ||
| Single conversions† | 77% | 78% | 82% |
| Single crossovers | 13% | 15% | 11% |
| Two events: Conversion and/or CO | 7% | 5% | 5% |
| Multiple events | 3% | 2% | 2% |
| Total of ad+ recombinants analyzed | 30 | 53 | 44 |
Classification of ad+ recombinants was done as in ref. 16.
adE8→+ or adE20→+.
We also tested both bimD6 and uvsC114 for intrachromosomal “gene conversion” within a duplication. We used a pyrG89 strain with a heterozygous direct repeat of benomyl resistant (benA22) and sensitive (benA+) alleles separated by intervening pyr-4 and plasmid sequences (17). On growth-reducing concentrations of benomyl, this heterozygous benA22/benA+ duplication strain shows intermediate resistance to benomyl; however, fully resistant sectors can arise by recombination (Fig. 7, www.pnas.org). When selection for pyr-4 function is maintained, all resistant sectors result from conversion of benA+ to benA22 without loss of the intervening pyr-4 segment. Such conversions can reflect intrachromatid and/or intersister events. Unexpectedly, bimD6 duplication strains were considerably more sensitive to benomyl than wild type or uvsC114 (see Table 3, www.pnas.org). Therefore, we assayed recombination at reduced benomyl concentrations for bimD6 (0.96 μg/ml) instead of 1.2 μg/ml used for wild-type/uvsC114. Under these conditions, bimD6 shows about as many resistant sectors as the control, whereas uvsC114 produced none (Fig. 7, www.pnas.org). Thus, whereas uvsC114 reduces conversion between interrupted benA22/benA+ repeats, bimD6 has no influence on this type of mitotic recombination.
The S. macrospora SPO76 Gene Complements the Heat and Methyl Methanesulfonate (MMS) Sensitivities of A. nidulans bimD6 but Not the Meiotic Defects.
Wild-type versions of either SPO76 or bimD were introduced into a bimD6, pyrG89 double mutant recipient by cotransformation with pyr+ plasmids. In each case, a considerable fraction of transformants exhibited complementation for both heat and MMS sensitivity: 26/77 for SPO76 and 34/54 for bimD. Analysis of six bimD transformants that exhibited complementation for mitotic defects revealed that meiotic defects were also complemented. In contrast, among 16 of the mitotically complemented SPO76 transformants, none showed complementation of bimD6 meiotic defects; in three of these cases, the presence of SPO76 was confirmed by Southern blotting. Analogously, in the reciprocal test, complementation of the meiotic defects of spo76–1 in S. macrospora was observed only after transformation with SPO76 and not after transformation with bimD (respectively, 25/51 and 0/786).
Discussion
The findings presented above confirm and extend the analogy between BIMD and Spo76p and provide information regarding the basic roles of the Spo76 family proteins.
Strong Analogies Between BIMD and Its Orthologs.
First, BIMD, like Spo76p, is abundant and chromosome-associated at all stages of the mitotic and meiotic programs except from end of prophase to telophase. Analogously, human PDS5 is chromatin associated during interphase and in mitosis, during prophase and telophase (11). In contrast, budding yeast Pds5p is seen on chromatin in G1 (low signal), S into mitosis (strong signals) until onset of chromatid separation (9, 10). Thus, in Aspergillus, Sordaria, and vertebrates, the protein dissociates from the chromosomes earlier than in budding yeast. An attractive hypothesis is to link this finding with the differences in chromosome condensation between these organisms (see refs. 9 and 11 for discussion). Second, SPO76 (4) and bimD (here) are required for cohesion during both mitotic and meiotic programs. Pds5p is found essential for mitotic sister cohesion establishment and maintenance (9, 10); a similar latter effect could explain why bimD6 is highly benomyl sensitive at permissive temperature (above). Third, Spo76p, BIMD, and Pds5p are also required for normal chromosome compactness during mitosis (refs. 4 and 9, here) and meiosis (ref. 4, here). Fourth, spo76–1 and bimD6 are sensitive to DNA-damaging agents (refs. 6 and 8, here). Finally, complementation analysis shows that SPO76 and bimD are functional homologs with respect to their roles in mitotic chromosome metabolism.
BIMD Is a Negative Regulator of Normal Mitotic Cell Cycle Progression.
We show that bimD6 cycles significantly faster than wild-type. Rapid cycling is observed for all aspects of germ tube development, with normal or nearly normal coupling retained between nuclear and extranuclear processes. Our results therefore imply that an abundant chromosome-associated protein is involved in modulating normal cell cycle progression. Moreover, because cycling is faster in bimD6 than in wild type, BIMD appears to be a negative regulator of cell cycle progression, i.e., BIMD is part of a mechanism that normally constrains cell cycle progression in circumstances where a slower rate of cycling is more appropriate. That overexpression of bimD causes a fully reversible arrest in G1 (6) further supports this interpretation. Because the exact nature of the bimD6 mutation is not known, its phenotype may, however, not reflect simply elimination of gene function.
We have not determined which stage(s) of the cell cycle are affected, but the only period when BIMD is not on the chromosomes, metaphase/anaphase, is normally so brief (13) that changes in these stages are clearly not the primary effect. We find it attractive to think that the major effect is during G1 or early S phase. First, overexpression of bimD in wild-type cells results in a block at the G1 or early S phase of the cell cycle (6). Second, a greater difference in Gts between bimD6 and wild type was seen in poorer medium, and in several organisms, variations in nutrient conditions affect specifically the duration of G1 (31–33). Third, cell cycle modulation at G1 would fit with the tumor suppressor role of the human ortholog AS3: in prostrate cells, up-regulated expression of the AS3 gene mediates androgen-induced G1 arrest (5). In accord with these possibilities, BIMD protein is seen in G1 nuclei. With respect to its essential function(s), however, BIMD is not required before the HU arrest point, as shown by experiments involving HU arrest in combination with temperature shifts. Likewise, yeast Pds5p function is not required during G1 (9). bimD6 may (also) influence progression through S phase. The most prominent difference in Gts between bimD6 and wild type was found when HU was used to synchronize germlings. The continued effect of HU (after removal) could be explained by a slowing of DNA replication, e.g., because the cell needs time to replicate over DNA lesions (34).
How could proteins like BIMD/Spo76p, whose primary role is to mediate chromosome morphogenesis, also be implicated in the cell cycle progression? They could remodel chromatin, for example, by providing (direct or indirect) intrachromatid crosslinks. This remodeling could define chromatin domains to be activated/repressed, for example in the vicinity of genes involved in cyclin D expression during G1 (33). Likewise, such crosslinks could define which chromosome region should be searched for repair from S through G2 (below) and/or mark at which chromosome regions sister-chromatid cohesion should be established during S phase (35).
BIMD Determines the Probability of Interhomolog Mitotic Recombination.
The absolute frequency of spontaneous allelic interhomolog recombination is strongly reduced in bimD6, but the distribution of recombinants into crossover and noncrossover classes among such recombinants is the same as in wild type. This phenotype suggests that bimD6 affects the probability that an interhomolog event will occur at all; once that decision has been made, the recombination process proceeds as usual. This interpretation is supported by the fact that intrachromosomal recombination is normal in bimD6, suggesting that BIMD may not be involved in the enzymology of recombinational repair per se.
Why is recombinational repair reduced in bimD6? Several other mutants defective in sister chromatid cohesion are also radiation sensitive and/or defective in DNA double strand break repair (reviewed in ref. 36). In budding yeast, cohesins bind to regularly spaced preferential positions that likely correspond to the AT-queue of the chromosome axis (37), and Pds5p binds to the same sites (9). We therefore suggest that BIMD may be required for recombination events that occur in the context of the chromosome axis. For example, BIMD could mark which chromatin loops have to be searched on the sister or the homolog as a template for recombinational repair. When some of those landmarks are missing, as for example in bimD6, safety mechanisms may prevent or reduce recombinational repair. On the other hand, intrachromosomal repair between repeated sequences apparently does not depend on those landmarks. Both bimD6 and the rad21–45 mutant of Schizosaccharomyces pombe (38, 39) are highly sensitive to DNA-damaging agents but have no defect in mitotic recombination between interrupted direct repeats. An analogous phenotype, decreased interhomolog recombination with no effect on intrachromosomal recombination, was also observed during budding-yeast meiosis in the hop1 and red1 mutants, which are defective in axis-associated components (40, 41). That direct repeat recombination is independent of Hop1 and Red1p (42) could imply that also in meiosis, this type of recombination does not occur in the context of the chromosome axis.
BIMD Localization and Function Mirror Atypical Features of Meiosis in A. nidulans.
Although BIMD and Spo76p play functionally analogous roles in mitosis, their meiotic roles may differ in accord with several “atypical” aspects of meiosis in A. nidulans. First, the meiotic localization of the two proteins is different. Spo76p is most abundant during Sordaria meiotic prophase and assembles in strong lines along chromosome axes during synapsis (4). BIMD, in contrast, is not or only slightly more abundant during Aspergillus meiosis, and it does not reveal defined axes during prophase I. These differences are likely related to differences in underlying chromosome structure and/or function and may also explain the lack of heterologous complementation of meiotic defects. Sordaria forms synaptonemal complexes (43), whereas A. nidulans appears to lack these structures (44). Moreover, with respect to meiotic chromosome function, A. nidulans differs from Sordaria (43) by its lack of positive crossover interference (review in ref. 18). On the basis of the proposition that crossover interference involves the imposition and relief of stress along the chromosomes (45), it was argued that Spo76p might be a transducer of the disruptive chromosomal forces that provide the necessary stress (4). If this hypothesis is correct, the need for a robust axis and a stress-transducing protein would be greater in an organism that exhibits crossover interference than in an organism in which this process is absent, thus explaining the differences between Spo76p and BIMD localization and, more generally, between axial development in organisms with and without interference.
Supplementary Material
Acknowledgments
We are grateful to Françoise James for expert technical assistance. We thank Greg May (University of Texas, Houston, TX) and Berl Oakley (The Ohio State University, Columbus) for the gift of strains and plasmids. We particularly thank Nancy Kleckner and the reviewers for helpful suggestions. This work was supported by the Netherlands Organisation for Scientific Research (D.v.H.), by European Economic Community contract CHRXCT 94 0511 (to C.H. and D.Z.), by a research grant from the Natural Science and Engineering Research Council of Canada (to E.K.), and by the Centre National de la Recherche Scientifique (UMR 8621 to D.Z.).
Abbreviations
- DAPI
4,6-diamidino-2-phenylindole
- HU
hydroxy urea
- Gts
calculated generation times
References
- 1.Yang J, Kornbluth S. Trends Cell Biol. 1999;9:207–210. doi: 10.1016/s0962-8924(99)01577-9. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- 3.Zou H, McGarry T J, Bernal T, Kirschner M W. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 4.van Heemst D, James F, Pöggeler S, Berteaux-Lecellier V, Zickler D. Cell. 1999;98:261–271. doi: 10.1016/s0092-8674(00)81020-x. [DOI] [PubMed] [Google Scholar]
- 5.Geck P, Maffini M V, Szelei J, Sonnenschein C, Soto A M. Proc Natl Acad Sci USA. 2000;97:10185–10190. doi: 10.1073/pnas.97.18.10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denison S H, Kafer E, May G S. Genetics. 1992;134:1085–1096. doi: 10.1093/genetics/134.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darwiche N, Freeman L A, Strunnikov A. Gene. 1999;233:39–47. doi: 10.1016/s0378-1119(99)00160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreau P J F, Zickler D, Leblon G. Mol Gen Genet. 1985;198:189–197. [Google Scholar]
- 9.Hartman T, Stead K, Koshland D, Guacci V. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- 11.Sumara I, Vorlaufer E, Gieffers C, Peters B, Peters J M. J Cell Biol. 2000;151:749–761. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt C L, May G S. Genetics. 1996;142:777–787. doi: 10.1093/genetics/142.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergen L G, Morris N R. J Bacteriol. 1983;156:155–160. doi: 10.1128/jb.156.1.155-160.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer R. FEMS Microbiol Rev. 1999;23:39–68. doi: 10.1111/j.1574-6976.1999.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 15.Pontecorvo G, Roper J A, Hemmons L M, MacDonald K D, Bufton A W J. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 16.Zhao P, Kafer E. Genetics. 1992;130:717–728. doi: 10.1093/genetics/130.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunne P W, Oakley B R. Mol Gen Genet. 1988;213:339–345. doi: 10.1007/BF00339600. [DOI] [PubMed] [Google Scholar]
- 18.Kafer E. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- 19.Debets F, Bos K. Fungal Genet News Lett. 1986;33:24. [Google Scholar]
- 20.Osmani S A, May G S, Morris N R. J Cell Biol. 1989;104:1495–1504. doi: 10.1083/jcb.104.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osiewacz H D. Curr Genet. 1994;26:87–90. doi: 10.1007/BF00326309. [DOI] [PubMed] [Google Scholar]
- 22.Oakley B R, Rinehart J E, Mitchell B L, Oakley C E, Carmona C, Gray G L, May G S. Gene. 1987;61:385–399. doi: 10.1016/0378-1119(87)90201-0. [DOI] [PubMed] [Google Scholar]
- 23.Waring R B, May G S, Morris N R. Gene. 1989;79:119–130. doi: 10.1016/0378-1119(89)90097-8. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan D. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 25.van Heemst D, Swart K, Holub E F, van Dijk R, Offenberg H H, Goosen T, van den Broek H W J, Heyting C. Mol Gen Genet. 1997;254:654–664. doi: 10.1007/s004380050463. [DOI] [PubMed] [Google Scholar]
- 26.Thompson-Coffe C, Zickler D. Dev Biol. 1984;165:257–271. doi: 10.1006/dbio.1994.1251. [DOI] [PubMed] [Google Scholar]
- 27.Lammers J H M, Offenberg H H, van Aalderen M, Vink A C G, Dietrich A J J, Heyting C. Mol Cell Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy J E G. Microbiol Mol Biol Rev. 1998;62:1496–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto A, Guacci V, Koshland D. J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kafer E, May G S. In: DNA Damage and Repair. Nickoloff J A, Hoekstra M F, editors. Vol. 1. Totowa, NJ.: Humana; 1998. pp. 477–502. [Google Scholar]
- 31.Silje H H W, Schure E G, Rommens A J M, Huls P G, Woldringh C L, Verkleij A J, Boonstra J, Verrips C T. J Bacteriol. 1997;179:6560–6565. doi: 10.1128/jb.179.21.6560-6565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlson C R, Grallert B, Stokke T, Boye E. J Cell Sci. 1999;112:939–946. doi: 10.1242/jcs.112.6.939. [DOI] [PubMed] [Google Scholar]
- 33.Riou-Khamlichi C, Menges M, Healy J M, Murray J A. Mol Cell Biol. 2000;20:4513–4521. doi: 10.1128/mcb.20.13.4513-4521.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulovitch A G, Hartwell L H. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Castano I B, DeLasPenas A, Adams C, Christman M F. Science. 2000;289:774–779. doi: 10.1126/science.289.5480.774. [DOI] [PubMed] [Google Scholar]
- 36.van Heemst D, Heyting C. Chromosoma. 2000;109:10–26. doi: 10.1007/s004120050408. [DOI] [PubMed] [Google Scholar]
- 37.Blat Y, Kleckner N K. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 38.Fortuno E A, Osman F, Subramani S. Mutat Res. 1996;364:147–160. [PubMed] [Google Scholar]
- 39.Subramani S. Mol Microbiol. 1991;5:2311–2314. doi: 10.1111/j.1365-2958.1991.tb02075.x. [DOI] [PubMed] [Google Scholar]
- 40.Hollingsworth N M, Byers B. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rockmill B, Roeder G S. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao-Draayer Y, Galbraith A M, Pittman D L, Cool M, Malone R E. Genetics. 1996;144:71–86. doi: 10.1093/genetics/144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zickler D, Moreau P J F, Huynh A D, Slezec A M. Genetics. 1992;132:135–148. doi: 10.1093/genetics/132.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egel-Mitani M, Olson L W, Egel R. Heriditas. 1982;97:179–187. doi: 10.1111/j.1601-5223.1982.tb00761.x. [DOI] [PubMed] [Google Scholar]
- 45.Kleckner N. Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.