Abstract
The ligand-activated transcription factor peroxisome proliferator-activated receptor (PPAR)-δ is highly expressed in colonic epithelial cells; however, the role of PPARδ ligands, such as fatty acids, in mucosal inflammation and malignant transformation has not been clarified. Recent evidence suggests that the anti-inflammatory/chemoprotective properties of fish oil (FO)-derived n-3 polyunsaturated fatty acids (PUFAs) may be partly mediated by PPARδ. Therefore, we assessed the role of PPARδ in modulating the effects of dietary n-3 PUFAs by targeted deletion of intestinal epithelial cell PPARδ (PPARδΔIEpC). Subsequently, we documented changes in colon tumorigenesis and the inflammatory microenvironment, i.e., local [mesenteric lymph node (MLN)] and systemic (spleen) T cell activation. Animals were fed chemopromotive [corn oil (CO)] or chemoprotective (FO) diets during the induction of chronic inflammation/carcinogenesis. Tumor incidence was similar in control and PPARδΔIEpC mice. FO reduced mucosal injury, tumor incidence, colonic STAT3 activation, and inflammatory cytokine gene expression, independent of PPARδ genotype. CD8+ T cell recruitment into MLNs was suppressed in PPARδΔIEpC mice. Similarly, FO reduced CD8+ T cell numbers in the MLN. Dietary FO independently modulated MLN CD4+ T cell activation status by decreasing CD44 expression. CD11a expression by MLN CD4+ T cells was downregulated in PPARδΔIEpC mice. Lastly, splenic CD62L expression was downregulated in PPARδΔIEpC CD4+ and CD8+ T cells. These data demonstrate that expression of intestinal epithelial cell PPARδ does not influence azoxymethane/dextran sodium sulfate-induced colon tumor incidence. Moreover, we provide new evidence that dietary n-3 PUFAs attenuate intestinal inflammation in an intestinal epithelial cell PPARδ-independent manner.
Keywords: peroxisome proliferator-activated receptor-δ, chronic inflammation, malignant transformation, T cell
inflammatory bowel disease (IBD), a chronic and recurring immunoinflammatory condition with unknown etiology, manifests as two overlapping phenotypes, i.e., ulcerative colitis and Crohn's disease (16, 74). In patients with chronic intestinal inflammation, the risk of developing colorectal cancer increases by ∼0.5–1% each year, 7 years after diagnosis (50, 77). Although complicating IBD only accounts for 2% of all colorectal cancer cases within the general population, it is considered a serious sequela of the disease, accounting for one in six of all deaths in IBD patients (68). Therefore, IBD patients represent a significant at-risk population for chronic inflammation-associated colorectal cancer development. Despite the functional link between inflammation and colon cancer, the overlapping regulatory pathways that drive inflammation-associated colonic tumor development remain poorly understood.
Long-chain n-3 polyunsaturated fatty acids (PUFAs) found in fish oil (FO), e.g., eicosapentaenoic acid (20:5Δ5,8,11,14,17) and docosahexaenoic acid (22:6Δ4,7,10,13,16,19), exhibit beneficial effects in IBD and colon carcinogenesis (8), in part due to their potent anti-inflammatory effects (13, 53). Additionally, the balance between colonic epithelial cell proliferation and apoptosis is favorably modulated by dietary n-3 PUFAs, thereby conferring resistance to carcinogenic agents (14, 18, 19). Moreover, n-3 PUFAs have been shown to modulate the important determinants that link inflammation to cancer development and progression (7, 19, 48, 75, 85). From a mechanistic perspective, the cellular incorporation of dietary n-3 PUFAs favorably affects a broad spectrum of physiological processes, including immune function, wound healing, cell membrane structure/function, eicosanoid signaling, macronutrient metabolism, and nuclear receptor activation (16). With respect to T cell function, dietary n-3 PUFAs have been shown to alter plasma membrane microorganization (lipid rafts) at the immunological synapse, ultimately suppressing signal transduction and nuclear translocation/activation of transcription factors (28, 55, 56, 98). However, the effect of n-3 PUFAs on mucosal immunoregulation has not been determined but is warranted, as ∼50% of IBD subjects utilize self-prescribed oral complementary alternative medicines/diets that include FO (58).
Independently, ligands for peroxisome proliferator-activated receptors (PPARs) PPARα, PPARδ (also referred to as PPARβ or PPARβ/δ), and PPARγ have been shown to inhibit IBD and colon carcinogenesis (16, 20, 73, 88). Ligand-activated PPAR complexes regulate the expression of PPAR-responsive genes and biological functions, including cell proliferation and differentiation, fatty acid metabolism, energy homeostasis, immune responses, and inflammation (2, 12, 36, 73, 103). Among the PPAR family members, functions of PPARα and PPARγ have been well characterized, whereas the physiological functions of PPARδ remain less clear. Although PPARδ mRNA and protein are ubiquitously expressed, among anatomic sites in rodents, expression is highest in colonic epithelium (27, 34), and PPARδ plays an important role in the terminal differentiation of colonic epithelial cells (65, 73). Therefore, it is likely that this nuclear receptor plays an important regulatory role within the gastrointestinal tract.
Ligands for PPARδ are anti-inflammatory, and enhanced inflammation is observed in the absence of PPARδ expression (44). Anti-inflammatory activity of PPARδ may be attributed, at least in part, to its ability to interfere with NF-κB signaling (73). At high ligand concentrations, inhibition of colitis is associated with PPARδ activation (92), and PPARδ null mice exhibit increased sensitivity to dextran sodium sulfate (DSS)-induced colitis, wherein clinical symptoms are exacerbated and expression of inflammatory cytokines is increased (44). Additionally, outcomes of a porcine model of IBD suggest that activation of PPARδ may accelerate colonic regeneration and clinical remission (6). The role of PPARδ activation in colon tumorigenesis remains controversial and is reviewed in detail elsewhere (73). PPARδ null human colon cancer (HCT116) cells have a reduced tumorigenicity in a xenograft model (71). Specifically, in the colon, in the absence of PPARδ, colon carcinogenesis is exacerbated in genetic (APCmin mouse) and chemically induced [azoxymethane (AOM)] carcinogenesis models (5, 38, 76), whereas other studies indicate that activation of a functional PPARδ is required to inhibit AOM-induced colon carcinogenesis (65). Interestingly, n-3 PUFAs have been identified as ligands for PPARδ (32, 95); yet, it is not known whether the beneficial effects of n-3 PUFAs on intestinal inflammatory pathologies are mediated through a PPARδ-dependent mechanism.
In the present investigation, PPARδ was selectively deleted from intestinal epithelial cells utilizing a Cre-lox-mediated recombination strategy to disrupt the PPARδ locus. By generating an intestine-specific PPARδ knockout mouse, we were able to assess the contribution of dietary n-3 PUFAs and PPARδ to mucosa-generated immune responses in a chronic intestinal inflammation/carcinogenesis model. Cross talk between lymphocytes and intestinal epithelial cells is an important component of mucosal inflammatory immune responses (17), evidenced by intestinal epithelial cell expression of major histocompatibility complex class II antigens and the costimulatory molecule CD86 in the inflamed colon (69, 83). Furthermore, intestinal epithelial cells can present antigens to lymphocytes in a mixed leukocyte reaction (10, 67), and the processing and presentation of antigens by intestinal epithelial cells may be important for the induction of colonic inflammation (66). Thus it remains possible that intestinal epithelial cells may condition the mucosal environment and, therefore, influence T cell recruitment and activation. What remains unknown is the extent to which specific changes in the intestinal epithelial cell from the inflamed mucosa are able to impact the activation status of lymphocyte populations locally and systemically and whether this is achieved through PPARδ- and/or n-3 PUFA-dependent mechanisms.
In the current study, we determined the impact of chemoprotective dietary FO and intestinal epithelial cell-specific deletion of PPARδ on the colonic inflammatory microenvironment and on local [mesenteric lymph node (MLN)] and systemic (spleen) resident T cell populations. In addition, we assessed T cell activation markers functionally associated with trafficking to inflammatory sites.
MATERIALS AND METHODS
Animals and diets.
All experimental procedures were conducted in accordance with guidelines approved by the US Public Health Service and the Institutional Animal Care and Use Committee at Texas A & M University. C57BL/6 mice with the NH2-terminal portion of the DNA binding domain of PPARδ were targeted by CRE-lox methodology (PPARδF/F) (5) and crossed with Cre DNA recombinase under the control of the villin promoter (villin-Cre mice) (25). Progeny homozygous for the PPARδ-floxed allele, progeny hemizygous for the villin-Cre transgene (PPARδΔIEpC), or littermate control [wild-type (PPARδF/F)] mice were generated. Subsequently, PPARδΔIEpC and PPARδF/F mice were inbred to produce littermates on the same genetic background. Mice were genotyped prior to recruitment into the study, housed on a 12:12-h light-dark cycle, and fed ad libitum a 5% (wt/wt) corn oil (CO) or 4% FO + 1% CO diet for 2 wk prior to the initiation of the carcinogen and chronic mucosal inflammation (AOM/DSS) regimen. Males and females were equally represented from each genotype (PPARδΔIEpC and PPARδF/F) consuming either of the two experimental diets. At the start of the experiment, PPARδ deletion was assessed by PCR analysis of DNA extracted from tails using a Qiagen DNA tissue kit. PCR was performed using the Platinum Taq polymerase kit (GIBCO BRL). The following primers were used: loxP (5′GAGCCGCCTCTCGCCATCCTTTCAG-3′ and 5′-GGCGTGGGGATTTGCCTGCTTCA-3′) and Cre recombinase (5′-GCATTACCGGTCGATGCAACGAGTG-3′ and 5′-GAACGCTAGAGCCTGTTTTGCACGTTC-3′). After completion of the experimental treatment regimen, PPARδ deletion was confirmed in the target tissue (colon) by PCR and immunoblotting.
Colitis and carcinogen induction.
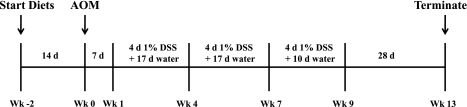
After a 2-wk diet intervention period, mice were injected with AOM (7.5 mg/kg body wt ip; Sigma-Aldrich). While the mice were maintained on the same diets, chronic inflammation was induced by exposure to three cycles of 1% (wt/wt) DSS (MP Biomedicals) in the drinking water (1 cycle = 4 days of DSS + 17 days of fresh tap water). Animals were euthanized after completion of the final DSS cycle (Fig. 1). At the time of euthanasia, colons were dissected at the junction of the cecum (proximally) and the anus (distally). The colon tissue was flushed with PBS, and the entire colon was processed by the Swiss-roll technique (n = 11–14 mice in each experimental group). Colon lesions were mapped and excised, and mucosal scrapings were subsequently collected from the remaining noninvolved tissue (n = 9–13 mice per experimental group) and snap-frozen for further analysis. Tissues were fixed in 4% paraformaldehyde, embedded in paraffin, stained with hematoxylin-eosin, and evaluated in a blinded manner by a board-certified pathologist (B. Weeks). Colon lesions were typed, and the degree of epithelial injury (score 0–3) on microscopic cross sections of the colon was graded as previously described (53).
Fig. 1.
Experimental dosing regimen. Wild-type (PPARF/F) mice and intestinal epithelial cell (IEpC) peroxisome proliferator-activated receptor (PPAR)-δ null (PPARδΔIEpC) mice consuming a 5% corn oil (CO) or a 1% corn oil + 4% fish oil (FO) diet were acclimated to experimental diets for 14 days prior to injection of azoxymethane (AOM, 7.5 mg/kg body wt ip). Subsequently, mice were exposed to 3 cycles of dextran sodium sulfate (DSS, 1% wt/wt via drinking water). Mice were euthanized 12 wk after completion of the final DSS cycle.
RNA isolation and quantitative real-time PCR.
RNA was isolated using the RNAqueous Total RNA kit (Ambion) and treated with DNase inactivation reagent (Ambion), its integrity was assessed using a bioanalyzer (model 2100, Agilent Technologies), and it was quantified and stored at −80°C. Reverse transcription of 1 μg of sample RNA was performed using Maloney's murine leukemia virus RT (Invitrogen). Expression of PPARδ in tissue-specific knockout mice was determined using mRNA isolated from colonic mucosa, duodenum, and kidney. Real-time PCR was performed using the AB 7900 PCR system (Applied Biosystems, Foster City, CA) and Taqman probes (Assay-on-Demand, Applied Biosystems) for PPARδ exon boundaries 4–5 (Mm01305435_m1) and PPARδ exon boundaries 7–8 (Mm00803186_g1).
For mucosal cytokine mRNA expression, Taqman gene expression kits (Applied Biosystems) were used for IL-6 (Mm00446190_m1), IL-17A (Mm00439618_m1), IL-17F (Mm00521423_m1), IL-21 (Mm00517640_m1), IL-23 (Mm00518984_m1), IL-23R (Mm00519943_m1), IL-27 (Mm004611664_m1), and IFN-γ (Mm01168134_m1). Amplification of mRNA (fluorescence) was recorded over 40 cycles, and the corresponding cycle numbers (Ct) were used to calculate mRNA expression as follows: 2(40 − Ct). Target gene expression was normalized to ribosomal 18S expression (Hs03928990_g1).
Immunoblotting.
Colonic mucosa was scraped from the underlying smooth muscle, and protein extraction and immunoblotting were conducted as described previously (89). Protein detection utilized primary monoclonal rabbit anti-mouse phosphorylated Stat3 antibody (Cell Signaling Technology, Danvers, MA) or polyclonal rabbit anti-mouse Stat3 antibody (sc-482, Santa Cruz Biotechnology, Santa Cruz, CA) diluted in PBS containing 1.5% BSA and 0.1% Tween 20. Membranes were washed with PBS containing 0.1% Tween 20 and incubated with secondary peroxidase-conjugated goat anti-rabbit IgG (Kirkegaard & Perry, Gaithersburg, MD) according to the manufacturer's instructions. Bands were developed using Pierce SuperSignal West Femto maximum-sensitivity substrate and subsequently scanned with a Fluor-S Max MultiImager system (Bio-Rad, Hercules, CA).
PPARδ protein detection was performed as described previously (34) using a primary antibody for PPARδ (no. 8099, 0.053 μg/ml in Tris-buffered saline + Tween 20) and secondary antibody, biotinylated anti-rabbit IgG (1:10,000 dilution) in Tris-buffered saline + Tween 20. Immunoreactive proteins were detected with 125I-labeled streptavidin using phosphorimaging analysis after 72 h of exposure. Hybridization signals for the proteins of interest were normalized to the hybridization signal of a housekeeping gene, lactate dehydrogenase. The positive control was a cell lysate from COS1 cells transfected with a mouse PPARδ expression vector, as previously described (34).
Flow cytometry analysis of lymphocytes.
MLNs and spleens were isolated and placed in sterile RPMI 1640 medium with 25 mmol/l HEPES (Irvine Scientific), supplemented with 5% FBS (Irvine Scientific), 2 mM GlutaMAX (GIBCO), and penicillin (100 U/ml) and streptomycin (0.1 mg/ml; GIBCO), henceforth, “complete medium.” Tissues were broken into single cell suspensions by filtering through a 70-μm-mesh cell strainer. Lymphocytes were subsequently enriched by density gradient centrifugation using Lympholyte-M (Cedarlane Laboratories). Single cells from MLNs or spleens were resuspended in flow cytometry staining buffer (eBioscience) and then incubated with anti-CD16/32 MAb according to the manufacturer's instructions (Fc block, eBiosciences). Subsequently, cells were labeled with anti-CD3-APC (eBioscience) + anti-CD4-FITC or anti-CD8a-FITC MAb (eBioscience). Markers for T cell activation status were assessed by additional staining with anti-CD11a-phycoerythrin (PE), anti-CD44-PE, or anti-CD62L-PE MAb (eBioscience) and analyzed using a flow cytometer (model C6, Accuricytometer). Cell characterization and expression status as determined by mean fluorescence intensity of marker proteins (i.e., CD11a, CD44, and CD62L) were assessed by CFlow Plus software (Accuricytometer). Cell viability was determined by propidium iodide staining and averaged 99%.
Statistics.
The predetermined upper limit of probability for statistical significance throughout this investigation was P ≤ 0.05, and analyses were performed using the SAS System for Windows (version 9.2). Data from all analyses were subjected to a two-way ANOVA, and the main effects were diet and genotype unless stated otherwise. If justified by the resulting probability value (i.e., P ≤ 0.05), analyses between means were performed using Tukey's Studentized range test. Moreover, if a statistically significant interaction term emerged from a two-way ANOVA, the permissible preplanned comparisons (i.e., equal in number to the treatment degrees of freedom) were made using the least-squares means procedure. Data sets not exhibiting a normal distribution were subjected to the Kruskal-Wallis test (χ2 approximation), followed, if justified by the statistical probability outcome (P < 0.05), by Wilcoxon two-sample testing.
RESULTS
Generation of intestine-specific PPARδ knockout mice.
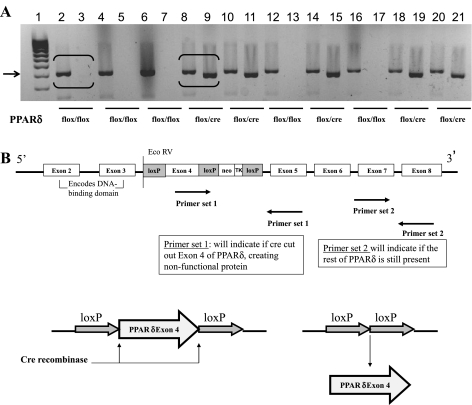
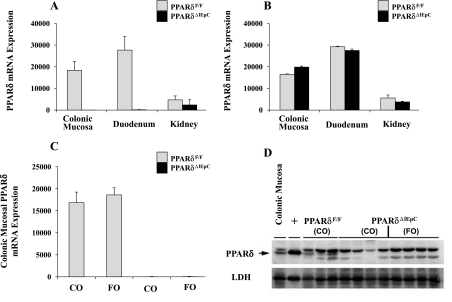
PPARδ was specifically deleted from intestinal epithelial cells utilizing the CRE-lox-mediated recombination strategy to disrupt the PPARδ locus, thereby generating PPARδ null (PPARδΔIEpC) and wild-type (PPARδF/F) mice (Fig. 2). Deletion of PPARδ exon 4 resulted in expression of a nonfunctional PPARδ, which was confirmed by mRNA expression in colon and duodenum of PPARδΔIEpC compared with PPARδF/F mice, whereas expression in the kidney, another anatomical site known to produce high levels of PPARδ, remained unchanged (34) (Fig. 3). Using primer sets that target PPARδ exon boundaries 4–5, we demonstrated the successful tissue-specific deletion of PPARδ in PPARδΔIEpC mice (Fig. 3A), whereas detection of PPARδ exon boundaries 7–8 demonstrated that a nonfunctional form of PPARδ was still expressed (Fig. 3B). Additionally, we confirmed the successful deletion of PPARδ in the colonic mucosa of Cre-homozygous floxed mice (PPARδΔIEpC) at mRNA and protein levels at the end of the AOM/DSS treatment regimen (Figs. 3, C and D). PPARδ mRNA and protein were readily detectable in the colonic mucosa of wild-type (PPARδF/F) mice, whereas PPARδ expression was undetectable, as expected, in the colonic mucosa of Cre-homozygous floxed (PPARδΔIEpC) mice. Cre recombinase has been shown to be transiently expressed in the target tissue in other model systems, highlighting the need to confirm deletion of the floxed gene of interest in the target tissue (81). Our results indicate that exposure to AOM/DSS did not alter expression of Cre recombinase in the target tissue (colon), validating the intestinal epithelial cell-specific deletion of PPARδ.
Fig. 2.
PPARδ deletion and genotyping strategy. A: DNA gel showing mouse genotypes that were assessed using 2 primer sets. Even-numbered lanes (2–20) detected loxP sites at 400 bp (arrow); odd-numbered lanes (3–21) detected Cre recombinase at 380 bp. B: primer set 1, used to detect specific deletion of PPARδ exon 4 by Cre recombinase, results in a nonfunctional form of PPARδ; primer set 2, used to detect PPARδ exon 7–8, indicates that the remainder of PPARδ is still intact.
Fig. 3.
Confirmation of an intestine-specific PPARδ knockout mouse. RNA was extracted from scraped colonic mucosa, duodenum, and kidney. A: deletion of exon 4 of PPARδ in PPARδΔIEpC mice, which was confirmed by detection of PPARδ mRNA expression using primer sets detecting exon boundaries 4–5 (n = 4–6 mice/genotype at each tissue site). B: intact PPARδ exons 7 and 8 (primer set detecting exon boundaries 7–8), indicating expression of a partial, nonfunctional protein (n = 4–6 mice/genotype at each tissue site). Values are means ± SE. C and D: confirmation of PPARδ deletion within colonic mucosa following completion of AOM/DSS treatment regimen. C: mRNA (n = 4–8 mice per experimental group). Values are means ± SE. D: protein expression level. Blots represent results from 8 PPARδΔIEpC and 3 PPARF/F mice. Protein expression was normalized to the housekeeping gene lactate dehydrogenase (LDH), and positive control (+) was a cell lysate from COS1 cells transfected with a mouse PPARδ expression vector. PPARF/F mice have a band at 52 kDa (arrow) that is absent in PPARδΔIEpC mice.
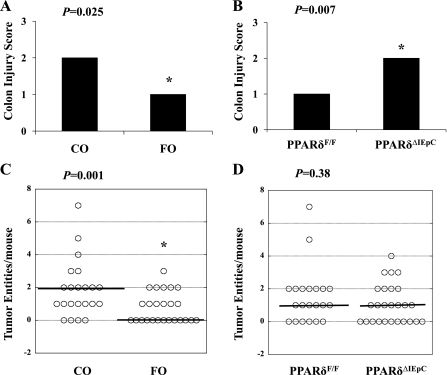
The effect of targeted intestinal epithelial PPARδ deletion and dietary FO on colon characteristics within the context of a carcinogenic and chronic inflammatory pathology was subsequently assessed. Colon injury scores in the middle region of the colon were ameliorated by dietary FO compared with CO (P = 0.03; Fig. 4A), whereas there was no difference between dietary groups in the most proximal (P = 0.67) and distal (P = 0.80) regions of the colon (results not shown). Similarly, there was no difference in the degree of colon injury in the proximal (P = 0.12) and distal (P = 0.26) regions of the colon between PPARδ null (PPARδΔIEpC) and wild-type (PPARδF/F) mice. However, an independent effect of genotype was apparent within the middle region of the colon, where the average colon injury score was higher (P = 0.007) in PPARδΔIEpC than PPARδF/F mice (Fig. 4B). Colon tumor entities (including adenocarcinomas and adenomas) were mapped to their specific region within the colon, excised, and typed by a board-certified pathologist (B.W.). Tumors did not develop in the proximal region of the colon in any of the experimental groups. In the most distal region (i.e., the most distal 2 cm) of the colon (46), there was no effect of genotype or diet on the total number of tumors (P = 0.81 and P = 0.78, respectively). However, in the middle region of the colon, an independent effect of diet emerged, wherein the total number of tumor entities was reduced by FO compared with CO (P = 0.01), whereas PPARδ deletion had no effect on total tumor number in the middle colon (P = 0.38; Fig. 4, C and D). Moreover, the percentage of animals that failed to develop colon tumors was higher in FO- than CO-fed mice (48% vs. 18%). Therefore, in a chronic inflammation and malignant transformation (AOM/DSS) model, intestinal epithelial cell-specific deletion of PPARδ affected the gross colonic phenotype by increasing colon injury scores, as seen previously in PPARδ null mice (44), but had no effect on tumor incidence. Interestingly, independent of PPARδ genotype, dietary FO had a beneficial effect on the gross colonic phenotype: it reduced colon injury and tumor incidence. However, these effects were site-specific and restricted to the middle region of the colon.
Fig. 4.
Identification of colonic phenotype in AOM/DSS-treated mice. PPARF/F and PPARδΔIEpC mice were fed CO (n = 11–12 per genotype) or FO (n = 13–14 per genotype) diet (n = 11–14 mice/treatment group) and euthanized 12 wk after completion of the final DSS cycle. Histological scoring (0–3) of colon epithelial injury and typing of tumor entities (total adenomas and adenocarcinomas) were carried out in a blinded manner by a board-certified pathologist (B. Weeks). A and B: independent effect of diet and genotype (IEpC PPARδ status) on colon injury in the middle region of the colon. Data were analyzed by Kruskal-Wallis test, and bars represent median values. *Statistical significance (P ≤ 0.05). C and D: effect of diet and genotype on tumor incidence in the middle region of the colon. Dot plots identify tumor distribution among treatment groups; solid black line denotes median value in each group.
Dietary FO decreases mucosal inflammatory biomarker expression.
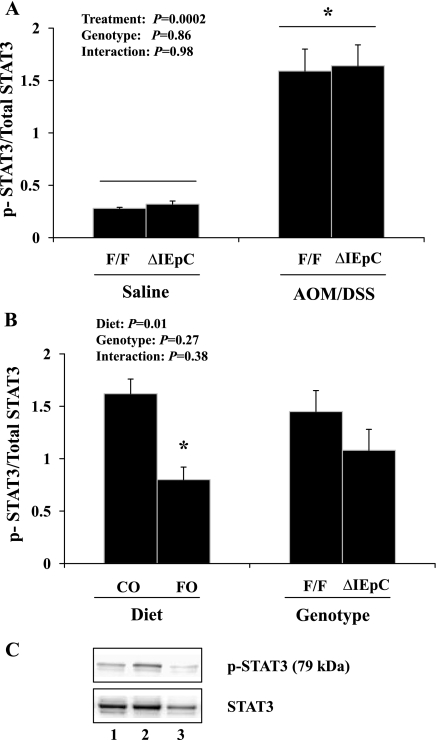
By examining STAT3 phosphorylation status, a well-accepted marker of mucosal inflammation (47), we verified that the AOM/DSS treatment established a subclinical chronic inflammatory microenvironment in the colon. Examination of phosphorylated (i.e., activated) STAT3 relative to total STAT3 expression within the colonic mucosa revealed a significant effect of the inflammation/carcinogen treatment regimen compared with saline-treated controls (P = 0.001; Fig. 5A) on the induction of this critical inflammatory biomarker, whereas PPARδ status had no effect (P = 0.86; Fig. 5A). Within the AOM/DSS-treated groups, we confirmed the anti-inflammatory effect of FO feeding (P = 0.002; Fig. 5B) in the context of chronic intestinal inflammation by decreasing colonic mucosal STAT3 activation. These findings were independent of intestinal epithelial cell PPARδ expression (genotype: P = 0.27; Fig. 5B).
Fig. 5.
Ratio of phosphorylated to total STAT3 expression in murine colonic mucosa as assessed by immunoblotting. Within each genotype (PPARδF/F and PPARδΔIEpC), mice were fed 5% corn oil (CO) diet and treated with AOM/DSS (n = 4) or received an equal volume of saline intraperitoneally without DSS (control) in drinking water (n = 2). Ratio of phosphorylated (i.e., activated) to total STAT3 protein expression (pSTAT3/STAT3) was assessed by 2-way ANOVA. Values are means ± SE. *P ≤ 0.05. A: effect of AOM/DSS treatment (P = 0.0002). B: effect of AOM/DSS in mice from each genotype fed CO (n = 4) or FO (n = 4) diet (P = 0.01). C: representative immunoblots from PPARδF/F mice (top) and phosphorylated and total STAT3 (bottom). Samples are from AOM/DSS-treated FO-fed PPARδF/F mice (lane 1), AOM/DSS-treated CO-fed PPARδF/F mice (lane 2), and saline-treated (no DSS treatment) CO-fed PPARδF/F mice (lane 3).
Dietary FO decreases colonic mucosal mRNA expression of critical inflammatory cytokines.
AOM/DSS increased mucosal mRNA expression of several key inflammatory cytokines above the baseline expression levels in saline-treated control animals in all groups. As expected, the average mRNA expression level of inflammatory cytokines was increased in AOM/DSS-treated mice compared with saline controls as follows: 2.5-fold for IL-6 (P = 0.01), 3.3-fold for IFN-γ (P = 0.04), 28-fold for IL-17A (P = 0.006), 10-fold for IL-17F (P = 0.0002), 9-fold for IL-21 (P = 0.0003), 3.3-fold for IL-27 (P = 0.03), 5.5-fold for IL-23 (P = 0.003), and 5.6-fold for IL-23R (P = 0.02).
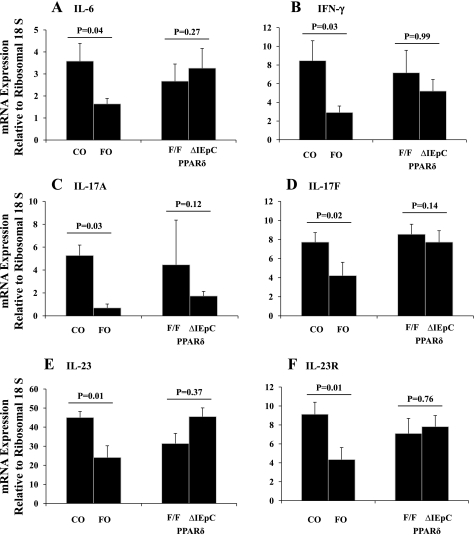
A distinct difference in the cytokine expression profile emerged between dietary groups. The colonic mucosal mRNA expression level of several critical inflammatory cytokines, including IL-6 (P = 0.04), IFN-γ (P = 0.03), IL-17A (P = 0.03), IL-17F (P = 0.02), IL-23 (P = 0.01), and IL-23R (P = 0.01), was consistently decreased in FO- compared with CO-fed animals (Fig. 6). There was no effect of diet on the mucosal mRNA expression of IL-21 (P = 0.31) or IL-27 (P = 0.15) (results not shown). Among the AOM/DSS-treated animals, there was no effect of genotype (i.e., intestinal epithelial cell PPARδ status) on colonic mucosal inflammatory cytokine gene expression: IL-6 (P = 0.27), IFN-γ (P = 0.99), IL-17A (P = 0.12), IL-17F (P = 0.14), IL-23 (P = 0.37), and IL-23R (P = 0.76) (Fig. 6) and IL-21 (P = 0.31) and IL-27 (P = 0.22) (results not shown).
Fig. 6.
Colonic mucosal cytokine mRNA expression. RNA was isolated from colonic mucosal scrapings from mice following exposure to carcinogen and 3 subsequent cycles of DSS. mRNA levels were determined by quantitative RT-PCR, and expression of each gene of interest was normalized to ribosomal 18S expression. Relative expression levels were analyzed for IL-6 (A), IFN-γ (B), IL-17A (C), IL-17F (D), IL-23 (E), and IL-23R (F). Data were analyzed by 2-way ANOVA (main effects: diet and genotype), and significance was at the level of P < 0.05. For all data sets, P (interaction) was not significant (P > 0.05); however, the outcome from each main effect is shown. Values are means ± SE.
Dietary FO and targeted deletion of intestinal epithelial cell PPARδ alters T cell populations locally and systemically.
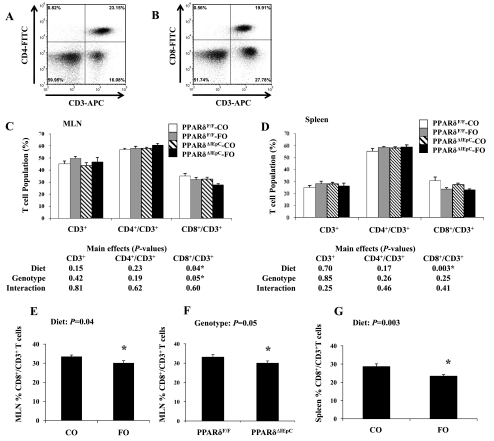
We measured the effect of dietary FO and/or the intestinal epithelial cell-specific deletion of PPARδ expression on T cell populations in the MLN and spleen. Representative histograms depicting double-stained lymphocytes (CD3+CD4+ or CD3+CD8+) from the MLN are presented in Fig. 7, A and B, respectively. Similar outcomes were obtained for double-positive lymphocyte populations isolated from the spleen (results not shown). The percentage of CD3+, CD4+/CD3+, and CD8+/CD3+ T cells residing in the MLN or spleen is depicted in Fig. 7, C and D, respectively. There was no effect of dietary FO and/or intestinal epithelial cell PPARδ deletion on the percentage of total T cells (CD3+) or within the CD4+ T cell population in the MLN or spleen. However, the percentage of CD8+ T cells in the MLN was decreased in FO-fed mice (P = 0.04; Fig. 7E). Interestingly, PPARδ deletion had a similar localized effect, decreasing the percentage of MLN CD8+ T cells relative to wild-type mice (P = 0.05), although these effects were not additive (interaction: P = 0.60; Fig. 7F). The aforementioned localized effects of FO consumption in the MLN were accompanied by a systemic effect, wherein the percentage of splenic CD8+ T cells was decreased in FO- compared with CO-fed animals (P = 0.05; Fig. 7G). Overall, a suppressive effect of dietary n-3 PUFAs was observed on the percentage of CD8+ T cells residing locally (MLN) and systemically (spleen) in both genotypes.
Fig. 7.
Characterization of mesenteric lymph node (MLN) cells by surface staining followed by flow cytometry. A and B: representative plots of CD3 vs. CD4 and CD3 vs. CD8 quadrants from MLN cells of CO-fed PPARδF/F mice. C and D: percentages of CD3+, CD4+/CD3+, and CD8+/CD3+ T cells in MLN and spleen calculated by summing the number of events in each quadrant (n = 5–9 mice per treatment). Within each separate surface marker analysis, resultant P values from a 2-way ANOVA are listed in tables below each graph. Values are means ± SE. *Statistical significance (P ≤ 0.05). E–G: significant main effects from 2-way analysis conducted on MLN and spleen data.
Dietary FO and targeted deletion of intestinal epithelial cell PPARδ alters T cell activation status locally and systemically.
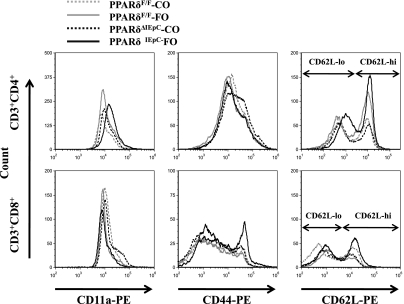
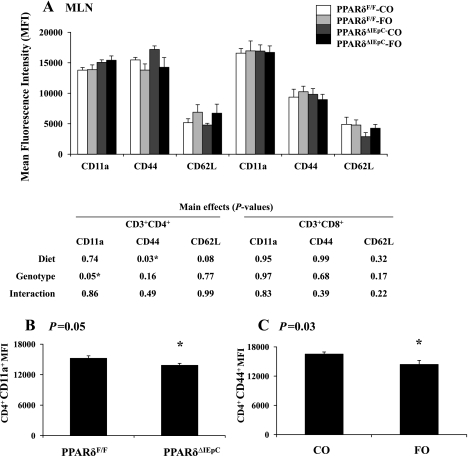
We determined if changes in the intestinal epithelium influence the activation status of T cell populations at local mucosal (MLN) and systemic (spleen) immunological sites. For this purpose, surface expression of the T cell activation markers CD11a, CD44, and CD62L was examined at each lymphoid tissue site. Representative histograms for surface markers expressed by the MLN CD4+ and CD8+ T cell populations are presented in Fig. 8; similar results were obtained in the spleen (results not shown). The mean fluorescence intensity of each surface marker assessed in the CD4+ and CD8+ T cell populations in the MLN is shown in Fig. 9A. Intestinal epithelial cell-specific deletion of PPARδ resulted in a decrease in surface expression of CD11a within the CD4+ T cell population of the MLN (Fig. 9B). Dietary FO also caused a decrease in CD44 expression within the CD4+ T cell population compared with cells isolated from animals fed the control (CO) diet (Fig. 9C). Total CD62L surface expression and bimodal expression (high vs. low) was unaffected by dietary FO or intestinal epithelial cell PPARδ deletion. These results are indicative of a reduction in T cell activation. Interestingly, neither FO consumption nor intestinal epithelial cell-specific deletion of PPARδ influenced the activation status of CD8+ T cells residing in the MLN.
Fig. 8.
Activation assessment of MLN T lymphocytes by surface staining. CD3+CD4+ or CD3+CD8+ double-positive cells were gated, and representative histograms of CD11a, CD44, and CD62L costaining are shown. PE, phycoerythrin.
Fig. 9.
A: quantitative analysis of T lymphocyte activation markers in MLN calculated by mean fluorescence intensity (MFI). Values are means ± SE. *Statistical significance (P ≤ 0.05). P values (main effects) were determined by 2-way ANOVA. B and C: significant main effects.
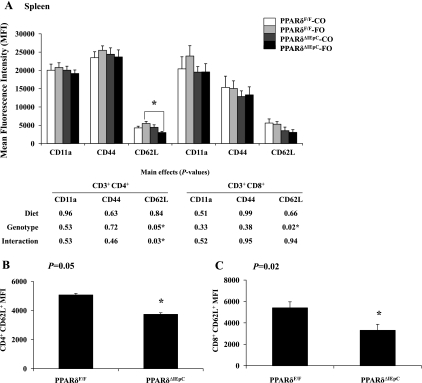
Splenic T cell activation status did not mirror the outcomes determined locally in the MLN (Fig. 10). In contrast to the effects observed in the MLN, there was no effect of diet or intestinal epithelial cell PPARδ genotype on the surface expression of CD11a or CD44 on splenic CD4+ T cells. Moreover, there was no effect of diet on CD62L expression in the CD4+ and CD8+ T cell populations (P = 0.84 and P = 0.66, respectively). Interestingly, the effect of intestinal epithelial cell PPARδ genotype on CD62L expression exhibited a bimodal distribution. Splenic T cells from wild-type PPARδF/F mice exhibited a high surface expression of CD62L (Fig. 10), while PPARδ null (PPARδΔIEpC) mice exhibited low expression of CD62L in the CD4+ (P = 0.05) and CD8+ (P = 0.02) T cell populations (Fig. 10, B and C). Therefore, intestinal epithelial cell deletion of PPARδ had a differential effect in local and systemic secondary lymphoid organs. MLN exhibited a surface maker expression pattern consistent with reduced T cell activation, whereas splenic T cells exhibited changes in surface marker expression consistent with recent activation. Additionally, the combined effect of FO consumption and PPARδ deletion in the intestinal epithelial cell decreased the number of CD4+ CD62L+-expressing lymphocytes in the spleen (P = 0.03), indicating that changes in the intestinal epithelium can impact T cell activation status systemically and can be further modified by diet.
Fig. 10.
A: quantitative analysis of T lymphocyte activation markers in the spleen calculated as mean fluorescence intensity. Values are means ± SE. *Statistical significance (P ≤ 0.05). P values (main effects) were determined by 2-way ANOVA. B and C: significant main effects.
DISCUSSION
In the context of a chronic intestinal inflammation/carcinogenesis model, we have examined the impact of dietary FO and targeted deletion of PPARδ within intestinal epithelial cells on the colonic microenvironment and the activation status of resident T cells in local (MLN) and systemic (spleen) secondary lymphoid organs. To our knowledge, this is the first study to utilize an intestinal epithelial cell-specific PPARδ knockout mouse to determine how alterations in the intestinal epithelial cells can impact adaptive immune competence following carcinogen exposure and the induction of chronic intestinal inflammation. This is noteworthy because T cells have been demonstrated to play a pathogenic role in IBD (80, 86). Since n-3 PUFAs are putative natural ligands for PPARδ (32, 95), we also determined if dietary n-3 PUFAs consumed at physiologically relevant levels for humans (57) would further impact T cell activation status. The contribution of the epithelium to intestinal pathologies is garnering greater appreciation as growing evidence implicates epithelial cell dysfunction as a primary cause of inflammatory pathologies arising in different tissues (87).
Dietary FO reduced colon injury and tumor incidence (adenomas and adenocarcinomas) following exposure to carcinogen and the induction of chronic colonic inflammation (Fig. 4, A and C). This confirms a previous report that n-3 PUFAs reduce colitis-associated colon tumor formation in a genetic model that produces n-3 PUFAs de novo (53) and extends this finding by showing that dietary intervention with n-3 PUFAs can also reduce colitis-associated colon tumor formation. In contrast to the effects of n-3 PUFAs on colon cancer, the role of PPARδ in colon tumorigenesis remains controversial (73). Some studies have shown that PPARδ protects against colon tumorigenesis, some studies have shown that PPARδ promotes colon tumorigenesis, and other studies have shown that PPARδ had no influence on APC-dependent colon tumorigenesis (for review see Ref. 73). In the present study, intestinal epithelial cell PPARδ status had no effect on tumor incidence (Fig. 4D). This is in contrast to a previous study where AOM-induced colon tumorigenesis was mitigated in mice when PPARδ expression was selectively deleted in intestinal epithelial cells (104). The reason for this difference cannot be explained from our results but could be due in part to the differences in the approach used to induce colon tumorigenesis (AOM alone vs. AOM/DSS). Interestingly, deletion of PPARδ within the colonic intestinal epithelium (PPARδΔIEpC) increased the degree of colon injury in response to the AOM/DSS treatment regimen (Fig. 4, B and D), which is consistent with a previous report showing that DSS-induced inflammation is exacerbated in PPARδ null mice (44). These findings collectively support a large body of evidence indicating that PPARδ has potent anti-inflammatory activities in multiple models (54). Additionally, while we found no evidence that PPARδ protects against colon tumorigenesis in the present study, exacerbation of AOM/DSS-induced colon injury when expression of PPARδ was deleted from intestinal epithelial cells is consistent with a protective role for PPARδ in colon tumorigenesis. This is in line with a recent retrospective study demonstrating that colorectal cancer patients with relatively low expression of PPARδ in primary tumors were nearly four times more likely to die from this disease than colorectal cancer patients with higher expression of PPARδ in primary tumors (96). To more definitely determine the role of PPARδ in colon tumorigenesis, future studies assessing the effect of PPARδ status within specific cell types in the inflamed colon and/or colon tumor microenvironment are required, as suggested elsewhere (73). While the mechanisms of colon tumorigenesis are complex, data from the present study do not support a role of intestinal epithelial cell PPARδ in this process.
Within the inflamed colonic mucosa, we confirmed that dietary FO decreases the expression of an inflammatory biomarker, phosphorylated (i.e., activated) STAT3 (52), following carcinogen exposure and the induction of chronic inflammation (Fig. 5), whereas intestinal epithelial cell PPARδ status had no effect on colonic STAT3 expression. Within the gastrointestinal tract, STAT3 resides at the nexus of multiple signaling inputs, the downstream targets of which ultimately link inflammation and tumorigenesis by mediating the activity of inflammatory cytokines and cancer-promoting inflammatory responses (51). Interestingly, STAT3 is constitutively activated in diverse types of cancer and plays a procarcinogenic role by promoting prooncogenic inflammatory pathways and enhancing the transcription of genes associated with cell cycle progression, cell survival, angiogenesis, and immune evasion (99–101). Moreover, STAT3 is often overexpressed in colon cancer, and the anti-inflammatory effects of PPARδ are believed to be mediated, at least in part, via inhibition of STAT3 and its downstream signaling, which is associated with antiapoptotic signaling and c-myc expression (43, 73). However, in the present study, there was no effect of intestinal epithelial cell PPARδ deletion on colon tumor incidence or mucosal STAT3 activation. Conversely, dietary FO suppressed colonic mucosal STAT3 activation and the mRNA expression of IL-6 and IFN-γ, which coincided with a reduced incidence of colon tumor entities and epithelial injury in FO-fed mice, thereby providing insight into a potential mechanism underlying the chemopreventive actions of FO (7, 19, 48, 53, 75, 85).
In T cells, STAT3 expression is essential for colitogenic activity and has a critical role in the differentiation of Th17 cells (22), which are characterized by their potent proinflammatory activities. Moreover, the IL-23/Th17 cell pathway is strongly involved in the pathogenesis of colitis (24, 26, 45, 97), and it is suggested that STAT3 promotes a procarcinogenic Th17 response (59, 94). Interestingly, in a chronic colitis model (3 cycles of DSS), n-3 PUFAs reduced the percentage of Th17 cells (CD4+ IL-17A+) within the inflamed colon lamina propria and reduced mucosal mRNA expression of critical Th17 cell-derived inflammatory cytokines, IL-17 and IL-21 (J. M. Monk et al., unpublished observations). Although the percentage of Th17 cells was not assessed in the present study, the outcome from mucosal mRNA expression supports an inhibitory effect of n-3 PUFAs on Th17 cell function, as dietary FO decreased mucosal mRNA expression of IL-17A and IL-17F (Fig. 6, C and D), cytokines predominantly produced by Th17 cells. The role of Th17 cells and IL-17 in the tumor microenvironment is unclear (64), although overexpression of IL-17 in tumors leads to increased angiogenesis and tumor growth (70), and IL-17−/− and IL-17R−/− mice exhibit reduced tumor growth (40, 91). Furthermore, the tumorigenic effects of IL-17 are mediated, at least in part, by IL-6 via a STAT3-dependent mechanism (91), and all three of these mediators were reduced by FO in the colon following AOM/DSS exposure.
IL-23 drives chronic intestinal inflammation by inducing inflammatory cytokine production and by promoting pathogenic Th1 and Th17 responses in the intestine (63). In addition to promoting tumor incidence and growth (64), IL-23 promotes the maintenance of differentiated Th17 cells and is required for providing Th17 cells with a pathogenic phenotype (79). IL-17 and IL-23 expression is also elevated in human colon cancer, and IL-23p19−/− mice are resistant to tumor induction (60). Interestingly, dietary FO decreased IL-23 and IL-23R colonic mucosal gene expression following AOM/DSS exposure (Fig. 6, E and F). Therefore, IL-23 represents an important molecular link between chronic intestinal inflammation and carcinogenesis, which may be beneficially augmented by dietary FO. Collectively, the aforementioned findings demonstrate a new and previously unappreciated beneficial role of n-3 PUFAs in a chronic inflammation carcinogenesis model, wherein tumor incidence, colonic injury, and inflammatory/protumorigenic mediators were depressed by dietary FO.
We showed that dietary FO decreased the percentage of CD8+ T cells residing in the MLN and spleen, an impact that was not apparent within the CD4+ T cell population at either organ site (Fig. 7). A similar localized effect within the MLN CD8+ T cell population was observed when PPARδ was deleted from the intestinal epithelial cell (Fig. 7). These changes were statistically significant, albeit modest; thus further studies are required to determine if CD8+ effector functions are similarly affected. Previously, consumption of a 4% FO diet elicited a similar modest change in the percentage of lung CD8+ T cells following influenza infection, whereas the CD4+ T cell population was unaffected (82). Despite decreasing cell numbers in secondary lymphoid organs, FO had no effect on CD8+ T cell expression of activation surface markers, indicating that function was unlikely compromised by n-3 PUFAs. In support of this interpretation, n-3 PUFA consumption was shown to have no effect on antigen-driven splenic CD8+ T cell proliferation (49). Expansion and differentiation of CD8+ T cells are critical for host defense against viral and intracellular bacterial infections, and during DSS-induced intestinal inflammation, the mucosa is structurally dysregulated, thereby reducing epithelial barrier integrity and increasing the exposure of the underlying mucosa to luminal bacteria and antigens. Future studies are required to assess the impact of n-3 PUFAs on antimicrobial functions of CD8+ T cells.
Adhesion molecules facilitate the interaction between T lymphocytes and either antigen-presenting cells (APCs) or the vascular endothelium, and efficient cell-mediated immune responses require appropriate surface expression of these molecules. Furthermore, the trafficking of lymphocytes between body compartments (i.e., into/out of lymphoid organs and into sites of immune or inflammatory reactivity) is also dependent on adhesion molecule expression (78). In this study, activation status of resident T lymphocytes in the MLN and spleen was identified on the basis of the surface expression pattern of three markers functionally associated with trafficking to inflammatory sites (3, 21, 72): CD11a (LFA-1), CD44, and CD62L (L-selectin). Typically, CD11a and CD44 expression is low, whereas CD62L expression is high on naive T cells compared with antigen-experienced T cells (effector and memory), which express high levels of CD11a and CD44 and reduced levels of CD62L (3, 9, 21, 49). Therefore, the expression pattern of these T cell activation markers was specifically chosen to detect antigen-experienced T lymphocytes as seen previously (9, 21, 23). CD62L initiates lymphocyte homing to lymph nodes (61, 93). Interaction between T cells and APCs is facilitated by CD11a (4, 72, 84), thereby promoting T cell activation (1, 4, 72). CD44 is involved in the recruitment of leukocytes to inflammatory sites (21) and plays a role in signaling to downstream target genes involved in orchestrating inflammatory responses (30); thus its expression is elevated within inflamed tissues (39).
With respect to lymphocyte activation status, FO consumption decreased CD44 expression on MLN CD4+ T cells (Fig. 9). Additionally, within the same lymphocyte population, intestinal epithelial cell PPARδ deletion resulted in decreased CD11a expression (Fig. 9). Both of these modest, but significant, changes in surface marker expression are consistent with reduced T cell activation in the MLN. Moreover, the effect of FO appears to be localized to the MLN, which is in close proximity to and drains the inflamed colon, as no impact of FO on T cell activation status was apparent within the spleen. This finding extends previous reports of FO reducing CD44 expression in human monocytes (62) and rat lymphocytes (78) to that in the mouse within the context of a chronic inflammation/malignant transformation model. Reducing CD4+ T cell activation may represent an additional mechanism through which FO is able to minimize the effects of pathogenic CD4+ T cells in DSS-induced IBD (80, 86). We previously demonstrated that n-3 PUFAs directly suppress CD4+ Th1 cell development (102) and CD4+ T cell numbers within the lamina propria following exposure to AOM/DSS (53). CD44 expression is important for the recruitment of leukocytes to inflammatory sites (21), and CD44 signaling is involved in the generation of inflammatory responses (30); therefore, the reduced expression in MLN CD4+ T cells from FO-fed animals is consistent with the general anti-inflammatory biological actions of n-3 PUFAs (13, 15, 16, 53). Previously, n-3 PUFAs have been shown to impair T cell activation at the immunological synapse (28, 29, 55, 56, 98) by altering the phospholipid and signaling protein composition of lipid rafts, i.e., specialized plasma membrane microdomains important for T cell-receptor signaling pathways (15, 35, 37). CD44 has been shown to mediate the cytoskeletal rearrangements that are required for the initiation of T cell activation (31). Therefore, engagement of CD44 on the T cell surface helps stabilize the immunological synapse by initiating F-actin bundle formation, which is accompanied by a redistribution of CD44 and the associated tyrosine kinases (lck and fyn) into lipid rafts at the immunological synapse (31). Interestingly, n-3 PUFAs have been shown to decrease key signaling proteins and F-actin recruitment into lipid rafts at the immunological synapse (55). Therefore, the aforementioned impairment in key aspects of T cell activation by FO may also include a disruption of CD44 localization into lipid rafts. Future studies are required to evaluate the effect of n-3 PUFAs and modulation of lipid rafts with respect to the contribution of CD44 and actin remodeling in T cells at the immunological synapse.
Within the MLN intestinal epithelial cell, PPARδ deletion resulted in a modest decrease in CD4+ T cell expression of CD11a (Fig. 9), a finding consistent with reduced T cell activation. Conversely, in the spleen, a bimodal CD62L surface expression pattern emerged in the CD4+ and CD8+ lymphocyte populations, wherein PPARδΔIEpC mice exhibited low expression and PPARδF/F mice exhibited high expression (Fig. 10). Therefore, intestinal epithelial cell PPARδ deletion promoted CD4+ and CD8+ T cell activation in the spleen. Lastly, combined FO consumption and intestinal epithelial cell PPARδ deletion resulted in decreased splenic CD4+ T cell expression of CD62L (Fig. 10), indicating a synergistic effect of dietary bioactive ingredient (n-3 PUFAs) and intestinal epithelial cell PPARδ status.
To our knowledge, we are the first to demonstrate that specific changes in the intestinal epithelial cell during chronic inflammation can impact T cell activation status and that this process can be further modulated by diet. Collectively, these findings demonstrate that, in a chronic intestinal inflammation/carcinogenesis model, alterations in the intestinal epithelial cell (via PPARδ deletion) can differentially impact T lymphocyte activation status in local vs. systemic secondary lymphoid organs, i.e., a modest depressive localized effect in the draining MLN and an enhanced systematic effect in the spleen. The data support the interpretation that intestinal epithelial cells have an active role in intestinal inflammatory processes and have the capacity to impact adaptive immune outcomes beyond the intestine. These findings provide a basis for future research initiatives directed toward identifying the specific contribution of intestinal epithelial cells to inflammatory pathologies.
Because of their centralized position in the intestinal mucosa, intestinal epithelial cells are a critical component of the mucosal immune system. Antigen presentation in the gut is not limited to classical APCs, as intestinal epithelial cells have been demonstrated to present luminal antigen directly to T cells in a polarized fashion with apical antigen uptake and basolateral antigen presentation to mucosal lymphocytes (41, 42) via major histocompatibility complex class II and costimulatory molecules (17, 69, 83, 90), the expression of which is upregulated in response to proinflammatory signals (11). Under normal intestinal conditions, intestinal epithelial cells lack classical costimulatory molecule expression, and interaction with naive CD4+ T cells would likely result in the induction of anergy, a mechanism necessary to support the state of mucosal immune hyporesponsiveness given the load of foreign antigen encountered in the intestine. Under conditions of intestinal inflammation, intestinal epithelial cells have the capacity to provide the second stimulus required to present antigen to naive T cells, thereby potentially contributing to exaggerated T cell activation often attributable to intestinal inflammatory pathologies.
When stress to the epithelium is relatively mild, epithelial cells secrete cytokines and other mediators that directly influence T cell responses and can elicit a range of functional immune outcomes that are essential for host protection and the limitation of immunopathology. When epithelial damage and dysregulation occur (i.e., AOM/DSS model), epithelial cell-derived mediators redirect the nature of T cell-mediated responses toward inflammatory (type 1) responses that are associated with disease (87). Further evidence suggests that the mucosal immune system is a system-wide organ, wherein studies have demonstrated that stimulation in one mucosal compartment can lead to changes in distal areas (33). However, the elements that link the mucosal immune compartments remain undetermined (33). Our findings that genetic and dietary perturbations in the intestinal epithelium can ultimately and differentially impact T cell activation status in local and systemic secondary lymphoid organs demonstrate a previously unidentified role of the intestinal epithelial cell in chronic intestinal inflammation. In addition, it is now evident that the mucosal epithelium can ultimately impact immune cell function in anatomically distant sites. The immunosuppressive effects of dietary FO within the chronically inflamed colon following carcinogen exposure on mucosal inflammatory responses and T cell activation support a growing number of studies indicating that bioactive food components can favorably modulate the clinical course of IBD and colorectal cancer.
GRANTS
This study was supported by National Institutes of Health Grants DK-071707 and CA-59034, US Department of Agriculture Vegetable Fruit Improvement Center Grant 2009-34402-19831, and Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship PDF-388466-2010 (to J. M. Monk).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.M.M., W.K., R.C.A., D.N.M., and R.S.C. are responsible for conception and design of the research; J.M.M., W.K., E.S.C., H.F.T., and J.E.F. performed the experiments; J.M.M., W.K., J.M.P., and B.W. analyzed the data; J.M.M., J.M.P., B.W., and R.S.C. interpreted the results of the experiments; J.M.M. prepared the figures; J.M.M. drafted the manuscript; J.M.M., J.E.F., J.M.P., D.N.M., and R.S.C. edited and revised the manuscript; J.M.M., W.K., E.S.C., H.F.T., J.E.F., J.M.P., W.H., B.W., R.C.A., D.N.M., and R.S.C. approved the final version of the manuscript.
ACKNOWLEDGMENTS
PPARδF/F mice were generously provided by Dr. Ronald Evans (The Salk Institute). Villin-Cre mice were kindly provided by Dr. Sylvie Robine (Institut Curie).
REFERENCES
- 1. Abraham C, Griffith J, Miller J. The dependence for leukocyte function-associated antigen-1/ICAM-1 interactions in T cell activation cannot be overcome by expression of high density TCR ligand. J Immunol 162: 4399–4405, 1999 [PubMed] [Google Scholar]
- 2. Akiyama TE, Meinke PT, Berger JP. PPAR ligands: potential therapies for metabolic syndrome. Curr Diab Rep 5: 45–52, 2005 [DOI] [PubMed] [Google Scholar]
- 3. Baaten BJ, Li CR, Deiro MF, Lin MM, Linton PJ, Bradley LM. CD44 regulates survival and memory development in Th1 cells. Immunity 32: 104–115, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bachmann MF, McKall-Faienza K, Schmits R, Bouchard D, Beach J, Speiser DE, Mak TW, Ohashi PS. Distinct roles for LFA-1 and CD28 during activation of naive T cells: adhesion versus costimulation. Immunity 7: 549–557, 1997 [DOI] [PubMed] [Google Scholar]
- 5. Barak Y, Liao D, He W, Ong ES, Nelson MC, Olefsky JM, Boland R, Evans RM. Effects of peroxisome proliferator-activated receptor-δ on placentation, adiposity, and colorectal cancer. Proc Natl Acad Sci USA 99: 303–308, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bassaganya-Riera J, Hontecillas R. CLA and n-3 PUFA differentially modulate clinical activity and colonic PPAR-responsive gene expression in a pig model of experimental IBD. Clin Nutr 25: 454–465, 2006 [DOI] [PubMed] [Google Scholar]
- 7. Belluzzi A, Boschi S, Brignola C, Munarini A, Cariani G, Miglio F. Polyunsaturated fatty acids and inflammatory bowel disease. Am J Clin Nutr 71: 339S–342S, 2000 [DOI] [PubMed] [Google Scholar]
- 8. Belluzzi A, Brignola C, Campieri M, Pera A, Boschi S, Miglioli M. Effect of an enteric-coated fish-oil preparation on relapses in Crohn's disease. N Engl J Med 334: 1557–1560, 1996 [DOI] [PubMed] [Google Scholar]
- 9. Bingaman AW, Patke DS, Mane VR, Ahmadzadeh M, Ndejembi M, Bartlett ST, Farber DL. Novel phenotypes and migratory properties distinguish memory CD4 T cell subsets in lymphoid and lung tissue. Eur J Immunol 35: 3173–3186, 2005 [DOI] [PubMed] [Google Scholar]
- 10. Bland PW, Warren LG. Antigen presentation by epithelial cells of the rat small intestine. I. Kinetics, antigen specificity and blocking by anti-Ia antisera. Immunology 58: 1–7, 1986 [PMC free article] [PubMed] [Google Scholar]
- 11. Buning J, Schmitz M, Repenning B, Ludwig D, Schmidt MA, Strobel S, Zimmer KP. Interferon-γ mediates antigen trafficking to MHC class II-positive late endosomes of enterocytes. Eur J Immunol 35: 831–842, 2005 [DOI] [PubMed] [Google Scholar]
- 12. Burdick AD, Kim DJ, Peraza MA, Gonzalez FJ, Peters JM. The role of peroxisome proliferator-activated receptor-β/δ in epithelial cell growth and differentiation. Cell Signal 18: 9–20, 2006 [DOI] [PubMed] [Google Scholar]
- 13. Calder PC. n-3 polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am J Clin Nutr 83: 1505S–1519S, 2006 [DOI] [PubMed] [Google Scholar]
- 14. Chang WC, Chapkin RS, Lupton JR. Predictive value of proliferation, differentiation and apoptosis as intermediate markers for colon tumorigenesis. Carcinogenesis 18: 721–730, 1997 [DOI] [PubMed] [Google Scholar]
- 15. Chapkin RS, Davidson LA, Ly L, Weeks BR, Lupton JR, McMurray DN. Immunomodulatory effects of (n-3) fatty acids: putative link to inflammation and colon cancer. J Nutr 137: 200S–204S, 2007 [DOI] [PubMed] [Google Scholar]
- 16. Chapkin RS, McMurray DN, Lupton JR. Colon cancer, fatty acids and anti-inflammatory compounds. Curr Opin Gastroenterol 23: 48–54, 2007 [DOI] [PubMed] [Google Scholar]
- 17. Dahan S, Roth-Walter F, Arnaboldi P, Agarwal S, Mayer L. Epithelia:lymphocyte interactions in the gut. Immunol Rev 215: 243–253, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davidson LA, Lupton JR, Jiang YH, Chapkin RS. Carcinogen and dietary lipid regulate ras expression and localization in rat colon without affecting farnesylation kinetics. Carcinogenesis 20: 785–791, 1999 [DOI] [PubMed] [Google Scholar]
- 19. Davidson LA, Nguyen DV, Hokanson RM, Callaway ES, Isett RB, Turner ND, Dougherty ER, Wang N, Lupton JR, Carroll RJ, Chapkin RS. Chemopreventive n-3 polyunsaturated fatty acids reprogram genetic signatures during colon cancer initiation and progression in the rat. Cancer Res 64: 6797–6804, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daynes RA, Jones DC. Emerging roles of PPARs in inflammation and immunity. Nat Rev Immunol 2: 748–759, 2002 [DOI] [PubMed] [Google Scholar]
- 21. DeGrendele HC, Estess P, Picker LJ, Siegelman MH. CD44 and its ligand hyaluronate mediate rolling under physiologic flow: a novel lymphocyte-endothelial cell primary adhesion pathway. J Exp Med 183: 1119–1130, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, O'Shea JJ. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32: 605–615, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dutton RW, Bradley LM, Swain SL. T cell memory. Annu Rev Immunol 16: 201–223, 1998 [DOI] [PubMed] [Google Scholar]
- 24. Eastaff-Leung N, Mabarrack N, Barbour A, Cummins A, Barry S. Foxp3+ regulatory T cells, Th17 effector cells, and cytokine environment in inflammatory bowel disease. J Clin Immunol 30: 80–89, 2010 [DOI] [PubMed] [Google Scholar]
- 25. el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39: 186–193, 2004 [DOI] [PubMed] [Google Scholar]
- 26. Elson CO, Cong Y, Weaver CT, Schoeb TR, McClanahan TK, Fick RB, Kastelein RA. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology 132: 2359–2370, 2007 [DOI] [PubMed] [Google Scholar]
- 27. Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology 142: 4195–4202, 2001 [DOI] [PubMed] [Google Scholar]
- 28. Fan YY, Ly LH, Barhoumi R, McMurray DN, Chapkin RS. Dietary docosahexaenoic acid suppresses T cell protein kinase C-θ lipid raft recruitment and IL-2 production. J Immunol 173: 6151–6160, 2004 [DOI] [PubMed] [Google Scholar]
- 29. Fan YY, Spencer TE, Wang N, Moyer MP, Chapkin RS. Chemopreventive n-3 fatty acids activate RXRα in colonocytes. Carcinogenesis 24: 1541–1548, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fitzgerald KA, O'Neill LA. Characterization of CD44 induction by IL-1: a critical role for Egr-1. J Immunol 162: 4920–4927, 1999 [PubMed] [Google Scholar]
- 31. Foger N, Marhaba R, Zoller M. Involvement of CD44 in cytoskeleton rearrangement and raft reorganization in T cells. J Cell Sci 114: 1169–1178, 2001 [DOI] [PubMed] [Google Scholar]
- 32. Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors-α and -δ. Proc Natl Acad Sci USA 94: 4312–4317, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gill N, Wlodarska M, Finlay BB. The future of mucosal immunology: studying an integrated system-wide organ. Nat Immunol 11: 558–560, 2010 [DOI] [PubMed] [Google Scholar]
- 34. Girroir EE, Hollingshead HE, He P, Zhu B, Perdew GH, Peters JM. Quantitative expression patterns of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) protein in mice. Biochem Biophys Res Commun 371: 456–461, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Giurisato E, McIntosh DP, Tassi M, Gamberucci A, Benedetti A. T cell receptor can be recruited to a subset of plasma membrane rafts, independently of cell signaling and attendantly to raft clustering. J Biol Chem 278: 6771–6778, 2003 [DOI] [PubMed] [Google Scholar]
- 36. Grimaldi PA. Regulatory role of peroxisome proliferator-activated receptor-δ (PPAR-δ) in muscle metabolism. A new target for metabolic syndrome treatment? Biochimie 87: 5–8, 2005 [DOI] [PubMed] [Google Scholar]
- 37. Harder T, Engelhardt KR. Membrane domains in lymphocytes—from lipid rafts to protein scaffolds. Traffic 5: 265–275, 2004 [DOI] [PubMed] [Google Scholar]
- 38. Harman FS, Nicol CJ, Marin HE, Ward JM, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-δ attenuates colon carcinogenesis. Nat Med 10: 481–483, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Haynes BF, Hale LP, Patton KL, Martin ME, McCallum RM. Measurement of an adhesion molecule as an indicator of inflammatory disease activity. Up-regulation of the receptor for hyaluronate (CD44) in rheumatoid arthritis. Arthritis Rheum 34: 1434–1443, 1991 [DOI] [PubMed] [Google Scholar]
- 40. He D, Li H, Yusuf N, Elmets CA, Li J, Mountz JD, Xu H. IL-17 promotes tumor development through the induction of tumor promoting microenvironments at tumor sites and myeloid-derived suppressor cells. J Immunol 184: 2281–2288, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hershberg RM, Cho DH, Youakim A, Bradley MB, Lee JS, Framson PE, Nepom GT. Highly polarized HLA class II antigen processing and presentation by human intestinal epithelial cells. J Clin Invest 102: 792–803, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hershberg RM, Framson PE, Cho DH, Lee LY, Kovats S, Beitz J, Blum JS, Nepom GT. Intestinal epithelial cells use two distinct pathways for HLA class II antigen processing. J Clin Invest 100: 204–215, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hodge DR, Hurt EM, Farrar WL. The role of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer 41: 2502–2512, 2005 [DOI] [PubMed] [Google Scholar]
- 44. Hollingshead HE, Morimura K, Adachi M, Kennett MJ, Billin AN, Willson TM, Gonzalez FJ, Peters JM. PPARβ/δ protects against experimental colitis through a ligand-independent mechanism. Dig Dis Sci 52: 2912–2919, 2007 [DOI] [PubMed] [Google Scholar]
- 45. Holtta V, Klemetti P, Sipponen T, Westerholm-Ormio M, Kociubinski G, Salo H, Rasanen L, Kolho KL, Farkkila M, Savilahti E, Vaarala O. IL-23/IL-17 immunity as a hallmark of Crohn's disease. Inflamm Bowel Dis 14: 1175–1184, 2008 [DOI] [PubMed] [Google Scholar]
- 46. Hong MY, Chapkin RS, Morris JS, Wang N, Carroll RJ, Turner ND, Chang WC, Davidson LA, Lupton JR. Anatomical site-specific response to DNA damage is related to later tumor development in the rat azoxymethane colon carcinogenesis model. Carcinogenesis 22: 1831–1835, 2001 [DOI] [PubMed] [Google Scholar]
- 47. Hruz P, Dann SM, Eckmann L. STAT3 and its activators in intestinal defense and mucosal homeostasis. Curr Opin Gastroenterol 26: 109–115, 2010 [DOI] [PubMed] [Google Scholar]
- 48. Hudert CA, Weylandt KH, Lu Y, Wang J, Hong S, Dignass A, Serhan CN, Kang JX. Transgenic mice rich in endogenous ω-3 fatty acids are protected from colitis. Proc Natl Acad Sci USA 103: 11276–11281, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Irons R, Fritsche KL. n-3 PUFA fail to affect in vivo, antigen-driven CD8+ T-cell proliferation in the spleen of naive mice. Br J Nutr 95: 838–844, 2006 [DOI] [PubMed] [Google Scholar]
- 50. Itzkowitz SH, Yio X. Inflammation and cancer. IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol 287: G7–G17, 2004 [DOI] [PubMed] [Google Scholar]
- 51. Jarnicki A, Putoczki T, Ernst M. Stat3: linking inflammation to epithelial cancer—more than a “gut” feeling? Cell Div 5: 14, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jia Q, Ivanov I, Zlatev Z, Alaniz RC, Weeks BR, Callaway E, Goldsby JS, Davidson LA, Fan YY, Zhou L, Lupton JR, McMurray DN, Chapkin RS. Dietary fish oil and cucumin combine to modulate colonic cytokinetics and gene expression in dextran sodium sulphate-treated mice. Br J Nutr 106: 519–529, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jia Q, Lupton JR, Smith R, Weeks BR, Callaway E, Davidson LA, Kim W, Fan YY, Yang P, Newman RA, Kang JX, McMurray DN, Chapkin RS. Reduced colitis-associated colon cancer in Fat-1 (n-3 fatty acid desaturase) transgenic mice. Cancer Res 68: 3985–3991, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kilgore KS, Billin AN. PPARβ/δ ligands as modulators of the inflammatory response. Curr Opin Investig Drugs 9: 463–469, 2008 [PubMed] [Google Scholar]
- 55. Kim W, Fan YY, Barhoumi R, Smith R, McMurray DN, Chapkin RS. n-3 polyunsaturated fatty acids suppress the localization and activation of signaling proteins at the immunological synapse in murine CD4+ T cells by affecting lipid raft formation. J Immunol 181: 6236–6243, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim W, Khan NA, McMurray DN, Prior IA, Wang N, Chapkin RS. Regulatory activity of polyunsaturated fatty acids in T-cell signaling. Prog Lipid Res 49: 250–261, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim W, McMurray DN, Chapkin RS. n-3 polyunsaturated fatty acids—physiological relevance of dose. Prostaglandins Leukot Essent Fatty Acids 82: 155–158, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kong SC, Hurlstone DP, Pocock CY, Walkington LA, Farquharson NR, Bramble MG, McAlindon ME, Sanders DS. The incidence of self-prescribed oral complementary and alternative medicine use by patients with gastrointestinal diseases. J Clin Gastroenterol 39: 138–141, 2005 [PubMed] [Google Scholar]
- 59. Kortylewski M, Xin H, Kujawski M, Lee H, Liu Y, Harris T, Drake C, Pardoll D, Yu H. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell 15: 114–123, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, Basham B, McClanahan T, Kastelein RA, Oft M. IL-23 promotes tumour incidence and growth. Nature 442: 461–465, 2006 [DOI] [PubMed] [Google Scholar]
- 61. Ley K, Kansas GS. Selectins in T-cell recruitment to non-lymphoid tissues and sites of inflammation. Nat Rev Immunol 4: 325–335, 2004 [DOI] [PubMed] [Google Scholar]
- 62. Madden J, Shearman CP, Dunn RL, Dastur ND, Tan RM, Nash GB, Rainger GE, Brunner A, Calder PC, Grimble RF. Altered monocyte CD44 expression in peripheral arterial disease is corrected by fish oil supplementation. Nutr Metab Cardiovasc Dis 19: 247–252, 2009 [DOI] [PubMed] [Google Scholar]
- 63. Maloy KJ, Kullberg MC. IL-23 and Th17 cytokines in intestinal homeostasis. Mucosal Immunol 1: 339–349, 2008 [DOI] [PubMed] [Google Scholar]
- 64. Maniati E, Soper R, Hagemann T. Up for mischief? IL-17/Th17 in the tumour microenvironment. Oncogene 29: 5653–5662, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Marin HE, Peraza MA, Billin AN, Willson TM, Ward JM, Kennett MJ, Gonzalez FJ, Peters JM. Ligand activation of peroxisome proliferator-activated receptor-β inhibits colon carcinogenesis. Cancer Res 66: 4394–4401, 2006 [DOI] [PubMed] [Google Scholar]
- 66. Mayer L, Eisenhardt D, Salomon P, Bauer W, Plous R, Piccinini L. Expression of class II molecules on intestinal epithelial cells in humans. Differences between normal and inflammatory bowel disease. Gastroenterology 100: 3–12, 1991 [DOI] [PubMed] [Google Scholar]
- 67. Mayer L, Shlien R. Evidence for function of Ia molecules on gut epithelial cells in man. J Exp Med 166: 1471–1483, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Munkholm P. The incidence and prevalence of colorectal cancer in inflammatory bowel disease. Aliment Pharmacol Ther 18 Suppl 2: 1–5, 2003 [DOI] [PubMed] [Google Scholar]
- 69. Nakazawa A, Watanabe M, Kanai T, Yajima T, Yamazaki M, Ogata H, Ishii H, Azuma M, Hibi T. Functional expression of costimulatory molecule CD86 on epithelial cells in the inflamed colonic mucosa. Gastroenterology 117: 536–545, 1999 [DOI] [PubMed] [Google Scholar]
- 70. Numasaki M, Fukushi J, Ono M, Narula SK, Zavodny PJ, Kudo T, Robbins PD, Tahara H, Lotze MT. Interleukin-17 promotes angiogenesis and tumor growth. Blood 101: 2620–2627, 2003 [DOI] [PubMed] [Google Scholar]
- 71. Park BH, Vogelstein B, Kinzler KW. Genetic disruption of PPARδ decreases the tumorigenicity of human colon cancer cells. Proc Natl Acad Sci USA 98: 2598–2603, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pavlick KP, Ostanin DV, Furr KL, Laroux FS, Brown CM, Gray L, Kevil CG, Grisham MB. Role of T-cell-associated lymphocyte function-associated antigen-1 in the pathogenesis of experimental colitis. Int Immunol 18: 389–398, 2006 [DOI] [PubMed] [Google Scholar]
- 73. Peters JM, Hollingshead HE, Gonzalez FJ. Role of peroxisome-proliferator-activated receptor β/δ (PPARβ/δ) in gastrointestinal tract function and disease. Clin Sci (Lond) 115: 107–127, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Podolsky DK. Inflammatory bowel disease. N Engl J Med 347: 417–429, 2002 [DOI] [PubMed] [Google Scholar]
- 75. Prescott SM, Stenson WF. Fish oil fix. Nat Med 11: 596–598, 2005 [DOI] [PubMed] [Google Scholar]
- 76. Reed KR, Sansom OJ, Hayes AJ, Gescher AJ, Winton DJ, Peters JM, Clarke AR. PPARδ status and Apc-mediated tumourigenesis in the mouse intestine. Oncogene 23: 8992–8996, 2004 [DOI] [PubMed] [Google Scholar]
- 77. Rubin DT, Kavitt RT. Surveillance for cancer and dysplasia in inflammatory bowel disease. Gastroenterol Clin North Am 35: 581–604, 2006 [DOI] [PubMed] [Google Scholar]
- 78. Sanderson P, Calder PC. Dietary fish oil diminishes lymphocyte adhesion to macrophage and endothelial cell monolayers. Immunology 94: 79–87, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sarra M, Pallone F, Macdonald TT, Monteleone G. IL-23/IL-17 axis in IBD. Inflamm Bowel Dis 16: 1808–1813, 2010 [DOI] [PubMed] [Google Scholar]
- 80. Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol 3: 390–407, 2006 [DOI] [PubMed] [Google Scholar]
- 81. Schulz TJ, Glaubitz M, Kuhlow D, Thierbach R, Birringer M, Steinberg P, Pfeiffer AF, Ristow M. Variable expression of Cre recombinase transgenes precludes reliable prediction of tissue-specific gene disruption by tail-biopsy genotyping. PLos One 2: e1013, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schwerbrock NM, Karlsson EA, Shi Q, Sheridan PA, Beck MA. Fish oil-fed mice have impaired resistance to influenza infection. J Nutr 139: 1588–1594, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Selby WS, Janossy G, Mason DY, Jewell DP. Expression of HLA-DR antigens by colonic epithelium in inflammatory bowel disease. Clin Exp Immunol 53: 614–618, 1983 [PMC free article] [PubMed] [Google Scholar]
- 84. Springer TA. Adhesion receptors of the immune system. Nature 346: 425–434, 1990 [DOI] [PubMed] [Google Scholar]
- 85. Stenson WF, Cort D, Rodgers J, Burakoff R, DeSchryver-Kecskemeti K, Gramlich TL, Beeken W. Dietary supplementation with fish oil in ulcerative colitis. Ann Intern Med 116: 609–614, 1992 [DOI] [PubMed] [Google Scholar]
- 86. Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu Rev Immunol 20: 495–549, 2002 [DOI] [PubMed] [Google Scholar]
- 87. Swamy M, Jamora C, Havran W, Hayday A. Epithelial decision makers: in search of the “epimmunome.” Nat Immunol 11: 656–665, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tanaka T, Kohno H, Yoshitani S, Takashima S, Okumura A, Murakami A, Hosokawa M. Ligands for peroxisome proliferator-activated receptors-α and -γ inhibit chemically induced colitis and formation of aberrant crypt foci in rats. Cancer Res 61: 2424–2428, 2001 [PubMed] [Google Scholar]
- 89. Turk HF, Kolar SS, Fan YY, Cozby CA, Lupton JR, Chapkin RS. Linoleic acid and butyrate synergize to increase Bcl-2 levels in colonocytes. Int J Cancer 128: 63–71, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Vainer B, Horn T, Nielsen OH. Colonic epithelial cell expression of ICAM-1 relates to loss of surface continuity: a comparative study of inflammatory bowel disease and colonic neoplasms. Scand J Gastroenterol 41: 318–325, 2006 [DOI] [PubMed] [Google Scholar]
- 91. Wang L, Yi T, Kortylewski M, Pardoll DM, Zeng D, Yu H. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J Exp Med 206: 1457–1464, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Welch JS, Ricote M, Akiyama TE, Gonzalez FJ, Glass CK. PPARγ and PPARδ negatively regulate specific subsets of lipopolysaccharide and IFN-γ target genes in macrophages. Proc Natl Acad Sci USA 100: 6712–6717, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wirth TC, Badovinac VP, Zhao L, Dailey MO, Harty JT. Differentiation of central memory CD8 T cells is independent of CD62L-mediated trafficking to lymph nodes. J Immunol 182: 6195–6206, 2009 [DOI] [PubMed] [Google Scholar]
- 94. Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 15: 1016–1022, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Xu HE, Lambert MH, Montana VG, Parks DJ, Blanchard SG, Brown PJ, Sternbach DD, Lehmann JM, Wisely GB, Willson TM, Kliewer SA, Milburn MV. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell 3: 397–403, 1999 [DOI] [PubMed] [Google Scholar]
- 96. Yang L, Zhang H, Zhou ZG, Yan H, Adell G, Sun XF. Biological function and prognostic significance of peroxisome proliferator-activated receptor-δ in rectal cancer. Clin Cancer Res 17: 3760–3770, 2011 [DOI] [PubMed] [Google Scholar]
- 97. Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, Murphy E, Sathe M, Cua DJ, Kastelein RA, Rennick D. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest 116: 1310–1316, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yog R, Barhoumi R, McMurray DN, Chapkin RS. n-3 polyunsaturated fatty acids suppress mitochondrial translocation to the immunologic synapse and modulate calcium signaling in T cells. J Immunol 184: 5865–5873, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer 4: 97–105, 2004 [DOI] [PubMed] [Google Scholar]
- 100. Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9: 798–809, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Yu QT, Saruta M, Avanesyan A, Fleshner PR, Banham AH, Papadakis KA. Expression and functional characterization of FOXP3+ CD4+ regulatory T cells in ulcerative colitis. Inflamm Bowel Dis 13: 191–199, 2007 [DOI] [PubMed] [Google Scholar]
- 102. Zhang P, Kim W, Zhou L, Wang N, Ly LH, McMurray DN, Chapkin RS. Dietary fish oil inhibits antigen-specific murine Th1 cell development by suppression of clonal expansion. J Nutr 136: 2391–2398, 2006 [DOI] [PubMed] [Google Scholar]
- 103. Zhang X, Young HA. PPAR and immune system—what do we know? Int Immunopharmacol 2: 1029–1044, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zuo X, Peng Z, Moussalli MJ, Morris JS, Broaddus RR, Fischer SM, Shureiqi I. Targeted genetic disruption of peroxisome proliferator-activated receptor-δ and colonic tumorigenesis. J Natl Cancer Inst 101: 762–767, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]