Background: NucS, new bipolar nuclease acting on branched DNA repair substrates, interacts with proliferating cell nuclear antigen (PCNA).
Results: PCNA and NucS form a stable 1:1 complex and PCNA directs the activity of NucS toward single-stranded/double-stranded DNA junctions in branched DNA substrates.
Conclusion: PCNA regulates NucS activity, preventing the nonspecific cleavage of NucS on the chromatin.
Significance: This study will help understand how PCNA regulates its client enzymes.
Keywords: Archaea, DNA Enzymes, DNA Repair, DNA-Protein Interaction, X-ray Scattering, Fluorescence Anisotropy, Surface Plasmon Resonance, Protein Complexes
Abstract
Pyrococcus abyssi NucS is the founding member of a new family of structure-specific DNA endonucleases that interact with the replication clamp proliferating cell nuclear antigen (PCNA). Using a combination of small angle x-ray scattering and surface plasmon resonance analyses, we demonstrate the formation of a stable complex in solution, in which one molecule of the PabNucS homodimer binds to the outside surface of the PabPCNA homotrimer. Using fluorescent labels, PCNA is shown to increase the binding affinity of NucS toward single-strand/double-strand junctions on 5′ and 3′ flaps, as well as to modulate the cleavage specificity on the branched DNA structures. Our results indicate that the presence of a single major contact between the PabNucS and PabPCNA proteins, together with the complex-induced DNA bending, facilitate conformational flexibility required for specific cleavage at the single-strand/double-strand DNA junction.
Introduction
To counteract the deleterious effects of the large variety of DNA lesions caused by endogenous or environmental factors, living organisms have evolved a multitude of biochemical strategies to maintain genomic integrity. DNA can be repaired either by directly reversing damage or, alternatively, by excision of abnormal DNA elements prior to DNA repair. Removal of these elements requires many nucleases that in a highly controlled fashion resolve irregular DNA structures like flaps, loops, splayed arms, replication forks, and Holliday junctions formed during DNA repair and/or recombination. Many of these abnormal structures contain highly toxic single-stranded regions. Nucleases function either autonomously or in complex with additional proteins, such as DNA polymerases that work in close association with nucleases to increase the fidelity of the replication machinery. One of the most studied nucleases is the RecB protein that is closely associated with the RecBCD complex involved in the recovery of double strand breaks using homologous recombination (1–4). Structural studies (1, 2) indicate that the RecB protein has a helicase activity that separates the two strands of damaged DNA. The resulting “fork” structure is then processed by the RecB nuclease activity (5).
Although Archaea appear to lack the RecBCD complex, many archeal genomes nevertheless, encode proteins that have been annotated as “predicted RecB family nuclease” (6, 7). The structure and biochemical characterizations of Pab2263, one of the RecB-like nucleases in Pyrococcus abyssi, have been described (8–10). This protein binds to ssDNA4 at nanomolar concentrations and possesses a nuclease activity specific for ssDNA and was thus dubbed “NucS.” Qualitative experiments have indicated that both 3′ and 5′ extremities of ssDNA are cleaved by P. abyssi NucS (PabNucS). Although bipolar endonucleases are not common, human Dna2 endonuclease and Mus81 can cleave both 5′ and 3′ tailed ssDNAs with comparable efficiency (11, 12). Little is currently known about how the activity of these bipolar nucleases is regulated on their various substrates.
The crystal structure of PabNucS revealed a self-assembled dimer with a dumbbell-like organization that does not resemble any known protein structures (8, 9). PabNucS is composed of 2 domains. The amino-terminal domain displays a half-closed β-barrel and hosts a noncatalytic ssDNA binding site, whereas the carboxyl-terminal domain carries a minimal RecB-like domain, with its classical α/β structure, and contains the active site with a sequence motif conserved in the family of the RecB-like nucleases (7). Cleavage specificity of PabNucS is modulated by poorly understood interactions with dsDNA and the PCNA replication clamp (9). In cell-free extracts of P. abyssi, PabNucS forms a high affinity complex with PCNA (KD = 15 nm) (9). This interaction with the replication clamp is of particular interest as the toroidal sliding clamp encircles DNA and functions as a central coordinator for a large number of DNA replication and repair proteins (13, 14). For instance, the processivity of the P. abyssi DNA polymerases is increased by PCNA (15–19), which also improves the binding affinity of eukaryotic and archaeal flap endonucleases (FEN-1) toward DNA (20–22). Most PCNA-interacting proteins contain short canonical PCNA interaction peptides (PIPs) that bind to the interdomain connecting loop (IDCL), a region linking the two similar domains of a PCNA monomer. The PCNA-binding motif has been identified in a large number of proteins involved in DNA metabolic processes ranging from DNA replication, DNA repair, to cell cycle control (23, 24). The PIP motif corresponding to the carboxyl terminus of P. abyssi NucS interacts with the replication clamp PCNA in an array screening (25). As the PIP/IDCL interface is a distinctive feature of the interactions of these enzymes with PCNA, regulatory mechanisms must exist to ensure the spatial and temporal coordination of these fundamentally distinct PCNA-dependent processes. Additional protein-protein interfaces may confer specificity on PCNA interactions and regulate the activity of the associated proteins (16, 26–28). PCNA coordinates the handoff of a DNA substrate between different enzymes during DNA replication, repair, or recombination (20). For instance, the replication clamp regulates the activities of DNA pol δ and FEN-1 during lagging strand synthesis (29), eukaryotic DNA ligase binds to the PCNA trimer and its interaction prevents FEN-1 to interact with PCNA (30–33). Moreover, in the absence of DNA ligase the trimer of PCNA can simultaneously bind 3 FEN-1 molecules (34).
In crenarchaeal organisms, the heterotrimeric PCNA protein is composed of subunits that display distinct client specificities. This feature has been shown, in Sulfolobus solfataricus, to allow the recruitment, in vitro, of different interacting partners on the same ring to form a large macromolecular complex composed of PCNA, DNA polymerase, Fen1, and DNA ligase. This arrangement may allow coordination of sequential activities of these enzymes during the Okazaki fragment maturation process (35). In euryarchaeal organisms, as in Eucaryotes, the picture is different; the PCNA ring is homotrimeric and it is unclear how PCNA temporally regulates sequential interactions of these distinct partners with DNA.
What is the overall structural organization of the NucS-PCNA complex? How does PCNA influence the affinity and activity of NucS toward DNA substrates? To answer these questions, we present SAXS, fluorescence resonance energy transfer (FRET), and anisotropy analyses of the P. abyssi NucS and homotrimeric PCNA (PabPCNA) using both, the individual proteins and their complexes. We found that the PabNucS solution structure is modulated through interactions with ssDNA and that the distinct NucS-PCNA complexes are assembled on the 5′ and 3′ flap substrates. Altogether our studies reveal that the structural flexibility of the NucS-PCNA complex mediated by the presence of one major contact point between the two proteins not only facilitates conformational flexibility during DNA binding, but also regulates the cleavage specificity of NucS proteins.
EXPERIMENTAL PROCEDURES
Preparation of Protein Samples for SAXS and SPR Analyses
PabNucS (Pab2263) and PabPCNA were purified as described earlier (8, 9, 25), followed by additional treatment with polyimine P to eliminate any DNA contamination. Polyimine P was removed after ammonium sulfate precipitation (60%), followed by a gel filtration step. The PabNucS-PCNA complex was purified using Superdex 200 size exclusion chromatography in buffer A (30 mm Tris-HCl, pH 7.5, 500 mm NaCl). All samples were adjusted to 250 mm NaCl using buffer B: 30 mm Tris-HCl, pH 7.5, 250 mm NaCl. Where indicated, 250 μm ssDNA oligonucleotide (sequences of oligonucleotides used in this work are indicated in supplemental Table S1) was added to the proteins to form either PabNucS-ssDNA or PabPCNA-NucS-ssDNA complexes (maximum concentration of NucS was 230 μm). 30 mm EDTA was added to the protein samples to suppress the Mg2+- or Mn2+-dependent nuclease activity of the protein. Immediately before SAXS measurements, the samples were centrifuged (10,000 × g, 1/2 h) to remove potential protein aggregates.
SPR Experiments
Data were obtained using a Reichert SR7000DC spectrometer instrument (Reichert Inc., Buffalo, NY). The running buffer was 50 mm Tris-HCl, pH 7.4, 500 mm NaCl and the flow rate was 25 μl min−1. 550 μRIU of PabPCNA was immobilized on a mixed self-assembled monolayer (1 C11-(OEG)6-COOH:10 C11-(OEG)3-OH) (Reichert Inc.) via classical amine coupling chemistry and the chip was regenerated after a 30-s injection of 10 mm NaOH. Each curve displayed was double referenced with a set of 6 blank buffer injections. For the contact inhibition experiment, a PabNucS peptide corresponding to the PIP motif (Ac-KRSKQKTLDFFTP-NH2) was synthesized and HPLC purified (purity >90% (Innobiochips, France)). 300 nm PabNucS was mixed with various PIP PabNucS peptide concentrations, ranging from 0 to 30 μm, before injection on the PabPCNA chip. When the interaction of the peptide alone on PabPCNA was studied, a concentration range from 81 nm to 1 μm of the PIP-peptide and 10 μm of a control peptide (Ac-KEVKEEYKRFLEE-NH2) were injected on the same PabPCNA chip at 25 °C. Data were then fitted using a global analysis method with Scrubber 2.0a software (Biologic Software, Australia). For PabNucS interaction with PabPCNA, a concentration range from 100 nm to 2.7 μm was injected on the same PabPCNA chip at 45 °C, the same temperature used for SAXS experiments on the complex. As the data did not fit a simple equilibrium system, we introduced a system composed of two consecutive simple equilibria: NucS + PCNA ⇋ (NucS:PCNA) ⇋ (NucS:PCNA), where kON1, kOFF1, KD1, and kON2, kOFF2, KD2 refer, respectively, to the kinetic and equilibrium constants of the first and second reaction. Data were then fitted using a global analysis with Clamp99 3.30. All kinetic constants are reported in supplemental Table S2.
SAXS Experiments
SAXS experiments were performed: a, at the European Synchrotron Radiation Facility (ESRF, Grenoble, France), on beamline ID14-eh3 (λ = 0.931 Å; q ranging from 0.1 to 5 nm−1), and b, at the Deutsches Elektronen Synchrotron (DESY, Hamburg, Germany), on beamline X33 (λ = 1.5 Å; q ranging from 0.08 to 4.5 nm−1).
The scattering vector is defined as q = 4 π/λsin θ, where 2θ is the scattering angle. Experiments were performed both at 15 °C (a) and 45 °C (b).
SAXS data were evaluated using the Primus software, as implemented in the ATSAS 2.3 suite (36). For each sample, scattering profiles at all concentrations superimposed well, and the experimental SAXS data for all samples were linear in a Guinier plot of the low q region. These observations indicated that samples did not aggregate. The radius of gyration (Rg) was derived from the Guinier approximation I(q) = I(0) exp (−q2RG2/3) for qRg < 1.0. The radii of gyration Rg, calculated for different protein concentrations, displayed a slight concentration dependence arising from particle interference in solution. Data collected at high q using high protein concentrations and at low q using low protein concentrations were then merged to minimize interference occurring at low q and maximize the signal/noise ratio at high q. Molecular mass of samples were estimated using both I(0) and the Porod volume calculation, using a BSA sample as a reference. The program GNOM (37) was used to compute the distance-distribution function P(r). This approach also features the maximum dimension of the macromolecule, Dmax, and offers an alternative calculation of Rg, which is based on the entire scattering spectrum. Dmax was scrupulously chosen to minimize differences between the two calculated Rg results and maximize the “total estimate,” which indicates the agreement of the values of each criterion to their “ideal” ones.
Ab Initio Modeling
The overall shapes of the proteins in the various samples were obtained from the experimental data using the programs GASBOR (38) and DAMMIF (39). The scattering profiles were fitted up to a qmax = 0.4, 0.5, and 0.55 Å−1, respectively, for PabNucS, PabPCNA, and the complex between PabNucS and PabPCNA.
In DAMMIF, a particle is represented as a collection of a large number of beads inside a search volume (for example, a sphere of a diameter equal to Dmax), each bead belonging to the protein or the solvent. GASBOR searches a chain-compatible spatial distribution of a number of dummy residues, this number being the exact number of residues in the protein. Ten models obtained from different runs were superimposed and averaged using SUPCOMB (40) and DAMAVER (41). This allowed to identify the general structural features of each reconstruction and to assess the consistency of ab initio solutions by the normalized spatial discrepancy (41).
Atomic Model of Proteins
For visual representation, the shape of the protein was represented as spheres or a mesh surrounding the dummy atoms using the visualization program PyMOL (PyMOL Molecular Graphics System), and the atomic structures of the individual modules were positioned into the low resolution model. The crystallographic structure of PabPCNA is not available and the structure of a Pyrococcus furiosus ortholog carries an unstructured IDCL that is not visible in the crystal (42). Consequently, we built a model of the P. abyssi PCNA structure with Modeler, using crystallographic structures of the P. furiosus PCNA (42) and the human PCNA in complex with Fen1 (34), for the core of the protein construction and IDCL loop representation, respectively. Various models of PabNucS were generated with different positions of the N-terminal domain relative to the C-terminal domains, but keeping the same interdomain distance. SASREF (43) was used to model the position of the various proteins onto the complex using the SAXS data and atomic coordinate models. No distance restraints were applied. The resulting models had consistent features and no steric clashes. Structures were compared using a homemade program and mean square displacement (<R2>) was plotted on the Cα atoms to visualize mobile areas.
Normal Mode Analysis
We submitted the crystallographic structure of PabNucS to the World Wide Web interface of the Elastic Network modeling (44) that provides a fast and simple tool to analyze low frequency normal modes of large protein assemblies (45). To analyze the overall motion of our protein, we examined the mean square displacement of Cα atoms in the 100 lowest normal modes. Final models were systematically compared with those generated following the procedure described in the previous section (see above).
Construction of Fluorescent DNA Probes
Synthetic deoxyoligonucleotides labeled either on the 5′ end or the 3′ end with either fluorescein (FAM) or rhodamine (TAMRA) were purchased from Eurogentec (supplemental Table S1). Complementary oligonucleotides were annealed by heating at 80 °C for 5 min followed by slow cooling in a buffer containing 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl. The purity of the probes was checked by polyacrylamide gel electrophoresis in 50 mm Tris-HCl, pH 7.5, and 100 nm EDTA, pH 8, 8 mm glycine.
Fluorescent Labeling of PabNucS
PabNucS was labeled at the amino terminus using the Alexa Fluor® 488 5-SDP ester in 100 mm sodium bicarbonate buffer, pH 8.3, 150 mm NaCl at room temperature according to the manufacturer's instructions (Invitrogen). The reaction was allowed to proceed for 3 h in darkness and then the labeled PabNucS was purified using Bio-Gel P-6 (polyacrylamide gel matrice) chromatography columns (Bio-Rad) equilibrated with 20 mm Tris-HCl, pH 7.5, 150 mm NaCl.
Fluorescence Anisotropy Measurements
Steady-state and kinetic fluorescence anisotropy measurements were performed with a Cary Eclipse fluorescence spectrophotometer (Varian) in L-format configuration equipped with a polarization accessory and a single cell Peltier accessory (Varian). Fluorescence titrations, using TAMRA-labeled DNA probes, were performed at an excitation wavelength of 560 nm with a vertical polarization filter and by monitoring the emission spectra from 570 to 630 nm with the polarization filter both parallel and perpendicular with respect to the excitation light polarization. Fluorescence anisotropy measurements, using FAM-labeled DNA probes, were performed as above using an excitation wavelength of 495 nm and by monitoring the emission from 505 to 570 nm. Fluorescence anisotropies were calculated from the fluorescence intensities detected, according to,
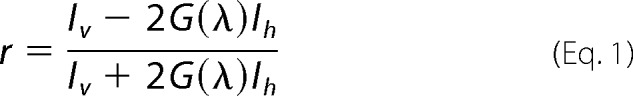 |
where r is the fluorescence anisotropy, Iv is the fluorescence emission intensity detected with vertical polarization, Ih is the fluorescence emission intensity detected with horizontal polarization, and G(λ) is the correction factor experimentally determined measuring the ratio Iv/Ih with a horizontally polarized excitation (46). Measurements were conducted at 20 °C in buffer containing 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl unless otherwise stated in the figure legends. Typically, 50 nm labeled DNA probes were used for each assay. Data processing was done using MATLAB and KD values were determined by nonlinear regression using GraphPad Prism 4 software as described earlier (47). The effect of ionic strength on binding of the PabNucS-PCNA complex to the 5′ flap substrate and the relative contributions of electrostratic and nonelectrostratic components to the free binding energy substrate were analyzed as described earlier and using 0.71 as the thermodynamic extent ψ of counterion “binding” (48, 49).
PabNucS Activity Tests
Nuclease assays at 46 °C were performed using substrates described in supplemental Table S1 using 20 min incubation time. Reaction mixtures (20 μl in all experiments) contained P. abyssi NucS and labeled oligonucleotides in 20 mm HEPES, pH 8, 50 mm NaCl, and 5 mm MnCl2. Reactions were analyzed on 18% denaturing polyacrylamide gels. Where indicated, PabPCNA (different concentrations) and 12 μm bovine serum albumin (BSA) were included in reaction mixtures.
Fluorescence Resonance Energy Transfer (FRET)
A fixed amount (50 nm) of double labeled, donor-only labeled or acceptor-only labeled DNA was incubated with both 2 μm PabNucS and 2 μm PabPCNA at 20 °C in buffer containing 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl. Steady-state fluorescence spectra were collected at magic angle from 500 to 650 nm after excitation with vertically polarized light of 485 nm. Increase of the acceptor fluorescence upon adding the donor label was used to monitor the FRET process. In this way only molecules that are involved in FRET are taken into account (50). Data processing was done using MATLAB. The spectra under FRET conditions were fitted to sums of separately measured spectra of the donor and acceptor at the same experimental conditions. The efficiency of energy transfer (E) was calculated as follows,
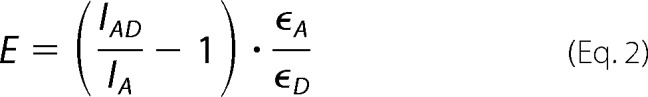 |
where IAD is the emission of the acceptor in the presence of the donor (consisting of fluorescence arising from energy transfer and from direct excitation of the acceptor) and IA is the fluorescence of the acceptor-only labeled sample (consisting of fluorescence arising from direct excitation only); ϵA and ϵD are the molar extinction coefficients of the acceptor and donor, respectively, at the wavelength of excitation (51). The Förster distances (R0) for the TAMRA-FAM and TAMRA-Alexa 488 pairs are 55 and 53.2 Å, respectively (20, 52), using 2/3 as the orientation factor. These R0 distances were used in combination with measured E to calculate the distance between the donor-acceptor dye pair according to,
 |
where E is the efficiency of energy transfer, R0 is the Förster distance, and r is the distance between the dye pair. DNA bending angles were calculated based on the length of the double-stranded B form DNA assuming that the vertex of the bend was positioned at the phosphate opposite of the nick. The length of the dye linker and the helical position of the fluorescently labeled base were determined using previously described helical models for nucleic acids (53). Where indicated, 5 mm CaCl2 was included in FRET experiments.
RESULTS
ssDNA Binding Modulates Solution Structure of P. abyssi NucS
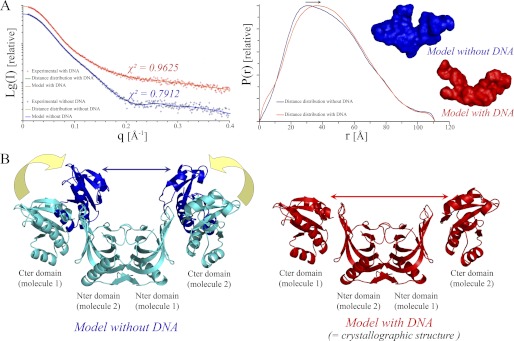
We used SAXS analyses to address the oligomeric state and conformational plasticity of PabNucS in solution (Fig. 1). The slope and intensity at zero angle of the experimental scattering data revealed that the protein adopts an extended conformation. These data also allowed determination of a Rg of 35.3 ± 0.4 Å (Table 1). Similar values were obtained using several distinct calculation methods (Guinier linearization around the origin, 35.0 Å; Porod volume computation, 35.6 Å; distance distribution computation, 35.6 Å). The distance distribution function P(r) displayed a biphasic pattern, also indicating an elongated shape with a maximum diameter (Dmax) of 111 ± 5 Å (Fig. 1A). The molecular mass determined from the scattering intensity extrapolated to zero angle indicated a molecular mass of ∼60 kDa (Table 1), in agreement with our observations that the PabNucS protein exists as a homodimer in solution (9). The theoretical scattering curve was calculated using the PabNucS dimer structure. Although the deduced scattering profile displayed a similar shape as the experimental scattering profile (data not shown) and the theoretical Rg = 35.0 Å was still in good agreement with the experimental value (Table 1), the χ2 value of 4.01 describing the fit between experimental and theoretical profiles suggests slight structural differences between solution and crystallographic structures. In agreement with this notion, we found that when the crystallographic structure was docked onto the SAXS envelop, the volume corresponding to the catalytic carboxyl-terminal domains was larger than expected (data not shown). This may reflect conformational flexibility of the catalytic domains mediated by the P. abyssi NucS interdomain loop that was not observed in the crystallographic structure (Fig. 1B). In contrast, the hydrophobic patches exposed on the six-stranded N-terminal β-sheet of the N-terminal domains that assemble to form the dimer in the crystal, with a large buried accessible surface area (2880 Å2, almost 20% of the total surface area of the subunit) (9), form an extremely rigid core. When the program OLIGOMER (54) was used to fit our experimental scattering curves using a multicomponent mixture of protein (conformers), best results were obtained using a mixture of 10 models (χ2 = 1.61).
FIGURE 1.
SAXS results suggest that flexibility of PabNucS is reduced upon ssDNA binding. A, scattering curves and P(r) functions for PabNucS in the absence (blue) and presence (red) of a 27-mer ssDNA oligonucleotide. Average SAXS envelopes are shown according to the same color code. B, model illustrating the putative movements of the C-terminal domains of PabNucS in the DNA-free form.
TABLE 1.
Overall parameters of PabNucS, PabPCNA, and the complex PabNucS-PCNA determined by SAXS
| Proteins | Mr | Concentration | Rga | MassGuinierb | Quality of the fit | RgPoroda | MassPorodb | RgP(r)a | Dmaxc | Estimate | MassP(r)ab | Rgatomica | Dmaxatomicc | Best χ2 | NSD between 10 models |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mg ml−1 | Å | kDa | % | Å | kDa | Å | Å | kDa | Å | Å | |||||
| NucS | 59.2 | 4.31 | 35.0 | 60.7 | 86 | 35.6 | 59.2 | 35.6 | 111 | 0.908 | 55.0 | 34.5 | 111.8 | 0.7912 | 1.510 |
| NucS + DNA | 59.2 | 3.56 | 35.0 | 63.2 | 91 | 35.7 | 62.4 | 35.6 | 111 | 0.935 | 55.5 | 34.6 | 112.4 | 0.9625 | 1.717 |
| PCNA | 86.4 | 5.3 | 33.5 | 84.7 | 85 | 33.7 | 84.1 | 34.1 | 99 | 0.968 | 82.5 | 33.6 | 95.6 | 0.8060 | 1.157 |
| NucS + PCNA | 145.6 | 4.6 | 38.7 | 148.9 | 89 | 38.8 | 132.8 | 39.4 | 130 | 0.889 | 137.5 | 39.9 | 131.2 | 1.242 | 0.833 |
a Rg, RgPorod, RgP(r), and Rgatomic given by the Guinier approximation, deduced from the Porod calculation, derived from the distance distribution function, and calculated for the final atomic model using the program CRYSOL, respectively.
b MassGuinier, MassPorod, MassP(r), mass of the protein given by the Guinier approximation, deduced from the Porod calculation, derived from the distance distribution function, and calculated for the final atomic model using the program CRYSOL, respectively.
c Dmax and Dmaxatomic correspond to the values deduced, respectively from the distance distribution function and the final model. The programs developed by Svergun and co-workers (65) minimize the normalized discrepancy function χ2.
The experimental P(r) function from SAXS collected in the presence of a ssDNA oligonucleotide (27 nucleotides) showed a shape change in comparison to the DNA-free form, indicating a more elongated conformation in the presence of ssDNA (Fig. 1A). It is also noteworthy that the scattering curve deduced from the dimer of the crystallographic structure of PabNucS was in better agreement (χ2 ∼ 2.0) with the experimental data in the presence of DNA than in its absence. This suggests that the solved crystal structure of PabNucS may be “locked” in the conformation that mimics the DNA bound configuration (Fig. 1B).
Solution Structure of PabPCNA
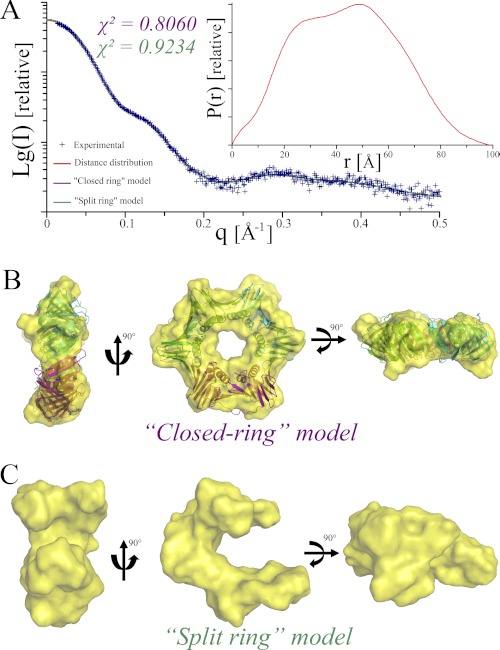
SAXS measurements for PabPCNA (Fig. 2) revealed that the protein adopts a globular conformation and extrapolation to zero angle indicated a molecular mass of 85.8 kDa in accordance with a homotrimeric assembly (Table 1). The approximated Rg was 33.7 Å, which is in good agreement with the theoretical Rg value of 32.1 Å that was calculated from the atomic coordinates of the model. The P(r) distance distribution (Fig. 2A) is bimodal and reflects the dimension of the protein ring with an approximate Dmax of 99 Å, consistent with the theoretical value of 95.9 Å. Assuming type 3 symmetry for the homotrimeric PabPCNA, the envelope obtained showed a ring structure with a hole at the center (Fig. 2B). The inner diameter of the torus of 22 Å is large enough to accommodate a dsDNA. The model of PabPCNA constructed using crystallographic information (see ”Experimental Procedures“) superimposed very well with the calculated envelope. Notably, the conformation of the flexible loop that was modeled using the atomic structure of the human PCNA-Fen1 complex fitted well within the obtained SAXS envelope (Fig. 2B). Considering the fact that the open state of P. furiosus PCNA has been observed using electron microscopy (55) and yet aware of known artifact associated with the ab initio modeling of discoidal particles (41), we also attempted ab initio modeling without constraining the program to type 3 symmetry. The latter protocol revealed an open U-shaped model for PabPCNA (Fig. 2C). In this model, the “open arms” of PabPCNA are not contained in the same plane. Thus, both the closed and open conformations of PabPCNA may exist in solution, although we cannot fully rule out the possibility that the split ring structure observed might be erroneously generated.
FIGURE 2.
Solution structure of the PabPCNA. A, fits of the calculated SAXS profiles of closed (purple, χ2 = 0.806) and split (green, χ2 = 0.9324) ring models for PabPCNA to the experimental (blue) SAXS data. P(r) function is shown in the inset. Ab initio shape predictions for closed (B) and split (C) ring models of PabPCNA. The crystallographic model of PabPCNA (see ”Experimental Procedures“) is overlaid on the molecular envelope calculated for the closed ring model.
PabPCNA Is Required for Loading of PabNucS on the 5′ and 3′ Flaps
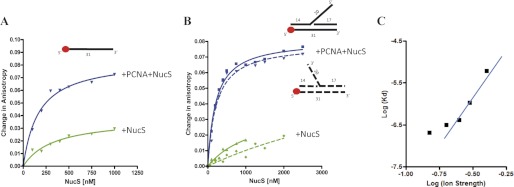
We then investigated the effect of PabPCNA on loading of PabNucS on ssDNA and branched DNA substrates. Using fluorescence anisotropy, PabNucS was titrated in the absence (Fig. 3, green lines) or presence (blue lines) of 80 nm PabPCNA and the increase in anisotropy of the ssDNA (50 nm) oligonucleotide was labeled with TAMRA at the 5′ extremity (31 nucleotides, supplemental Table S1), indicated binding to the ssDNA substrate (Fig. 3A). Nonlinear regression analyses using a single site binding model indicated that PabPCNA markedly increased the affinity of PabNucS toward ssDNA, as the measured KD values were ≈330 and ≈170 nm in the absence and presence of PabPCNA, respectively. As expected, in the presence of PabPCNA, the observed change in anisotropy is 2–3 times higher than observed for the binding of PabNucS alone, reflecting the formation of a considerably larger complex. This confirms that PabNucS and PabPCNA bind simultaneously to the used ssDNA substrate.
FIGURE 3.
Fluorescent anisotropy measurements of PabNucS binding to DNA. A, binding curves of PabNucS for a single-stranded oligonucleotide either in the presence (blue line) or absence of PabPCNA (green line). B, binding curves of PabNucS for both 3′ flap (dotted line) and 5′ flap (continuous line) substrates in the presence (blue line) or absence (green line) of PabPCNA. C, log-plot of the Kd of PabNucS-PCNA complex on the 5′ flap as a function of ionic strength (from 150 to 400 mm NaCl). The line corresponds to linear regression of data points from 200 to 400 mm NaCl.
We also assembled the branched DNA substrates carrying either 5′ (continuous lines, Fig. 3B) or 3′ (dotted lines, Fig. 3B) flaps and used them in further binding experiments where BSA served as the negative control. In the absence of the PabPCNA, anisotropy curves did not saturate. However, when PabPCNA was included in the binding reactions, we observed strong binding that allowed determination of KD values of 208 ± 20 and 225 ± 32 nm for 5′ and 3′ flaps, respectively. These results clearly indicate that PabPCNA markedly increases affinity of PabNucS toward branched DNA substrates. It is of note that formation of specific ternary complexes was observed under stoichiometric protein concentrations (1:1, PabNucS dimer:PabPCNA trimer). Repeating these binding experiments with the 5′ flap substrate using salt concentrations ranging from 150 to 400 mm NaCl resulted in increased KD values at higher ionic strength (Fig. 3C). This effect, resulting from weakening of electrostatic interactions between the PabNucS-PCNA complex and DNA was particularly clear starting from 250 mm NaCl. A plot of log KD as a function of ionic strength (log I) was linear with a slope of 4.4 between 250 and 400 mm NaCl (Fig. 3C), in agreement with formation of up to 6 salt bridges between DNA and the PabNucS-PCNA complex (48, 49). We estimate that these electrostatic interactions correspond approximately to 21 kJ/mol of a free energy of binding at 293 K in 150 mm NaCl, whereas nonelectrostatic contributions contribute to 15 kJ/mol under identical conditions.
Solution Structure of NucS-PCNA Complex
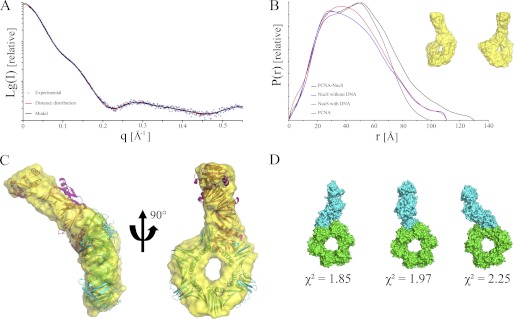
To better define the molecular basis of the influence of PabPCNA on the PabNucS affinity on its preferential substrates, we performed SAXS analyses on the isolated P. abyssi NucS-PCNA complex (Fig. 1A). A Porod volume of the complex indicates a molecular mass of 140 ± 10 kDa (Table 1), which is in excellent agreement with the predicted mass of 145.6 kDa for the complex formed by the PabNucS dimer and PabPCNA trimer (Table 1). The Dmax for the complex is estimated to be 130 Å from this analysis (Fig. 4B), with a Rg = 39 ± 0.4 Å (Table 1). The P(r) distance distribution function was trimodal and showed a peak at 51 Å with a shoulder on the side of high r values, which is typical for elongated proteins (Fig. 4B). These new features in the 90–130 Å range of the P(r) distribution corresponds to the interatomic distances between the two proteins. The Dmax value of about 130 Å was larger than the Dmax values obtained from the individual PabNucS and PabPCNA proteins (111 and 99 Å, respectively), indicating that the two proteins form an elongated complex, where PabNucS does not engage in extensive interactions with the side of the PabPCNA. This observation is similar to the previously characterized Fen1-PCNA complex (56). The overall shape of the complex was calculated and repetitive runs yielded superimposable models with similar overall structure and robustness of fit (χ2 ∼ 1.20, Fig. 1A, Table 1). The solution structure of the protein revealed a torus displaying an appendix that extends from the circumference and lies almost 45 degrees out of the plane of the ring (Fig. 4C). Only one such extension is seen, reflecting that only a single PabNucS dimer is bound to the PabPCNA trimer.
FIGURE 4.
Solution structure of the PabNucS-PCNA complex. A, scattering curve, and B, P(r) of the complex PabPCNA-NucS (black) in comparison to PabNucS, in the absence (blue) and presence (red) of DNA and PabPCNA (purple). The average ab initio shape predicted is shown in the inset. C, the average SAXS envelope is superimposed on the atomic models of PabNucS and PabPCNA. D, SAXS-directed docking of PabNucS (blue) onto PabPCNA (green). Only models satisfying the experimental SAXS data were kept, low χ2 values indicates very good agreement of the models to the experimental data.
Because the low resolution of the SAXS data did not allow determining whether PabNucS adopts a “closed” or an “open” configuration in the complex with PabPCNA, we used the crystallographic model of PabNucS and the homology-based calculated model of PabPCNA in docking experiments. These models were easily docked into the molecular shell derived from the SAXS measurements (Fig. 4C). The theoretical scattering curve was calculated for this docking model of the complex. The deduced overall dimensions of the docking model agreed well with those of the ab initio SAXS model (Table 1). Although PabPCNA was modeled as a split ring in some ab initio solutions for the complex with PabNucS (data not shown), the average of multiple independent models generates a closed ring, suggesting that this conformation of PabPCNA is adopted in the complex with PabNucS. To account for the possibility of steric clashes occurring for the proposed docking model, we used SASREF to model higher order structures of the PabPCNA-NucS complex. This approach uses the atomic structures of two individual components to fit our SAXS data set. SASREF calculations produced consistent and similar models as the docking experiments for the association of the two proteins (Fig. 4D), which agree well with the experimental SAXS data (χ2 ∼ 1.8, Fig. 4D).
PIP Motif Is Critical for PCNA-NucS Interaction
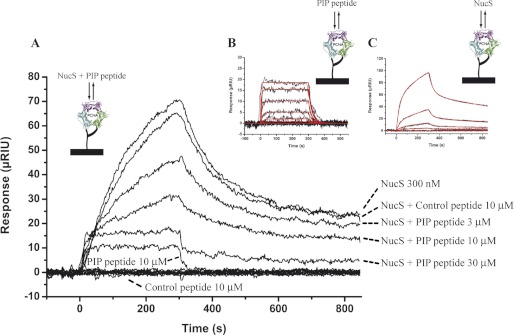
All structural models that were tested consistently indicated that one molecule of the dimeric PabNucS sits on the outside of the PabPCNA ring, albeit the resolution of our models did not allow precise determination of the docking site on the PabPCNA. To investigate whether the carboxyl-terminal PIP motif of PabNucS mediates critical interactions with the IDCL loop of PabPCNA, we used SPR experiments in the presence of the PIP-peptide. We found that this peptide interacted with immobilized PabPCNA (Fig. 5B). Global analysis of the data, considering 3 eq binding sites on the PabPCNA, indicated that 3 molecules of PIP PabNucS bind simultaneously to one molecule of PabPCNA with an affinity of 9.12 ± 0.09 μm at 25 °C, whereas a negative control peptide did not bind (Fig. 5A). Using the same PabPCNA chip, we also injected a fixed concentration of PabNucS with increasing amounts of the peptide corresponding to the PIP motif of this protein (Fig. 5A). Addition of this peptide substantially decreased the rate of complex formation, whereas the nonbinding control peptide had no effect. In the presence of 30 μm PIP-peptide, the signal observed nearly corresponded to that measured with the peptide alone, indicating that the carboxyl-terminal PIP motif of PabNucS is the major contact point between the two proteins. We have also analyzed the interaction between PabNucs and PabPCNA immobilized on the chip and at the same temperature used for acquisition of SAXS data (Fig. 5C). In contrast with the response observed using the PIP-peptide of PabNucS, the stoichiometry deduced from the interaction of the entire protein with PabPCNA indicated binding of a single dimer of PabNucS to the PabPCNA trimer. In addition, and still in contrast with the response obtained with the PIP-peptide, the shape of the curves obtained differed from a single exponential and suggested a complex interaction that is not explained using a 1:1 model. To explain these data, we constructed a model of interaction describing two consecutive binding events. The kinetic parameters of the first reaction, set to initiate the fitting process, were fixed in the same range as the kinetic constants measured from the interaction with the peptide, whereas the kinetic parameters for the second reaction were not constrained until completion of the fit. The theoretical curves obtained fitted very well the experimental data and allow determination of the kinetic parameters for the two reactions (supplemental Table S2). Overall, these SPR data suggest that a single PabNucS dimer interacts with the PabPCNA. In addition, the PIP/IDCL interface is the major contact point between the two proteins, albeit we cannot fully exclude the possibility that additional weaker contact points may occur between the two proteins during the binding event.
FIGURE 5.
The PIP motif of PabNucS is critical for the interaction with PabPCNA. A, for the competition experiment, sensorgrams were obtained after a 300-s injection of 300 nm PabNucS mixed with a range of PIP-peptide concentrations from 3 to 30 μm or with 10 μm control peptide. Controls consisted in the injection of 10 μm PIP-peptide and 10 μm of a noninteracting control peptide alone. B, fits of the calculated sensorgrams (red) of the interaction between immobilized PabPCNA and PIP-peptide, according to a simple 1:1 model, to the experimental data (black) obtained after injection of a concentration range from 81 nm to 1 μm PIP-peptide. C, fits of the calculated sensorgrams (red) of the interaction between immobilized PabPCNA and PabNucS, according to a complex model (see ”Experimental Procedures“), to the experimental data (black) obtained after injection of a concentration range from 100 nm to 2.7 μm PabNucS.
NucS-PCNA Complex Bends DNA
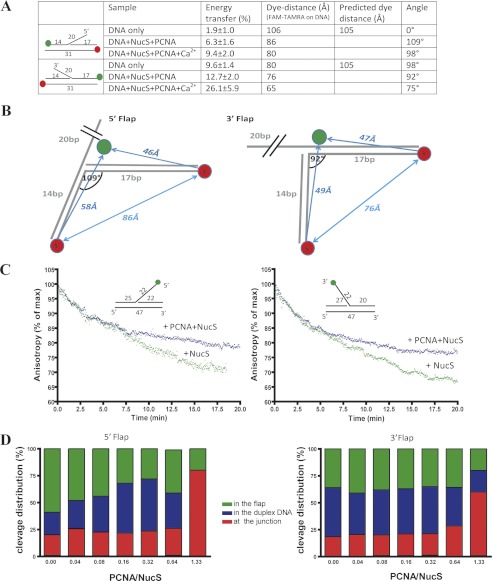
We also investigated the DNA conformation of the 5′ and 3′ flaps in the PabNucS-PCNA complex using FRET as previously described for Fen1 nuclease (20). Our data revealed that in the absence of divalent cation required for catalytic activity, the addition of PabNucS and PabPCNA decreased the end-to-end distance of both the 5′ and 3′ flap substrates (Fig. 6A), thus indicating that DNA substrates are bound at the bent configuration to the PabNucS-PCNA complex. Assuming that a kink is located in the vicinity of the ssDNA/dsDNA junction, we estimate that DNA structures carrying 5′ or 3′ flaps form angles of 109° and 92°, respectively, when bound to the PabNucS-PCNA complex. Further bending was observed in the presence of 5 mm CaCl2 (Fig. 6A). We also performed intermolecular FRET experiments between DNA substrates and Alexa 488-labeled (at the N terminus) PabNucS. The fact that our anisotropy data demonstrated that PabNucS is bound to DNA under these conditions allows us to project the obtained distances onto two-dimensional models that suggest binding of PabNucS “downstream” of the flap on the 5′ flap substrate and “upstream” of the flap on the 3′ flap substrate (Fig. 6B).
FIGURE 6.
Cleavage dynamics of PabNucS-PCNA complex on both 5′ flap and 3′ flap. A, summary of FRET measurements between 5′ and 3′ extremities of duplex DNA in both 5′ flap and 3′ flap substrates. B, FRET measurements define kinked DNA binding to the PabNucS-PCNA complex for both 5′ flap and 3′ flap. PabNucS binds upstream from the 5′ flap, whereas it binds downstream from the 3′ flap. C, cleavage activity of PabNucS in the presence (blue triangle) or absence (green triangle) of PabPCNA on both 3′ flap (right) and 3′ flap (left) detected by a decrease in anisotropy. D, cleavage activity of PabNucS on 5′ (right) and 3′ (left) flaps with and without PabPCNA. The reactions were performed using 1 μm PabNucS and the PabPCNA/PabNucS molar ratio was varied. The incubation time was 20 min. Cleavage products were analyzed and quantified using 18% denaturing polyacrylamide gels (9).
To investigate how PabPCNA influences activity of PabNucS on 5′ or 3′ flaps, we used a kinetic assay for measuring PabNucS cleavage in the presence and absence of the replication clamp PabPCNA. In these experiments, the FAM-fluorophore was placed at the terminal nucleotide of the 5′ or 3′ flaps. These experiments allowed detecting nuclease activity of PabNucS by measuring a decrease in anisotropy that reflects a release of the flap from DNA constructs. We found a time-dependent decrease in the anisotropy signal that was independent of PabPCNA during 4–5 min. After this period of time, PabPCNA appeared to inhibit PabNucS activity (Fig. 6C). When we analyzed reaction products of similar experiments using denaturating gel electrophoresis, we noticed that PabPCNA modulates PabNucS activity (Fig. 6D). On the 5′ and 3′ flaps, in the absence of PabPCNA, PabNucS cleaves in the ssDNA region of the flap, ∼14 nucleotides from the 5′ extremity of the flap (data not shown) or within the duplex region, whereas PabPCNA directed PabNucS activity toward the ss/dsDNA junction and inhibited nonspecific cleavage outside of the junction (Fig. 6D).
DISCUSSION
In a previous study (9), we have reported the biochemical characterization and crystallographic structure of PabNucS, the prototype of a new family of structure-specific DNA endonucleases. Up to date, the physiological function of bacterial and archaeal NucS orthologs remains unclear because we have not been able to identify a phenotype for Haloferax volcanii strains deleted for nucS. However, the enzyme tightly associates with the replication clamp PabPCNA and can process both 3′ and 5′ ssDNA extremities of branched DNA structures. To get further insight into the structural assembly of the PabPCNA-NucS complex in the presence or absence of ssDNA, we have performed SAXS analyses of the complex and its different constitutive components.
The solution structure of PabNucS revealed that the C-terminal domains of the two subunits of the dimer, encompassing the nuclease active sites, are found in different conformations in solution and crystal structures. Upon binding with the ssDNA ligand, the proteins do not undergo drastic conformational changes, but adopt a rigid and elongated conformation, where the individuals are similar to that observed in the crystal, which is consistent with the fact that movements in enzymes upon substrate binding are generally small (57). These fluctuations of the protein in the ligand-free state could allow it to reach a conformation close to that of the ligand-bound state. As the ligand spans both the N-terminal binding site and the C-terminal active site, another possibility could be that the reduced flexibility of the ligand-bound state induces a distortion of the ssDNA prior to cleavage.
Using fluorescence anisotropy, we investigated DNA binding properties of the PabNucS-PCNA complex. These studies indicate that the PabPCNA increases the binding affinity of PabNucS toward ssDNA as well as branched DNA substrates carrying either 5′ or 3′ flaps (Fig. 3B). The obtained results clearly indicate that PabPCNA is required for optimal loading of PabNucS on the 5′ and 3′ flaps and is able to increase the cleavage specificity of PabNucS proteins toward ss/ds junctions on 5′ and 3′ flaps (Fig. 6). To our knowledge, this is the first study demonstrating that PCNA is capable of loading an endonuclease to the 3′ and 5′ flaps and prevents nonspecific cleavage on both substrates. Similar experiments using salt concentrations in the range from 150 to 400 mm NaCl resulted in increased KD values at higher ionic strength (Fig. 3C). A plot of log KD as a function of ionic strength (log I) was linear with a slope of 4.4 between 250 and 400 mm NaCl, indicating the formation of many (up to 6) salt bridges between DNA and the PabNucS-PCNA complex (48, 49). This observation is in agreement with our earlier structural data indicating that the ssDNA binding cleft of the PabNucS protein is rather basic (9).
Our characterization of the solution structure of PabPCNA raises the question of whether PabPCNA adopts both or only one of the open and solid ring conformations. When compared with the bacterial DNA polymerase III β-subunit and eukaryotic PCNA, the trimeric form of P. furiosus PCNA is mainly stabilized by a network of ion pairs at the molecular interface rather than by main chain hydrogen bonds and hydrophobic contacts (42, 58). This difference, which is conserved at the primary sequence level among PCNAs from thermophilic archaea, could reflect a weaker intermolecular interaction between the subunits of archaeal thermophilic PCNAs and account for the self-loading process observed for homotrimeric euryarchaeal PCNA (16–19). In addition, a recent molecular dynamic study of the role of the eukaryotic replication factor C at the initial step of the clamp loading cycle has raised the hypothesis that the clamp loader could trap and stabilize the open conformation of PCNA (59), thus strengthening the idea that the open ring conformation of PCNA also exists in solution. This might be explained by the weak interface between monomers of Pyroccocus PCNA, considering that it contains fewer hydrogen bonds than eukaryotic ones, and that hydrogen bonds are weakened at the optimal growth temperature of the organism (95 °C).
The combination of SAXS and SPR data allows us to characterize, for the first time, the general three-dimensional organization of the PabPCNA-NucS complex and indicates that one molecule of PabNucS dimer binds to the PabPCNA homotrimer, forming a stable 1:1 complex. We stress that formation of this complex has direct functional consequences, because PabPCNA is able to increase the cleavage specificity of PabNucS proteins toward ss/ds junctions on 5′ and 3′ flaps. Furthermore, the PIP/IDCL contact is critical for the interaction. This single major contact point between the carboxyl terminus PIP motif of PabNucS and the IDCL of PabPCNA likely facilitates conformational flexibility of the protein-DNA substrate complexes. A similar structural organization was observed for the archaeal PCNA-DNA ligase complex without DNA (31), where only one molecule of DNA ligase extends from the PCNA ring in the same plane, and the interaction is mediated by the PIP-IDCL interaction. This particular binding stoichiometry results from the distinct specificity displayed by the different subunits of the heterotrimeric crenarchaeal PCNA (31, 35). The organization for the PCNA from P. abyssi is different, it is composed of three identical subunits and the three binding sites are thus considered to be equivalent. In this respect, it might be possible that the fixation of one molecule of PabNucS could lead to spatial rearrangements on the PabPCNA preventing subsequent binding events or at least decreasing the kinetics of additional NucS/PCNA binding events. It is known that PCNA may provide more protein-protein interfaces, in addition to the general PIP-IDCL site, which confer different specificities to PCNA and regulate the diverse activities of associated enzymes (20). These contact sites reside on subunits distinct from that engaged in the IDCL-PIP interaction. In particular, these additional contacts may even block access of other active clients to PCNA (60). The SPR data indicate that the IDCL/PIP PabNucS contact alone is not sufficient to generate long range effects affecting the PabNucS-PCNA complex stoichiometry, but rather additional contacts, close to this main interaction surface and implicating residues located on the same or adjacent PabPCNA subunits. It is also possible that DNA bending induced by the PabNucS-PCNA complex plays a key role in regulating the cleavage specificity of NucS proteins. In this respect, the interaction between PabNucS and PabPCNA resembles that of SsoFen1 with SsoPCNA in that both PabNucS and SsoFen1 display a PIP box motif at the extreme C-terminal of their sequences, thus preventing the formation of an extended β-sheet with the IDCL. Doré and co-workers (56) have shown that SsoFen1 engages extra contacts with the C-terminal part of PCNA, most likely to compensate for the C-terminal truncation of the enzyme. The same might also hold true for PabNucS, this still open question awaits more resolutive crystallographic data of the PCNA-NucS complex.
These potential rearrangements regulating the binding stoichiometry of PabNucS could allow or even promote other binding partners to be recruited by the PabPCNA and reflect the requirement of temporal and spatial coordination of their respective activities. In this regard, it is worth noting that PabNucS was found to be associated with Hef nuclease in pull-down experiments (9). Recent genetic analyses have shown that Hef is involved in numerous DNA repair pathways in the archaeal cells, including resolution of stalled replication forks, interstrand cross-links, and NER (61, 62). Up to now, there is no evidence of direct interaction between Hef and PCNA, however, crenarchaeal XPF, the shorter form of Hef lacking the helicase segment, was shown to be dependent on the PCNA for its nuclease activity, activating the catalytic step by 4 orders of magnitude (22, 63, 64). It is thus tempting to postulate that the activities of NucS and Hef could be coordinated by PCNA in a branched DNA repair pathway.
In summary, we have shown that the sliding clamp PCNA of P. abyssii can modulate the activity of PabNucS. Our data confirm that PabPCNA increases the binding affinity of PabNucS toward ssDNA as well as branched DNA substrates carrying either 5′ or 3′ flaps and that PabPCNA is required for optimal loading of PabNucS on these substrates. In addition, PabPCNA appeared to direct PabNucS activity toward the ss/dsDNA junction, thus inhibiting nonspecific cleavage outside of the junction. This choreography between PabPCNA and PabNucS can be viewed as a form of regulation of PabNucS activities, which will only take place on the appropriate substrate where PabPCNA is already loaded, thus preventing the potentially deleterious nonspecific cleavage activity of PabNucS on the chromatin. At the structural level, our data also indicate that the PabNucS-PCNA complex distorts DNA by inducing a kink located in the vicinity of the ssDNA/dsDNA junction and that PabNucS binds downstream of the flap on the 5′ flap substrate and upstream of the flap on the 3′ flap substrate. Finally, the solution structure of the PabPCNA-NucS complex indicates that one molecule of PabNucS dimer binds to the PabPCNA homotrimer, and that the PIP/IDCL contact is critical for the interaction. The regulation of this particular stoichiometry could be triggered by additional contacts between PabNucS and PabPCNA inducing long range conformational changes on PabPCNA that in turn could favor the recruitment of additional partners of PabNucS acting in the same DNA repair pathways. More resolutive crystallographic data of the PabPCNA-NucS complex and in vivo characterization of the function of PabNucs will help answer these still open questions.
Supplementary Material
Acknowledgments
We thank the staff on the SAXS beamlines (ID14-EH3, ESRF, Grenoble; SWING, SOLEIL, Saint Aubin; and X33, DESY, Hamburg) for their warm welcome, and especially Clement Blanchet (EMBL-Hamburg) for valuable help during data collection. We also thank members of our laboratories for thoughtful discussions.
This work was supported by Agence Nationale de la Recherche Grants ANR-07-BLAN-0371 (to C. C., M. C., H. M., and D. F.), ANR-09-PIRI-0019-03 (to M. V. and H. M.), and the CREAtivité & Thématiques Exploratoires program of the Brittany regional council, REPAR number 4852 (to S. L. and D. F.).

This article contains supplemental Tables S1 and S2.
- ssDNA
- single-strand DNA
- dsDNA
- double-strand DNA
- PCNA
- proliferating cell nuclear antigen
- FEN-1
- flap endonuclease 1
- PIP
- PCNA interaction peptide
- IDCL
- interdomain connecting loop
- SAXS
- small angle x-ray scattering
- TAMRA
- carboxytetramethylrhodamine.
REFERENCES
- 1. Dillingham M. S., Kowalczykowski S. C. (2008) RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Singleton M. R., Dillingham M. S., Gaudier M., Kowalczykowski S. C., Wigley D. B. (2004) Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432, 187–193 [DOI] [PubMed] [Google Scholar]
- 3. Wang J., Chen R., Julin D. A. (2000) A single nuclease active site of the Escherichia coli RecBCD enzyme catalyzes single-stranded DNA degradation in both directions. J. Biol. Chem. 275, 507–513 [DOI] [PubMed] [Google Scholar]
- 4. Yu M., Souaya J., Julin D. A. (1998) The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 981–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shevelev I. V., Hübscher U. (2002) The 3′-5′ exonucleases. Nat. Rev. Mol. Cell Biol. 3, 364–376 [DOI] [PubMed] [Google Scholar]
- 6. Kinch L. N., Ginalski K., Rychlewski L., Grishin N. V. (2005) Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 33, 3598–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aravind L., Makarova K. S., Koonin E. V. (2000) Survey and summary. Holliday junction resolvases and related nucleases. Identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28, 3417–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ren B., Kuhn J., Meslet-Cladiere L., Myllykallio H., Ladenstein R. (2007) Crystallization and preliminary X-ray analysis of a RecB-family nuclease from the archaeon Pyrococcus abyssi. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 406–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ren B., Kühn J., Meslet-Cladiere L., Briffotaux J., Norais C., Lavigne R., Flament D., Ladenstein R., Myllykallio H. (2009) Structure and function of a novel endonuclease acting on branched DNA substrates. EMBO J. 28, 2479–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Creze C., Lestini R., Kühn J., Ligabue A., Becker H. F., Czjzek M., Flament D., Myllykallio H. (2011) Structure and function of a novel endonuclease acting on branched DNA substrates. Biochem. Soc. Trans. 39, 145–149 [DOI] [PubMed] [Google Scholar]
- 11. Ehmsen K. T., Heyer W. D. (2008) Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 36, 2182–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J. H., Kim H. D., Ryu G. H., Kim D. H., Hurwitz J., Seo Y. S. (2006) Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res. 34, 1854–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J. (1994) Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 14. Pan M., Kelman L. M., Kelman Z. (2011) The archaeal PCNA proteins. Biochem. Soc. Trans. 39, 20–24 [DOI] [PubMed] [Google Scholar]
- 15. Castrec B., Laurent S., Henneke G., Flament D., Raffin J. P. (2010) The glycine-rich motif of Pyrococcus abyssi DNA polymerase D is critical for protein stability. J. Mol. Biol. 396, 840–848 [DOI] [PubMed] [Google Scholar]
- 16. Castrec B., Rouillon C., Henneke G., Flament D., Querellou J., Raffin J. P. (2009) Binding to PCNA in euryarchaeal DNA replication requires two PIP motifs for DNA polymerase D and one PIP motif for DNA polymerase B. J. Mol. Biol. 394, 209–218 [DOI] [PubMed] [Google Scholar]
- 17. Henneke G., Flament D., Hübscher U., Querellou J., Raffin J. P. (2005) The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 350, 53–64 [DOI] [PubMed] [Google Scholar]
- 18. Henneke G., Gueguen Y., Flament D., Azam P., Querellou J., Dietrich J., Hübscher U., Raffin J. P. (2002) Replication factor C from the hyperthermophilic archaeon Pyrococcus abyssi does not need ATP hydrolysis for clamp-loading and contains a functionally conserved RFC PCNA-binding domain. J. Mol. Biol. 323, 795–810 [DOI] [PubMed] [Google Scholar]
- 19. Rouillon C., Henneke G., Flament D., Querellou J., Raffin J. P. (2007) DNA polymerase switching on homotrimeric PCNA at the replication fork of the euryarchaea Pyrococcus abyssi. J. Mol. Biol. 369, 343–355 [DOI] [PubMed] [Google Scholar]
- 20. Chapados B. R., Hosfield D. J., Han S., Qiu J., Yelent B., Shen B., Tainer J. A. (2004) Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell 116, 39–50 [DOI] [PubMed] [Google Scholar]
- 21. Gomes X. V., Burgers P. M. (2000) Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19, 3811–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hutton R. D., Roberts J. A., Penedo J. C., White M. F. (2008) PCNA stimulates catalysis by structure-specific nucleases using two distinct mechanisms. Substrate targeting and catalytic step. Nucleic Acids Res. 36, 6720–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsurimoto T. (1999) PCNA binding protein. Front Biosci. 4, 849–858 [DOI] [PubMed] [Google Scholar]
- 24. Warbrick E. (1998) PCNA binding through a conserved motif. Bioessays 20, 195–199 [DOI] [PubMed] [Google Scholar]
- 25. Meslet-Cladiére L., Norais C., Kuhn J., Briffotaux J., Sloostra J. W., Ferrari E., Hübscher U., Flament D., Myllykallio H. (2007) A novel proteomic approach identifies new interaction partners for proliferating cell nuclear antigen. J. Mol. Biol. 372, 1137–1148 [DOI] [PubMed] [Google Scholar]
- 26. Kiyonari S., Takayama K., Nishida H., Ishino Y. (2006) Identification of a novel binding motif in Pyrococcus furiosus DNA ligase for the functional interaction with proliferating cell nuclear antigen. J. Biol. Chem. 281, 28023–28032 [DOI] [PubMed] [Google Scholar]
- 27. Rolef Ben-Shahar T., Castillo A. G., Osborne M. J., Borden K. L., Kornblatt J., Verreault A. (2009) Two fundamentally distinct PCNA interaction peptides contribute to chromatin assembly factor 1 function. Mol. Cell. Biol. 29, 6353–6365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu H., Zhang P., Liu L., Lee M. Y. (2001) A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry 40, 4512–4520 [DOI] [PubMed] [Google Scholar]
- 29. Garg P., Burgers P. M. (2005) DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40, 115–128 [DOI] [PubMed] [Google Scholar]
- 30. Subramanian J., Vijayakumar S., Tomkinson A. E., Arnheim N. (2005) Genetic instability induced by overexpression of DNA ligase I in budding yeast. Genetics 171, 427–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pascal J. M., Tsodikov O. V., Hura G. L., Song W., Cotner E. A., Classen S., Tomkinson A. E., Tainer J. A., Ellenberger T. (2006) A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol. Cell 24, 279–291 [DOI] [PubMed] [Google Scholar]
- 32. Pascal J. M., O'Brien P. J., Tomkinson A. E., Ellenberger T. (2004) Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 432, 473–478 [DOI] [PubMed] [Google Scholar]
- 33. Levin D. S., Bai W., Yao N., O'Donnell M., Tomkinson A. E. (1997) An interaction between DNA ligase I and proliferating cell nuclear antigen. Implications for Okazaki fragment synthesis and joining. Proc. Natl. Acad. Sci. U.S.A. 94, 12863–12868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sakurai S., Kitano K., Yamaguchi H., Hamada K., Okada K., Fukuda K., Uchida M., Ohtsuka E., Morioka H., Hakoshima T. (2005) Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 24, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dionne I., Nookala R. K., Jackson S. P., Doherty A. J., Bell S. D. (2003) A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Cell 11, 275–282 [DOI] [PubMed] [Google Scholar]
- 36. Petoukhov M. V., Svergun D. I. (2007) Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr. Opin. Struct. Biol. 17, 562–571 [DOI] [PubMed] [Google Scholar]
- 37. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 24, 495–503 [Google Scholar]
- 38. Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Franke D., Svergun D. I. (2009) AMMIF, a program for rapid ad initio shape determination in small angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kozin M. B., Svergun D. I. (2000) A software system for rigid body modeling of solution scattering data. J. Appl. Crystallogr. 33, 775–777 [Google Scholar]
- 41. Volkov V. V., Svergun D. I. (2003) Uniquenes of ad initio shape determination in small angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsumiya S., Ishino Y., Morikawa K. (2001) Crystal structure of an archaeal DNA sliding clamp. Proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci. 10, 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petoukhov M. V., Svergun D. I. (2005) Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 89, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Delarue M., Sanejouand Y. H. (2002) Simplified normal mode analysis of conformational transitions in DNA-dependent polymerases. The elastic network model. J. Mol. Biol. 320, 1011–1024 [DOI] [PubMed] [Google Scholar]
- 45. Suhre K., Sanejouand Y. H. (2004) ElNemo, a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 32, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lakowicz J. R. (2006) Principles of Fluorescence Spectroscopy, 3rd Ed., pp. 353–381, Springer, New York [Google Scholar]
- 47. LiCata V. J., Wowor A. J. (2008) Applications of fluorescence anisotropy to the study of protein-DNA interactions. Methods Cell Biol. 84, 243–262 [DOI] [PubMed] [Google Scholar]
- 48. Lohman T. M., Mascotti D. P. (1992) Thermodynamics of ligand-nucleic acid interactions. Methods Enzymol. 212, 400–424 [DOI] [PubMed] [Google Scholar]
- 49. Record M. T., Jr., Lohman M. L., De Haseth P. (1976) Ion effects on ligand-nucleic acid interactions. J. Mol. Biol. 107, 145–158 [DOI] [PubMed] [Google Scholar]
- 50. Laptenok S. P., Borst J. W., Mullen K. M., van Stokkum I. H., Visser A. J., van Amerongen H. (2010) Global analysis of Förster resonance energy transfer in live cells measured by fluorescence lifetime imaging microscopy exploiting the rise time of acceptor fluorescence. Phys. Chem. Chem. Phys. 12, 7593–7602 [DOI] [PubMed] [Google Scholar]
- 51. Selvin P. R. (1995) Fluorescence resonance energy transfer. Methods Enzymol. 246, 300–334 [DOI] [PubMed] [Google Scholar]
- 52. Hopkins B. B., Reich N. O. (2004) Simultaneous DNA binding, bending, and base flipping. Evidence for a novel M.EcoRI methyltransferase-DNA complex. J. Biol. Chem. 279, 37049–37060 [DOI] [PubMed] [Google Scholar]
- 53. Clegg R. M., Murchie A. I., Zechel A., Lilley D. M. (1993) Observing the helical geometry of double-stranded DNA in solution by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 90, 2994–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. (2003) PRIMUS, a Windows PC-based system for small angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 55. Miyata T., Suzuki H., Oyama T., Mayanagi K., Ishino Y., Morikawa K. (2005) Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc. Natl. Acad. Sci. U.S.A. 102, 13795–13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Doré A. S., Kilkenny M. L., Jones S. A., Oliver A. W., Roe S. M., Bell S. D., Pearl L. H. (2006) Structure of an archaeal PCNA1-PCNA2-FEN1 complex. Elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res. 34, 4515–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gutteridge A., Thornton J. (2005) Conformational changes observed in enzyme crystal structures upon substrate binding. J. Mol. Biol. 346, 21–28 [DOI] [PubMed] [Google Scholar]
- 58. Matsumiya S., Ishino S., Ishino Y., Morikawa K. (2003) Intermolecular ion pairs maintain the toroidal structure of Pyrococcus furiosus PCNA. Protein Sci. 12, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tainer J. A., McCammon J. A., Ivanov I. (2010) Recognition of the ring-opened state of proliferating cell nuclear antigen by replication factor C promotes eukaryotic clamp-loading. J. Am. Chem. Soc. 132, 7372–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mayanagi K., Kiyonari S., Nishida H., Saito M., Kohda D., Ishino Y., Shirai T., Morikawa K. (2011) Architecture of the DNA polymerase B-proliferating cell nuclear antigen (PCNA)-DNA ternary complex. Proc. Natl. Acad. Sci. U.S.A. 108, 1845–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fujikane R., Ishino S., Ishino Y., Forterre P. (2010) Genetic analysis of DNA repair in the hyperthermophilic archaeon, Thermococcus kodakaraensis. Genes Genet Syst 85, 243–257 [DOI] [PubMed] [Google Scholar]
- 62. Lestini R., Duan Z., Allers T. (2010) The archaeal Xpf/Mus81/FANCM homolog Hef and the Holliday junction resolvase Hjc define alternative pathways that are essential for cell viability in Haloferax volcanii. DNA Repair 9, 994–1002 [DOI] [PubMed] [Google Scholar]
- 63. Roberts J. A., Bell S. D., White M. F. (2003) An archaeal XPF repair endonuclease dependent on a heterotrimeric PCNA. Mol. Microbiol. 48, 361–371 [DOI] [PubMed] [Google Scholar]
- 64. Rouillon C., White M. F. (2011) The evolution and mechanisms of nucleotide excision repair proteins. Res. Microbiol. 162, 19–26 [DOI] [PubMed] [Google Scholar]
- 65. Svergun D., Barberato C., Koch M. H. J. (1995) CRYSOL, a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.