Abstract
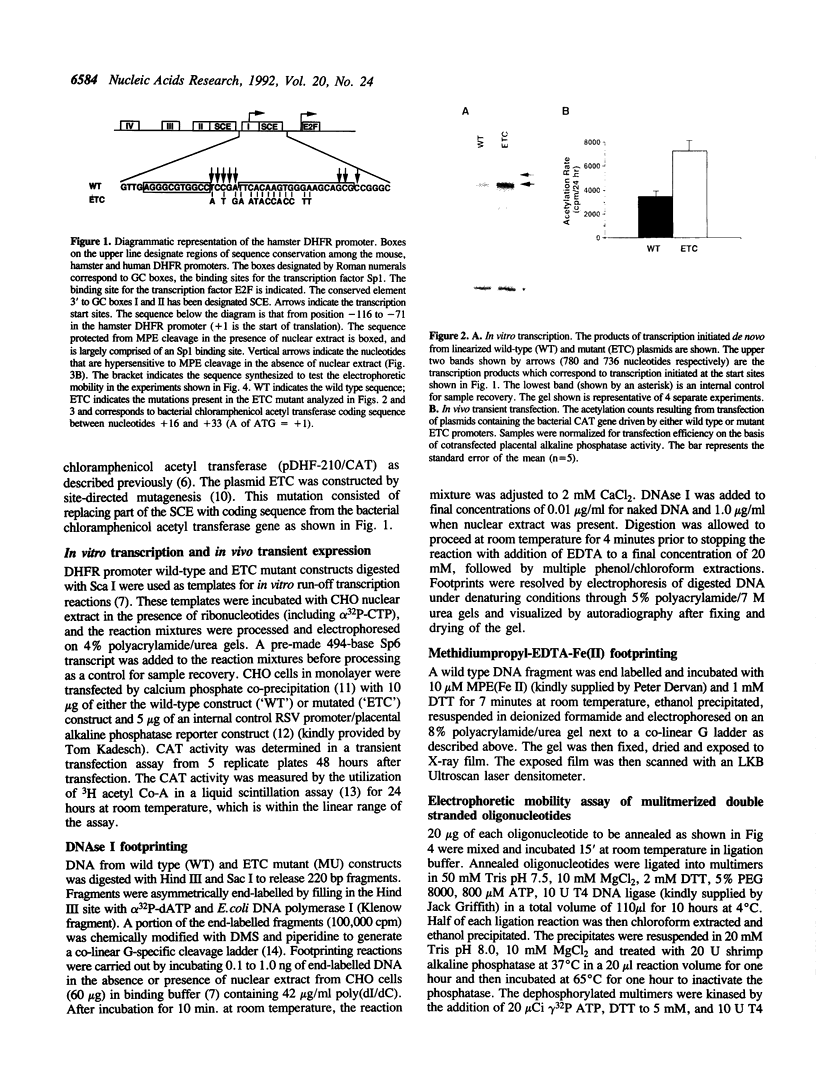
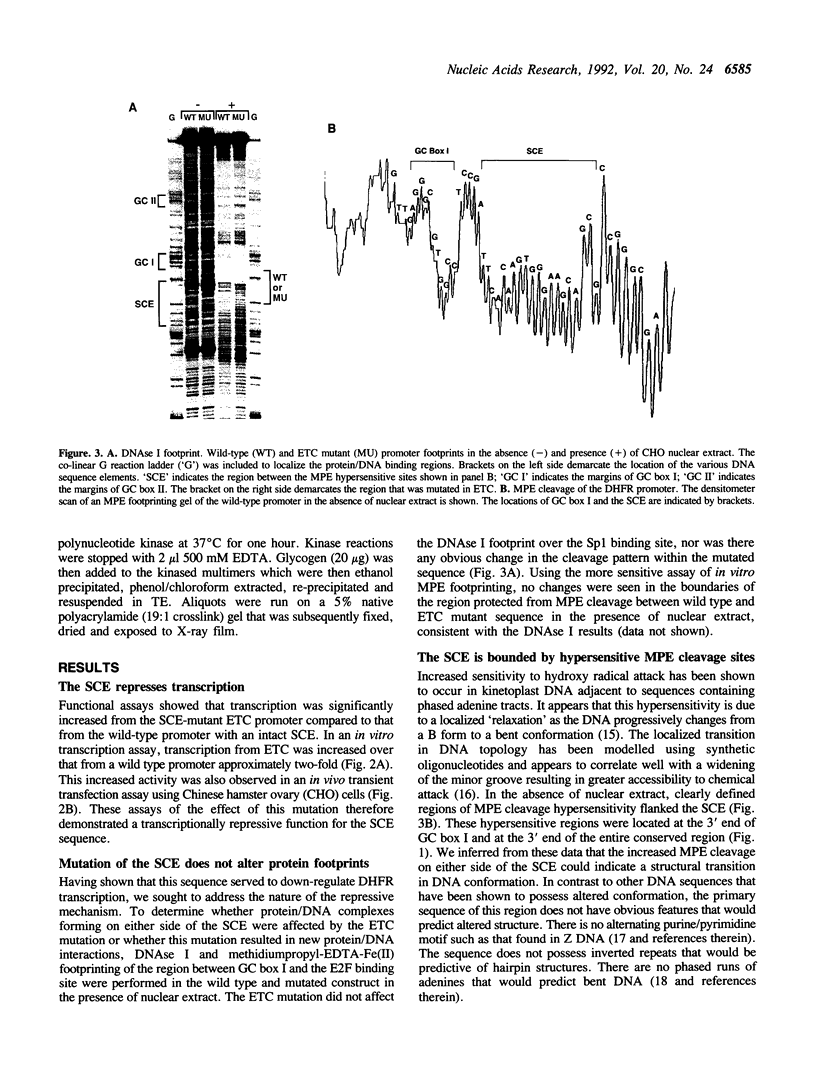
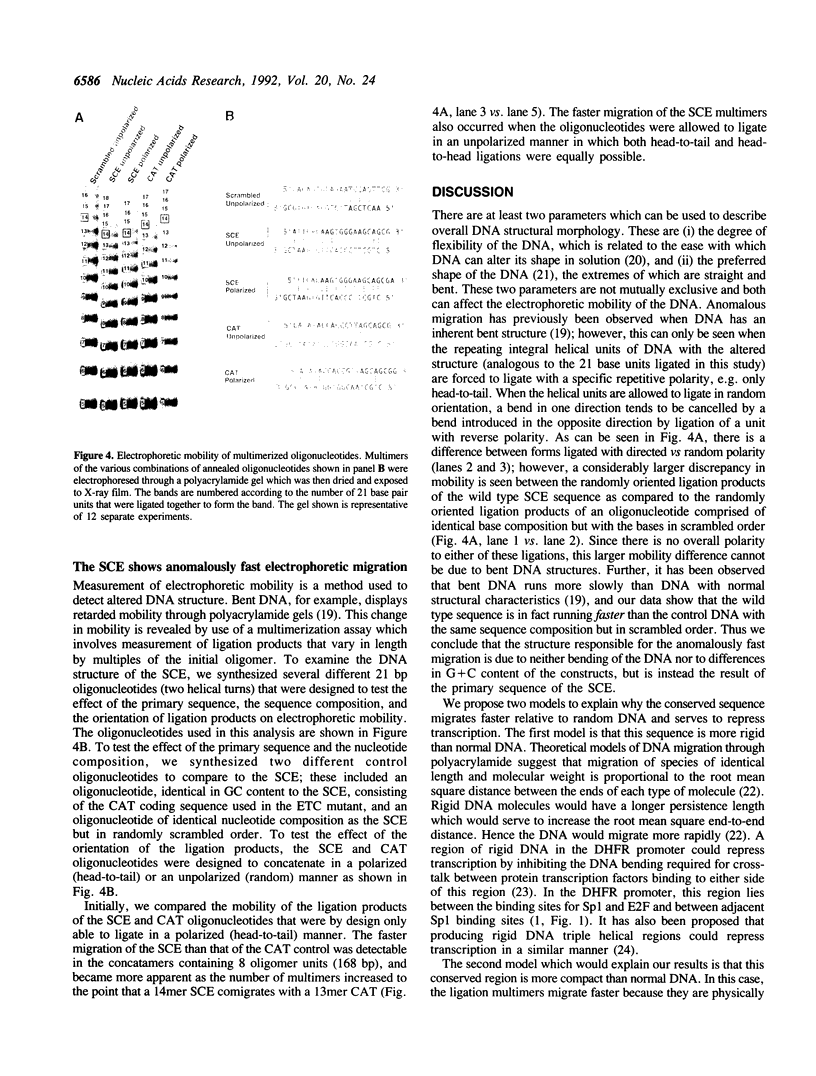
The mammalian dihydrofolate reductase (DHFR) gene promoters contain several conserved sequence elements which bind protein, and yet there are other conserved DNA sequences that do not footprint. We report here that mutation of one of these conserved non-footprinting regions increases transcription from this promoter both in vitro and in vivo. We show that this conserved region is flanked by sites hypersensitive to cleavage by methidiumpropyl-EDTA-Fe(II). Furthermore, multimers of a double-stranded oligonucleotide comprised of this region display faster migration through polyacrylamide than control DNA. The difference in mobility is not the result of bending, nor does the primary sequence contain features that would predict altered mobility. We propose that this 'Structural Control Element' is rigid and down-regulates transcription by inhibiting interactions between proteins binding adjacent to this region.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azizkhan J. C., Vaughn J. P., Christy R. J., Hamlin J. L. Nucleotide sequence and nuclease hypersensitivity of the Chinese hamster dihydrofolate reductase gene promoter region. Biochemistry. 1986 Oct 7;25(20):6228–6236. doi: 10.1021/bi00368a059. [DOI] [PubMed] [Google Scholar]
- Blake M. C., Azizkhan J. C. Transcription factor E2F is required for efficient expression of the hamster dihydrofolate reductase gene in vitro and in vivo. Mol Cell Biol. 1989 Nov;9(11):4994–5002. doi: 10.1128/mcb.9.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake M. C., Jambou R. C., Swick A. G., Kahn J. W., Azizkhan J. C. Transcriptional initiation is controlled by upstream GC-box interactions in a TATAA-less promoter. Mol Cell Biol. 1990 Dec;10(12):6632–6641. doi: 10.1128/mcb.10.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Chen M. J., Shimada T., Moulton A. D., Cline A., Humphries R. K., Maizel J., Nienhuis A. W. The functional human dihydrofolate reductase gene. J Biol Chem. 1984 Mar 25;259(6):3933–3943. [PubMed] [Google Scholar]
- Ciudad C. J., Morris A. E., Jeng C., Chasin L. A. Point mutational analysis of the hamster dihydrofolate reductase minimum promoter. J Biol Chem. 1992 Feb 25;267(6):3650–3656. [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R., Conner B. N., Wing R. M., Fratini A. V., Kopka M. L. The anatomy of A-, B-, and Z-DNA. Science. 1982 Apr 30;216(4545):475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- Eastman H. B., Swick A. G., Schmitt M. C., Azizkhan J. C. Stimulation of dihydrofolate reductase promoter activity by antimetabolic drugs. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8572–8576. doi: 10.1073/pnas.88.19.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith M. E., Beckman C. A., Cowan K. H. 5' Nucleotide sequences influence serum-modulated expression of a human dihydrofolate reductase minigene. Mol Cell Biol. 1986 Mar;6(3):878–886. doi: 10.1128/mcb.6.3.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Henthorn P., Zervos P., Raducha M., Harris H., Kadesch T. Expression of a human placental alkaline phosphatase gene in transfected cells: use as a reporter for studies of gene expression. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6342–6346. doi: 10.1073/pnas.85.17.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert S. W., Blake M., Azizkhan J., Nevins J. R. Role of E2F transcription factor in E1A-mediated trans activation of cellular genes. J Virol. 1991 Jul;65(7):3547–3552. doi: 10.1128/jvi.65.7.3547-3552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Crothers D. M. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lerman L. S., Frisch H. L. Why does the electrophoretic mobility of DNA in gels vary with the length of the molecule? Biopolymers. 1982 May;21(5):995–997. doi: 10.1002/bip.360210511. [DOI] [PubMed] [Google Scholar]
- Lumpkin O. J. Mobility of DNA in gel electrophoresis. Biopolymers. 1982 Nov;21(11):2315–2316. doi: 10.1002/bip.360211116. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Dervan P. B., Wold B. Analysis of promoter-specific repression by triple-helical DNA complexes in a eukaryotic cell-free transcription system. Biochemistry. 1992 Jan 14;31(1):70–81. doi: 10.1021/bi00116a012. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGrogan M., Simonsen C. C., Smouse D. T., Farnham P. J., Schimke R. T. Heterogeneity at the 5' termini of mouse dihydrofolate reductase mRNAs. Evidence for multiple promoter regions. J Biol Chem. 1985 Feb 25;260(4):2307–2314. [PubMed] [Google Scholar]
- Mitchell P. J., Carothers A. M., Han J. H., Harding J. D., Kas E., Venolia L., Chasin L. A. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5' region of the CHO dhfr gene. Mol Cell Biol. 1986 Feb;6(2):425–440. doi: 10.1128/mcb.6.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago C., Collins M., Johnson L. F. In vitro and in vivo analysis of the control of dihydrofolate reductase gene transcription in serum-stimulated mouse fibroblasts. J Cell Physiol. 1984 Jan;118(1):79–86. doi: 10.1002/jcp.1041180114. [DOI] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Effects of DNA sequence and histone-histone interactions on nucleosome placement. J Mol Biol. 1990 Nov 5;216(1):69–84. doi: 10.1016/S0022-2836(05)80061-0. [DOI] [PubMed] [Google Scholar]
- Su W., Jackson S., Tjian R., Echols H. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 1991 May;5(5):820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- Swick A. G., Blake M. C., Kahn J. W., Azizkhan J. C. Functional analysis of GC element binding and transcription in the hamster dihydrofolate reductase gene promoter. Nucleic Acids Res. 1989 Nov 25;17(22):9291–9304. doi: 10.1093/nar/17.22.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]





