Background: Specific phospholipid composition in mitochondria is essential for mitochondrial activities.
Results: Two intermembrane space proteins, Ups1p and Ups2p, antagonistically regulate conversion of phosphatidylethanolamine to phosphatidylcholine.
Conclusion: The endoplasmic reticulum-mitochondria tethering complex and Ups proteins have related functions in phospholipid metabolism and trafficking.
Significance: Deciphering regulation of phospholipid metabolism is vital for understanding the biogenesis of mitochondrial membranes and functions.
Keywords: Cardiolipin, Mitochondria, Mitochondrial Metabolism, Phospholipid, Yeast, Mdm31, Ups Proteins, Mitochondrial Morphology, Phospholipid Trafficking
Abstract
Mitochondrial membranes maintain a specific phospholipid composition. Most phospholipids are synthesized in the endoplasmic reticulum (ER) and transported to mitochondria, but cardiolipin and phosphatidylethanolamine are produced in mitochondria. In the yeast Saccharomyces cerevisiae, phospholipid exchange between the ER and mitochondria relies on the ER-mitochondria encounter structure (ERMES) complex, which physically connects the ER and mitochondrial outer membrane. However, the proteins and mechanisms involved in phospholipid transport within mitochondria remain elusive. Here, we investigated the role of the conserved intermembrane space proteins, Ups1p and Ups2p, and an inner membrane protein, Mdm31p, in phospholipid metabolism. Our data show that loss of the ERMES complex, Ups1p, and Mdm31p causes similar defects in mitochondrial phospholipid metabolism, mitochondrial morphology, and cell growth. Defects in cells lacking the ERMES complex or Ups1p are suppressed by Mdm31p overexpression as well as additional loss of Ups2p, which antagonizes Ups1p. Combined loss of the ERMES complex and Ups1p exacerbates phospholipid defects. Finally, pulse-chase experiments using [14C]serine revealed that Ups1p and Ups2p antagonistically regulate conversion of phosphatidylethanolamine to phosphatidylcholine. Our results suggest that Ups proteins and Mdm31p play important roles in phospholipid biosynthesis in mitochondria. Ups proteins may function in phospholipid trafficking between the outer and inner mitochondrial membranes.
Introduction
Within eukaryotic cells are elaborate membrane-enclosed organelles composed of characteristic sets of proteins and lipids, to establish specific shapes. For instance, mitochondria are bounded by the outer membrane (OM)4 and inner membrane (IM). Each membrane has a specific phospholipid composition that is essential for mitochondrial functions, such as oxidative phosphorylation for ATP production (1, 2), import of mitochondrial proteins from the cytosol (3–8), mitochondrial morphogenesis (9–12), and apoptosis (13–16). Most phospholipids, such as phosphatidic acid, phosphatidylinositol, phosphatidylserine (PS), and phosphatidylcholine (PC), are synthesized in the endoplasmic reticulum (ER) membrane and then imported into mitochondria. However, other phospholipids, including phosphatidylethanolamine (PE) and cardiolipin (CL), are produced in mitochondria. To produce PE, PS is transferred from the ER to the OM and then the IM, where Psd1p mediates its conversion to PE through decarboxylation (17–19). PE is then sent back to the OM and the ER, where PE is methylated and converted into PC (20). Phosphatidic acid is also moved from the ER through the OM to the IM and used to produce CL by sequential enzymatic modifications involving Pgs1p, Gep4p, and Crd1p (21–23). Clearly, phospholipid trafficking is important for phospholipid metabolism.
Recent studies have suggested that a protein complex that physically connects mitochondria to the ER, termed the ER-mitochondria encounter structure (ERMES) complex, facilitates phospholipid exchange between these two organelles in yeast (24, 25). The ERMES complex is composed of an integral protein in the ER, Mmm1p, and four mitochondrial proteins, Mmm2p, Mdm10p, Mdm12p, and Gem1p (24–26). Mmm1p, Mmm2p, and Mdm12p have sequence homology to the tubular lipid-binding protein superfamily and have been proposed to bind directly to hydrophobic lipids to facilitate phospholipid transport (27). Mmm1p, Mmm2p, Mdm10p, and Mdm12p are essential components of the ERMES complex because punctate structures of the ERMES complex no longer form when any of these proteins are absent (i.e. in ERMES-deleted cells). However, Gem1p has been proposed to play a regulatory role in phospholipid transport by controlling the number and size of the ERMES complex (25). All ERMES proteins were originally identified as essential factors for mitochondrial morphogenesis, as deletion of each ERMES protein individually causes severe mitochondrial morphological defects (28–33). These results demonstrate a close relationship between phospholipid metabolism and mitochondrial morphology.
Proteins responsible for lipid shuttling within mitochondria most likely reside in the intermembrane space (IMS). In addition, loss of intra-mitochondrial lipid trafficking would likely alter mitochondrial phospholipid composition and morphology, similar to ERMES-deleted cells. Our previous studies demonstrated that Ups1p and Ups2p, two conserved IMS proteins, control mitochondrial phospholipids (7, 34). Loss of Ups1p causes decreased CL, whereas loss of Ups2p leads to decreased PE (7, 35). Additional loss of Ups2p restores CL levels in ups1Δ cells, suggesting antagonistic roles for Ups1p and Ups2p in CL metabolism. Furthermore, ups1Δ cells accumulate PS, which is a precursor for PE (7). In addition to phospholipid defects, loss of Ups proteins alters mitochondrial shape (7, 35). The mitochondrial morphology defects appear only when ups-deleted mutants are grown in fermentable media but not in nonfermentable media (7, 34). In contrast, phospholipid defects occur regardless of growth media, indicating that phospholipid defects are not due to mitochondrial shape changes (7). Therefore, Ups1p and Ups2p are strong candidates for factors that facilitate intra-mitochondrial phospholipid transport.
Other candidates that may play a role in phospholipid transport are Mdm31p and Mdm32p, two IM proteins with a large IMS domain. Similar to Ups and ERMES proteins, Mdm31p and Mdm32p have been shown to affect phospholipid composition and mitochondria morphology (35, 36). Quantitative analyses for phospholipids revealed decreased levels of CL in mdm31Δ and mdm32Δ mitochondria (35). mdm31Δ and mdm32Δ cells exhibit large spherical mitochondria similar to those in ERMES-deleted cells (31, 36). Furthermore, combined loss of the ERMES complex with Mdm31p or Mdm32p results in synthetic lethality (36), suggesting their overlapping functions. Here, we further studied the involvement of Ups1p, Ups2p, and Mdm31p in mitochondrial phospholipid metabolism and morphology. Our analyses suggest that these proteins function in mitochondrial phospholipid biosynthesis.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, Media, and Genetic Methods
Yeast strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Complete disruption of the yeast gene was accomplished by PCR-mediated gene replacement (37) with a pair of primers listed in Table 1 and supplemental Table S1. The HIS3 (pRS303), TRP1 (pRS304), URA3 (pRS306), and kanMX4 (pRS400) genes were used as disruption markers. To obtain mmm1Δ, mmm2Δ, mdm10Δ, and mdm12Δ strains, the appropriate DNA cassette for the gene disruption was introduced into a diploid strain, FY833/FY834 (38). After introducing the URA3-containing single copy plasmid harboring the corresponding gene (MMM1, MMM2, MDM10, or MDM12), the diploid cells were subjected to sporulation and dissection to obtain haploids. Haploid strains that lack the MMM1, MMM2, MDM10, or MDM12 gene in chromosomes with the complementing plasmid were selected. These strains were also used to further disrupt the UPS1, UPS2, or MDM35 gene. To analyze complete deletion strains, the cells were cultured on 5′-fluoro-orotic acid (5′-FOA)-containing media to remove the complementing URA3 plasmid prior to analysis.
TABLE 1.
Yeast strains used in this study
| Name | Genotype | Source | Parential strains | Primers for gene disruption or chromosomal tagging |
|---|---|---|---|---|
| FY833 (haploid WT) | MATa his3 leu2 lys2 trp1 ura3 | Ref. 38 | ||
| FY833/FY834 (diploid WT) | MATa/α his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 | Ref. 38 | ||
| ups1Δ | MATa his3 leu2 lys2 trp1 ura3 ups1::kanMX4 | Ref. 34 | ||
| ups2Δ | MATa his3 leu2 lys2 trp1 ura3 ups2::HIS3 | Ref. 7 | ||
| mdm35Δ | MATa his3 leu2 lys2 trp1 ura3 mdm35::URA3 | Ref. 44 | ||
| mmm1Δ | MATα his3 leu2 lys2 trp1 ura3 mmm1::kanMX4 | This study | FY833/FY834 | PTM361/362 |
| mmm1Δups1Δ | MATα his3 leu2 lys2 trp1 ura3 mmm1::kanMX4 ups1::HIS3 | This study | mmm1Δ | 1729/1730 |
| mmm1Δups2Δ | MATα his3 leu2 lys2 trp1 ura3 mmm1::kanMX4 ups2::HIS3 | This study | mmm1Δ | 1744/1745 |
| mmm1Δmdm35Δ | MATα his3 leu2 lys2 trp1 ura3 mmm1::kanMX4 mdm35::HIS3 | This study | mmm1Δ | PTM243/244 |
| mmm2Δ | MATα his3 leu2 lys2 trp1 ura3 mmm2::HIS3 | This study | FY833/FY834 | PTM272/273 |
| mmm2Δups1Δ | MATα his3 leu2 lys2 trp1 ura3 mmm2::kanMX4 ups1::kanMX4 | This study | mmm2Δ | 1729/1730 |
| mmm2Δups2Δ | MATα his3 leu2 lys2 trp1 ura3 mmm2::kanMX4 ups2::kanMX4 | This study | mmm2Δ | 1744/1745 |
| mmm2Δmdm35Δ | MATα his3 leu2 lys2 trp1 ura3 mmm2::kanMX4 mdm35::kanMX4 | This study | mmm2Δ | PTM243/244 |
| mdm10Δ | MATa his3 leu2 lys2 trp1 ura3 mdm10::HIS3 | This study | FY833/FY834 | PTM363/364 |
| mdm10Δups1Δ | MATa his3 leu2 lys2 trp1 ura3 mdm10::kanMX4 ups1::kanMX4 | This study | mdm10Δ | 1729/1730 |
| mdm10Δups2Δ | MATa his3 leu2 lys2 trp1 ura3 mdm10::kanMX4 ups2::kanMX4 | This study | mmm2Δ | 1744/1745 |
| mdm10Δmdm35Δ | MATa his3 leu2 lys2 trp1 ura3 mdm10::kanMX4 mdm35::kanMX4 | This study | mmm2Δ | PTM243/244 |
| mdm12Δ | MATa his3 leu2 lys2 trp1 ura3 mdm12::kanMX4 | This study | FY833/FY834 | PTM365/366 |
| mdm12Δups1Δ | MATa his3 leu2 lys2 trp1 ura3 mdm12::kanMX4 ups1::HIS3 | This study | mdm12Δ | 1729/1730 |
| mdm12Δups2Δ | MATa his3 leu2 lys2 trp1 ura3 mdm12::kanMX4 ups2::HIS3 | This study | mdm12Δ | 1744/1745 |
| mdm12Δmdm35Δ | MATa his3 leu2 lys2 trp1 ura3 mdm12::kanMX4 mdm35::HIS3 | This study | mdm12Δ | PTM243/244 |
| psd2Δdpl1Δ | MATa his3 leu2 lys2 trp1 ura3 psd2Δ::HIS3 dpl1Δ::URA3 | This study | FY833 | PTM337/338, PTM740/741 |
| ups1Δpsd2Δdpl1Δ | MATa his3 leu2 lys2 trp1 ura3 ups1::kanMX4 psd2Δ::HIS3 dpl1Δ::URA3 | This study | ups1Δ | PTM337/338, PTM740/741 |
| ups2Δpsd2Δdpl1Δ | MATa his3 leu2 lys2 trp1 ura3 ups2::HIS3 psd2Δ::kanMX4 dpl1Δ::URA3 | This study | ups2Δ | PTM337/338, PTM740/741 |
| mdm31Δ | MATa his3 leu2 lys2 trp1 ura3 mdm31::kanMX4 | This study | FY833 | PTM316/317 |
| psd1Δ | MATa his3 leu2 lys2 trp1 ura3 psd1::HIS3 | This study | FY833 | PTM183/184 |
| crd1Δ | MATa his3 leu2 lys2 trp1 ura3 crd1::kanMX4 | This study | FY833 | PTM181/182 |
| gep4Δ | MATa his3 leu2 lys2 trp1 ura3 gep4::kanMX4 | This study | FY833 | PTM324/325 |
| MMM1GFP | MATa his3 leu2 lys2 trp1 ura3 MMM1GFP::TRP1 | This study | FY833 | PTM353/354 |
| MMM1GFPups1Δ | MATa his3 leu2 lys2 trp1 ura3 MMM1GFP::TRP1 ups1Δ::kanMX4 | This study | MMM1GFP | 1729/1730 |
| MMM1GFPups2Δ | MATa his3 leu2 lys2 trp1 ura3 MMM1GFP::TRP1 ups2Δ::HIS3 | This study | MMM1GFP | 1744/1745 |
| MMM1GFPmdm31Δ | MATa his3 leu2 lys2 trp1 ura3 MMM1GFP::TRP1 mdm31Δ::kanMX4 | This study | MMM1GFP | PTM316/317 |
TABLE 2.
Plasmids used in this study
NA means not applicable.
| Plasmid name | Replication origin/marker/protein expressed | Source | Primers for cloning | Template DNA |
|---|---|---|---|---|
| pRS314-MMM1 | CEN, TRP1, Mmm1p | This study | 374/375 | Yeast genomic DNA |
| pRS314-MMM2 | CEN, TRP1, Mmm2p | This study | 376/377 | Yeast genomic DNA |
| pRS314-MDM10 | CEN, TRP1, Mdm10p | This study | 371/372 | Yeast genomic DNA |
| pRS314-MDM12 | CEN, TRP1, Mdm12p | This study | 379/380 | Yeast genomic DNA |
| pRS314-MDM31 | CEN, TRP1, Mdm12p | This study | 460/461 | Yeast genomic DNA |
| pRS424-MDM31 | 2μ, TRP1, Mdm31p | This study | 460/461 | Yeast genomic DNA |
| pRS424-MDM32 | 2μ, TRP1, Mdm32p | This study | 462/463 | Yeast genomic DNA |
| pRS314-MDM31ΔTM | CEN, TRP1, Mdm31p | This study | 460/685 | Yeast genomic DNA |
| pRS424-MDM31ΔTM | 2μ, TRP1, Mdm31p | This study | 460/685 | Yeast genomic DNA |
| pRS314-FLAG-MDM31-TEV-FLAG | CEN, TRP1, FLAG-Mdm31p-Tev-FLAG | This study | 460/705, 706/712, 709/690 | pRS314-MDM31 |
| pRS424-UPS1 | 2μ, TRP1, Ups1p | This study | NA | pRS314-UPS1 (34) |
| pRS424-UPS2 | 2μ, TRP1, Ups2p | This study | 489/492 | Yeast genomic DNA |
| pRS314-Crd1 | CEN, TRP1, Crd1p | This study | 468/469 | Yeast genomic DNA |
| pRS314-Gep4 | CEN, TRP1, Gep4p | This study | 454/455 | Yeast genomic DNA |
| pRS313-Cox4-Tev | CEN, HIS3, fusion protein of the presequence of Cox4p and TEV protease | This study | 571/572 | pBG153.6 (a gift from S. Gould) |
| pRS313-Cyt c1-Tev | CEN, HIS3, fusion protein of the presequence of cytochrome c1 and TEV protease | This study | NA | pAC1-c1-Cox4 (40) |
| pMY3 | CEN, Leu2, fusion protein of Mmm2p and RFP | Ref. 33 | NA | NA |
For expression of FLAG-Mdm31-TEV-FLAG in yeast, three DNA fragments were amplified from yeast genomic DNA with primer sets PTM460/705, PTM706/712 and PTM709/690 and cloned into pRS314 digested with NotI and XhoI.
pRS313-Cox4-TEV, a HIS3-CEN plasmid that expresses a Cox4-TEV fusion protein from the ADH1 promoter, was constructed as follows. The TEV protease sequence was amplified from pBG153.6 (a gift of Dr. S. Gould) using primers 571 and 582. The PCR product was digested with XbaI and SacII and inserted into XbaI/SacII-digested pOK29 (39).
pRS313-Cyt c1-TEV, a HIS3-CEN plasmid that expresses a Cyt c1-TEV fusion protein from the ADH1 promoter, was constructed as follows. Cytochrome c1 presequence was digested from pRJ618 (40), using XbaI and EcoRI, and inserted into pRS313-Cox4-TEV digested with XbaI and EcoRI.
PCR primers, restriction enzymes, and vectors used for cloning MMM1, MMM2, MDM10, MDM12, MDM31, MDM32, UPS2, CRD1, and GEP4 genes are listed in Table 2 and supplemental Table S1. Expression of these genes was controlled by their own promoter and terminator.
Cells were grown in YPD (1% yeast extract, 2% polypeptone, and 2% glucose), YPGE (1% yeast extract, 2% polypeptone, 3% glycerol, and 3% ethanol), and SCD (0.67% yeast nitrogen base without amino acids, 0.5% casamino acid, 2% glucose). Cells that have the kanMX4 gene were selected on YPD containing 200 μg/ml G418 sulfate. To remove the URA3-containing plasmid, cells were grown on SCD (−Trp) containing 1 mg/ml 5′-FOA. Standard genetic techniques were used (41).
Recombinant Protein Expression
To generate yeast Psd1p containing an N-terminal His6 tag, the entire open reading frame was cloned into the pET28a vector (EMD) in-frame and downstream of the His6 tag. His6Psd1p was induced in BL21-CodonPlus(DE3)-RIL Escherichia coli (Agilent Technologies), and inclusion bodies were isolated from native protein extracts (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole, pH 8.0, and supplemented with 1% (v/v) Tween 20 and 0.5 mm EDTA) by centrifugation at 10,000 × g for 20 min at 4 °C. Inclusion bodies were solubilized with 3 ml of inclusion body solubilization buffer (1.67% (w/v) Sarkosyl, 0.1 mm EDTA, 10 mm DTT, 10 mm Tris-Cl, pH 7.4, and 0.05% polyethylene glycol 3350) by vigorous vortexing, incubated on ice for 20 min, and diluted with 6 ml of 10 mm Tris-Cl, pH 7.4, and the solubilized proteins were recovered after centrifugation for 10 min at 12,000 × g at 4 °C. After dialysis against phosphate-buffered saline (PBS), His6Psd1p was purified under native conditions using Ni2+-agarose (Qiagen) as per the manufacturer's instructions and quantitated by SDS-PAGE with Coomassie Blue staining and a BSA standard curve. Antibodies were raised in rabbits using His6Psd1p as the antigen.
Phospholipid Analysis
Yeast cells were diluted to an A600 = 0.02 in 2 ml of YPD in the presence of 10 μCi/ml 32Pi and cultivated to stationary phase. Phospholipids were extracted from crude mitochondrial fractions and separated by TLC as described (7, 42–44).
Pulse-Chase Experiment with [14C]Serine
Yeast cells were grown in SCD-Ura and diluted in YPD (A600 = 0.05) and further cultivated to log phase. The cells were pulse-labeled in PBS containing 3 μCi/ml [14C]serine for 15 min at 30 °C. After labeling, the cells were washed with water, and a small fraction of the cells was saved as the starting material (zero time point). The rest of the cells were suspended in YPD and cultivated for different time periods. Total phospholipids were extracted as described (24). Briefly, the cells were precipitated by centrifugation, resuspended in 330 μl of methanol, and kept on ice until all pulse-chase samples were collected. The samples were then vortexed with 100 μl of glass beads for 15 min. 660 μl of chloroform was added, and the samples were centrifuged at 12,000 × g for 10 min to remove cell debris. 200 μl of water was added to the supernatant and vortexed for 5 min. The organic phase was separated by centrifugation at 400 × g for 5 min, dried in a speedvac, and then resuspended in chloroform. All the samples were loaded and analyze by TLC.
Microscopy
Cells were observed using an Olympus BX61 microscope with a ×100, 1.3 NA, objective. Fluorescence and differential interference contrast images were captured with a Roper/Photometrics Cool Snap HQ using Slidebook software version 5.0 (3i) and processed with Photoshop software (Adobe).
Immunoblotting
For immunoblotting, proteins were visualized by fluorophores conjugated with secondary antibodies (Alexa Fluor 488 or 647 goat anti-mouse or rabbit IgG (H+L) (Invitrogen)) and analyzed using a PharosFX Plus Molecular Imager (Bio-Rad) and Quantity One (Bio-Rad) and Photoshop (Adobe) software.
Alkaline Treatment
Mitochondria were isolated from cells expressing FLAG-Mdm31-TEV-FLAG and Cyt c1-TEV grown in YPGE. 150 μg of mitochondria were suspended in 0.1 m Na2CO4. After vortexing, the sample was incubated on ice for 30 min and centrifuged at 45,000 rpm for 30 min in a TLA55 rotor (Beckman) at 4 °C. Extracted proteins in the supernatant fraction were precipitated with 10% trichloroacetic acid. The precipitated protein and the membrane-bound fractions were analyzed by SDS-PAGE and immunoblotting.
Mitochondrial OM and IM Vesicle Separation
Mitochondrial OM and IM vesicles were prepared as described (45) with some modifications. Briefly, 4 mg of mitochondria were suspended in EM buffer (1 mm EDTA, 20 mm HEPES-KOH, pH 7.4) for 30 min on ice to open the OM by osmotic shock. The resulting mitoplast was further incubated for 10 min on ice after addition of sucrose to 0.45 m from 60% sucrose in EM buffer. The sample was then sonicated three times for 10 s using Sonic Dismembrator Model 100 (Fisher) with power setting 8. Intact mitochondria were removed by centrifugation at 18,800 rpm with an SW55Ti rotor (Beckman) for 20 min at 4 °C. OM and IM vesicles in the supernatant were pelleted by centrifugation at 47,000 rpm in an SW55Ti rotor for 40 min at 4 °C. The resulting pellet was resuspended in EM buffer with 10 mm NaCl, placed onto a sucrose step gradient (26:31:35:40%), and then centrifuged at 39,000 rpm for 16 h with a SW55Ti rotor.
RESULTS
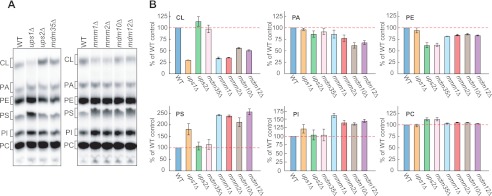
Loss of Ups1p and ERMES Proteins Leads to Similar Alterations in Phospholipid Composition
We compared the phospholipid composition of cells lacking Ups1p, Ups2p, and ERMES proteins (i.e. Mmm1p, Mmm2p, Mdm10p, and Mdm12p). Yeast cells were cultivated in 32Pi-containing media, and crude mitochondria fractions were isolated. Total phospholipids were extracted from the membranes and separated by thin layer chromatography (TLC) (Fig. 1). As we reported previously (7, 44), the amount of CL decreased, whereas PS levels increased in ups1Δ mitochondria (Fig. 1). In ups2Δ mitochondria, PE levels were decreased. In mitochondria isolated from ERMES-deleted cells, mmm1Δ, mmm2Δ, mdm10Δ, and mdm12Δ also contained reduced CL levels (Fig. 1), consistent with previous studies (22, 24). In addition, we found that PS was increased in all ERMES-deleted mitochondria, similar to ups1Δ mitochondria (Fig. 1). Thus, ups1Δ and ERMES-deleted cells exhibit similar alterations in steady state CL and PS levels. Our data suggest that Ups1p and ERMES proteins act at similar steps in phospholipid metabolism.
FIGURE 1.
Altered phospholipid composition in mitochondria lacking Ups proteins or ERMES subunits. A, indicated yeast cells were cultivated in YPD in the presence of 32Pi. Phospholipids were extracted from crude mitochondrial fractions and separated by TLC. PI, phosphatidylinositol. B, amounts of each lipid relative to total phospholipids were determined, and those detected in (WT) mitochondria were set to 100% (red dashed lines). Values are mean ± S.E. (n = 3).
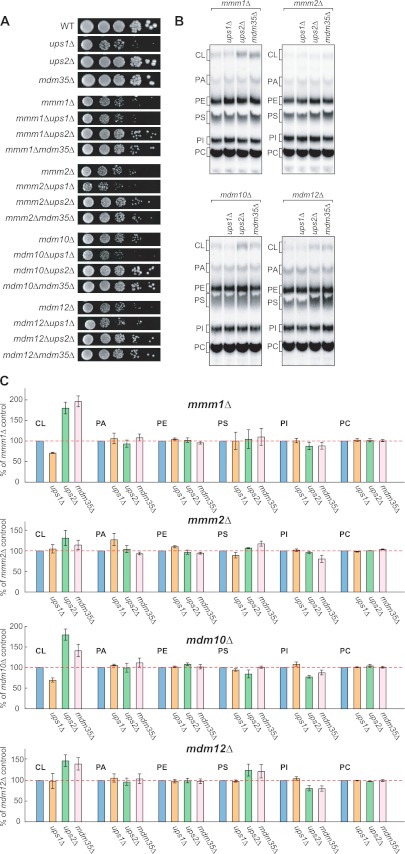
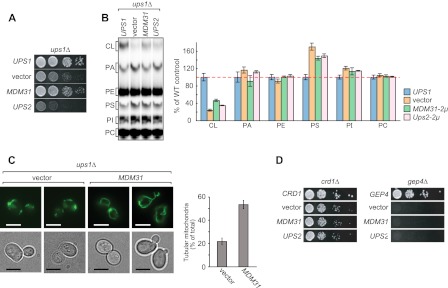
Loss of Ups1p Worsens Defects in Cell Growth and CL Levels in ERMES-deleted Cells although Loss of Ups2p Rescues These Phenotypes
We have previously reported that Ups1p and Ups2p affect CL levels antagonistically and that additional loss of Ups2p in ups1Δ cells restores CL levels and growth defects (7). In addition, we have shown that the IMS protein Mdm35p is required for importing Ups proteins into mitochondria (44). Not surprisingly, mdm35Δ cells exhibited decreased levels of both Ups proteins and normal CL quantities and cell growth (Figs. 1 and 2A), similar to ups1Δups2Δ cells (44). Because loss of ERMES proteins and Ups1p led to similar effects on phospholipids, we investigated whether loss of Ups1p exacerbates defects in cell growth and phospholipid composition in ERMES-deleted cells. We also assessed whether loss of Ups2p or Mdm35p suppresses these processes. To obtain double deletion mutants, we deleted the UPS1, UPS2, or MDM35 genes in ERMES-deleted cells carrying the URA3 plasmid expressing the appropriate ERMES genes (i.e. MMM1, MMM2, MDM10, or MDM12). These cells were grown on culture medium containing 5′-fluoro-orotic acid (5′-FOA) to allow cells to lose the complementing URA3 plasmid and generate the double deletion mutants. Cells lacking Ups1p and either Mmm2p or Mdm10p showed synthetic growth defects (Fig. 2A). In contrast, the growth defects of all ERMES-deleted cells were partially rescued by additional loss of Ups2p or Mdm35p (Fig. 2A).
FIGURE 2.
Loss of Ups2p improves cell growth and CL levels in ERMES-deleted cells. A, serial dilutions of the indicated yeast cells were spotted onto YPD and cultivated at 30 °C for 2 days. B, indicated yeast cells were cultivated in YPD in the presence of 32Pi. Phospholipids were extracted from crude mitochondria fractions and analyzed by TLC. C, amounts of each lipid relative to total phospholipids were determined, and those detected in ERMES-deleted mitochondria were set to 100% (e.g. mmm1Δ, mmm2Δ, mdm10Δ, or mdm12Δ). Values are mean ± S.E. (n = 3). PI, phosphatidylinositol.
Similar to cell growth, defects in CL were enhanced by additional loss of Ups1p and rescued by loss of Ups2 or Mdm35p in ERMES-deleted cells. As shown in Fig. 2, B and C, we found that loss of Ups1p further decreased CL levels in mmm1Δ and mdm10Δ cells, whereas CL levels in mmm1Δ, mdm10Δ, and mdm12Δ mitochondria were increased significantly in the absence of Ups2p or Mdm35p. In contrast, PS levels remained unchanged in these double mutants. Therefore, our data further suggest that the ERMES complex and Ups1p have a similar function.
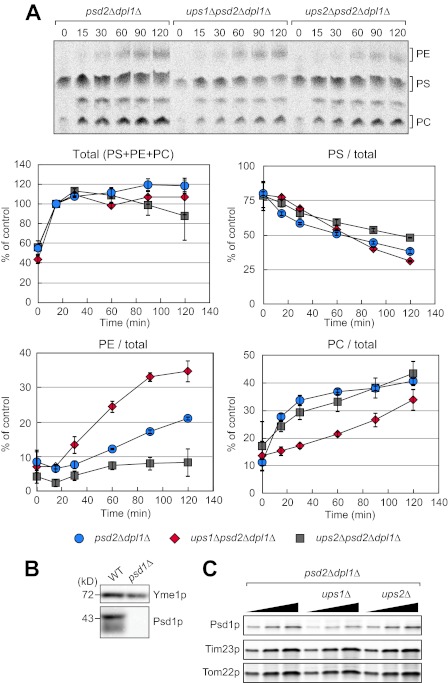
Loss of Ups1p and Ups2p Antagonistically Affects Conversion of PE to PC
We next examined whether loss of Ups1p or Ups2p alters phospholipid metabolism in pulse-chase experiments with [14C]serine. In this assay, we assessed incorporation of [14C]serine into PS, conversion of PS to PE, and conversion of PE to PC. Newly synthesized 14C-labeled PS is transported to the mitochondria from the ER and converted to PE by a mitochondrial PS decarboxylase Psd1p (17–19). The resulting PE then moves back to the ER and becomes PC through methylation by two ER-located enzymes, Cho2p and Opi3p (20). To directly assess this pathway, we blocked other pathways for PE production using psd2Δdpl1Δ cells as a parental strain. Psd2p is another PS decarboxylase located in the Golgi complex and vacuoles, although Dlp1p is a dihydrosphingosine phosphate lyase that generates phosphorylethanolamine, a precursor of PE through sphingolipid metabolism. We pulse-labeled psd2Δdpl1Δ, ups1Δpsd2Δdpl1Δ, and ups2Δpsd2Δdpl1Δ cells with [14C]serine and analyzed their total phospholipids by TLC at different time points (Fig. 3A). We found that incorporation of [14C]serine into PS, PE, and PC was similar in three strains (Fig. 3A, Total). We also found similar rate of consumption of PS in these cells (Fig. 3A, PS/total). These observations indicate that ups1Δ and ups2Δ mutations do not affect the incorporation of [14C]serine into PS or the stability of PS.
FIGURE 3.
Loss of Ups1p and Ups2p affects conversion of PE to PC. A, cells were pulse-labeled with [14C]serine for 15 min and further cultivated for the indicated periods of time. Total phospholipids were extracted from the cells and analyzed by TLC followed by radioimaging. Total amounts of PS, PE, and PC at 15 min were set to 100% (Total). The ratio of PS, PE, or PC relative to the total amounts was determined at each time point. Values are means ± S.E. (n = 3). B, whole cell extracts were prepared from wild-type and psd1Δ cells and analyzed by immunoblotting with antibodies against Yme1p and Psd1p. C, mitochondria were isolated from the indicated cells and analyzed by immunoblotting with antibodies to Psd1p, Tim23p, and Tom22p.
Interestingly, we found that ups1Δ and ups2Δ mutations led to opposite effects on PE accumulations. ups1Δ accumulated more PE, whereas ups2Δ decreased the PE amounts (Fig. 3A, PE/total). In addition, ups1Δ mutation slowed the production of PC, suggesting that conversion of PE to PC is slowed down in ups1Δ cells (Fig. 3A, PC/total). In contrast, although PE amounts are decreased in ups2Δ cells, production of PC was normal, suggesting that conversion of PE to PC is accelerated in ups2Δ cells (Fig. 3A, PC/total). Different amounts of PE are not due to differences in the level of the mitochondrial PS decarboxylase Psd1p. Immunoblotting of isolated mitochondria with anti-Psd1p antibodies showed similar amounts of Psd1p in psd2Δdpl1Δ, ups1Δpsd2Δdpl1Δ, and ups2Δpsd2Δdpl1Δ cells with a slight decrease in ups1Δpsd2Δdpl1Δ cells using immunoblotting with anti-Psd1p antibodies (Fig. 3, B and C).
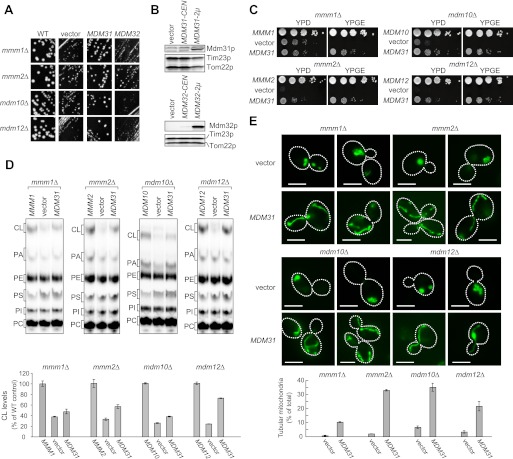
Overexpression of Mdm31p Rescues Defects in Growth, CL Levels, and Mitochondrial Morphology in ERMES-deleted Cells
Cells lacking Mdm31p or Mdm32p form large spherical mitochondria, similar to ERMES-deleted cells (31, 36). The combined loss of ERMES proteins and Mdm31p or Mdm32p results in synthetic lethality (36), suggesting that all of these proteins possess similar functions. To test this, we examined whether overexpression of Mdm31p or Mdm32p can compensate for the loss of ERMES proteins in cell growth, CL levels, and mitochondrial morphology. We introduced multicopy plasmids expressing either Mdm31p or Mdm32p into ERMES-deleted cells carrying the URA3-containing plasmid encoding the corresponding gene (MMM1, MMM2, MDM10, or MDM12). Cells containing an empty vector were also analyzed as a negative control. To eliminate the URA3-containing plasmid, the cells were streaked onto a 5′-FOA-containing plate (Fig. 4A). The colony sizes of all ERMES-deleted cells were larger when multicopy plasmid encoding the MDM31 gene is introduced (MDM31) compared with those carrying empty vector (vector) or 2 μm of plasmid encoding the MDM32 gene (MDM32). We confirmed overexpression of Mdm31p and Mdm32p by immunoblotting of whole cell extracts using antibodies against Mdm31p and Mdm32p (Fig. 4B). To more carefully compare growth of ERMES-deleted cells, serial dilutions of the cells were spotted onto growth media containing either a fermentable (YPD) or nonfermentable (YPGE) carbon source (Fig. 4C). Overexpression of Mdm31p partially rescued the growth defects observed in all ERMES-deleted cells grown on both YPD and YPGE (Fig. 4C). These results suggest that Mdm31p, but not Mdm32p, can at least partially replace ERMES proteins in cell growth.
FIGURE 4.
Overexpression of Mdm31p improves cell growth, CL abundance, and mitochondrial morphology in ERMES deletion cells. A, mmm1Δ, mmm2Δ, mdm10Δ, and mdm12Δ cells with URA3-single copy plasmid harboring the corresponding gene were co-transformed with TRP1 multicopy vector harboring MDM31 or MDM32 or an empty vector. Transformants were streaked onto SCD-Trp + FOA plates and cultivated for 3 days to remove the URA3 plasmid. B, whole cell extracts were prepared from mdm31Δ (upper panel) or mdm32Δ cells (lower panel) and analyzed by immunoblotting with the indicated antibodies. Cells were transformed with single copy (CEN) and multicopy (2μ) plasmids expressing Mdm31p or Mdm32p. An empty vector was used as a negative control. C, serial dilutions of the indicated yeast cells were spotted onto YPD and YPGE, and grown for 2 and 5 days, respectively. D, ERMES-deleted cells with either the single copy plasmid harboring the corresponding gene (MMM1, MMM2, MDM10, or MDM12), an empty vector (vector), or the multicopy plasmid harboring MDM31 (MDM31) were grown in YPD in the presence of 32Pi. Phospholipids were extracted from crude mitochondrial fractions and analyzed by TLC. Amounts of CL relative to total phospholipids were determined, and those detected in mitochondria were isolated from ERMES-deleted cells with the wild-type plasmid (e.g. MMM1, MMM2, MDM10, or MDM12) were set to 100%. E, mitochondria were visualized in the indicated cells using matrix-targeted Su9-GFP. Cells were grown to log phase in YPD and examined by fluorescence microscopy and differential interference microscopy. Cells containing tubular mitochondria were scored. Values are mean ± S.E. (n = 3). At least 100 cells were examined in each experiment. Bar, 5 μm.
Next, we assessed whether Mdm31p overexpression also suppresses CL defects in ERMES-deleted cells. We cultured ERMES-deleted cells with either a single copy plasmid encoding the corresponding gene (i.e. MMM1, MMM2, MDM10, or MDM12), a multicopy plasmid encoding the MDM31 gene (MDM31), or an empty vector (vector) in the presence of 32Pi, and we then isolated crude mitochondrial fractions. Phospholipids were extracted and analyzed by TLC. As shown in Fig. 4D, the amounts of CL were increased significantly in all ERMES-deleted mitochondria when Mdm31p was overexpressed. Therefore, in addition to cell growth, overexpression of Mdm31p functions to increase CL levels in ERMES-deleted cells.
Furthermore, Mdm31p overexpression also partially restored the tubular mitochondrial morphology in ERMES-deleted cells. To visualize mitochondrial shape, we expressed mitochondrion-targeted GFP (Su9-GFP) in ERMES-deleted cells with or without Mdm31p overexpression. We confirmed that virtually all ERMES-deleted cells exhibited abnormal mitochondrial morphology such as large spherical shapes or aggregations of small spheres (Fig. 4E). In contrast, when Mdm31p was overexpressed, the number of ERMES-deleted cells with tubular mitochondria increased significantly (Fig. 4E). Thus, overexpression of Mdm31p can partially rescue defects in cell growth, CL levels, and mitochondrial morphology in ERMES-deleted cells.
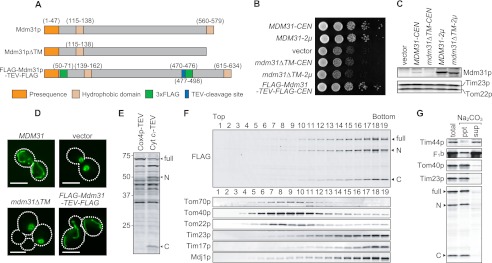
Mdm31p Spans the IM Twice and the Second Transmembrane Domain Is Required for Its Function
Mdm31p has an N-terminal matrix-targeting presequence and two putative transmembrane domains. Mdm31p spans the inner mitochondrial membrane at least once in the first hydrophobic region, and the portion between the first and second hydrophobic regions is exposed to the IMS (36). However, the exact topology of Mdm31p has not been determined. Therefore, we investigated the functional importance and topology of the second hydrophobic region. First, we introduced Mdm31p (MDM31), truncated Mdm31p lacking the second hydrophobic domain (MDM31ΔTM) (Fig. 4A), or an empty vector (vector) into mdm31Δ cells and observed their growth and mitochondrial shape (Fig. 5, B and D). We confirmed comparable expression levels of full-length and truncated Mdm31p by immunoblotting using anti-Mdm31p antibodies (Fig. 5C). mdm31Δ cells expressing Mdm31pΔTM were defective in cell growth and contained spherical mitochondria, similar to vector control cells, thereby demonstrating that the second hydrophobic domain is required for Mdm31p function (Fig. 5, B and D). To determine membrane topology, we marked Mdm31p by introducing 3×FLAG tags at two different locations along the protein. One tag was placed immediately after the presequence, and the second was inserted between the first and second hydrophobic domains. Moreover, a TEV cleavage site was introduced before the second 3×FLAG tag (FLAG-Mdm31p-TEV-FLAG, Fig. 4A). Expression of FLAG-Mdm31p-TEV-FLAG rescued defects in both cell growth and mitochondrial morphology in mdm31Δ cells, confirming that this FLAG-tagged protein is functional (Fig. 5, B and D). We then expressed IMS-targeted TEV protease (Cyt c1-TEV) or matrix-targeted TEV protease (Cox4p-TEV) into mdm31Δ cells expressing FLAG-Mdm31p-TEV-FLAG. Whole cell extracts were analyzed by immunoblotting with anti-FLAG antibodies (Fig. 5E). Although Cox4p-TEV-expressing cells only exhibited full-length FLAG-Mdm31p-TEV-FLAG (68 kDa), two additional bands corresponding to the N-terminal (Mdm31p-N; 50 kDa) and C-terminal halves (Mdm31p-C; 18 kDa) of Mdm31p were detected in Cyt c1-TEV-expressing cells (Fig. 5E). These results indicate that Cyt c1-TEV processes FLAG-Mdm31p-TEV-FLAG in the IMS.
FIGURE 5.
Analysis of the membrane topology of Mdm31p. A, to analyze the function of the second hydrophobic region of Mdm31p, this region was deleted (Mdm31pΔTM). To determine membrane topology of Mdm31p, 3×FLAG tags were introduced at two different locations along the protein. One tag was inserted after the presequence, and the second was placed between the first and second hydrophobic domains. In addition, a tobacco etch virus (TEV)-cleavage site was introduced before the second 3×FLAG tag (FLAG-Mdm31p-TEV-FLAG). B, serial dilution of mdm31Δ cells carrying the single (CEN) or multicopy (2μ) plasmid expressing Mdm31p, Mdm31pΔTM (mdm31ΔTM), FLAG-Mdm31p-TEV-FLAG (FLAG-MDM31-TEV-FLAG), or an empty vector (vector) were spotted onto YPD and incubated for 1 day. C, whole cells extracts prepared from the cells indicated in B were analyzed by immunoblotting with the indicated antibodies. D, mitochondria were visualized by matrix-targeted Su9-GFP in mdm31Δ cells expressing different forms of Mdm31p. Mdm31pΔTM (mdm31ΔTM) was expressed from the multicopy plasmid. E, whole cell extracts from cells expressing FLAG-Mdm31p-TEV-FLAG and either Cox4p-TEV or Cyt c1-TEV were analyzed by immunoblotting using anti-FLAG antibodies. F, mitochondria were isolated from cells expressing FLAG-Mdm31p-TEV-FLAG and Cyt c1-TEV. OM and IM vesicles were generated by osmotic shock followed by sonication. These vesicles were separated by sucrose density gradient centrifugation. Proteins from each fraction were analyzed by immunoblotting using antibodies against the indicated proteins. G, isolated mitochondria expressing FLAG-Mdm31p-TEV-FLAG and Cyt c1-TEV were treated with 0.1 m Na2CO4, and the supernatant and pellet fractions were separated by ultracentrifugation. Proteins were analyzed by immunoblotting using antibodies against the indicated proteins. C and N, C- and N-terminal halves of Mdm231p.
Next, we examined the localization of these TEV-cleaved fragments, Mdm31p-N and -C. Mitochondria were isolated from cells expressing FLAG-Mdm31p-TEV-FLAG and Cyt c1-TEV. OM- and IM-derived vesicles from the mitochondria were generated by sonication and separated on a sucrose density gradient. If Mdm31p spans both the OM and IM, then Mdm31p-N and Mdm31p-C would be present in the IM and OM, respectively. Alternatively, if Mdm31p spans the IM twice, then both Mdm31p-N and -C would be associated with the IM. Fractions from the sucrose gradients were analyzed using antibodies against OM proteins (i.e. Tom70p, Tom40p, and Tom22p), IM proteins (i.e. Tim23p, Tim17p, and Mdj1p), and FLAG. Tom70p, Tom40p, and Tom22p were predominantly present in fractions 7–10, although Tim23p, Tim17p, and Mdj1p were detected starting from fraction 14 (Fig. 5F). Full-length Mdm31p and both TEV-cleaved fragments showed localization patterns similar to those of IM proteins, indicating that the N- and C-terminal regions of Mdm31p are attached to the inner membrane (Fig. 5F).
We also performed alkaline extraction to test whether Mdm31p-N and -C are inserted into the IM. Mitochondria expressing FLAG-Mdm31p-TEV-FLAG and Cyt c1-TEV were treated with Na2CO3, and then peripheral membrane proteins were extracted. As shown in Fig. 5G, two peripherally associated IM proteins, Tim44p and F1β, were extracted into the supernatant, although two integral membrane proteins, Tom40p and Tim23p, were present in the pellet. Similar to Tom40p and Tim23p, Mdm31p-full-length, -N, and -C were resistant to alkaline treatment (Fig. 5G). Taken together, our results demonstrate that Mdm31p spans the IM twice, and that the second hydrophobic region is essential for its functions.
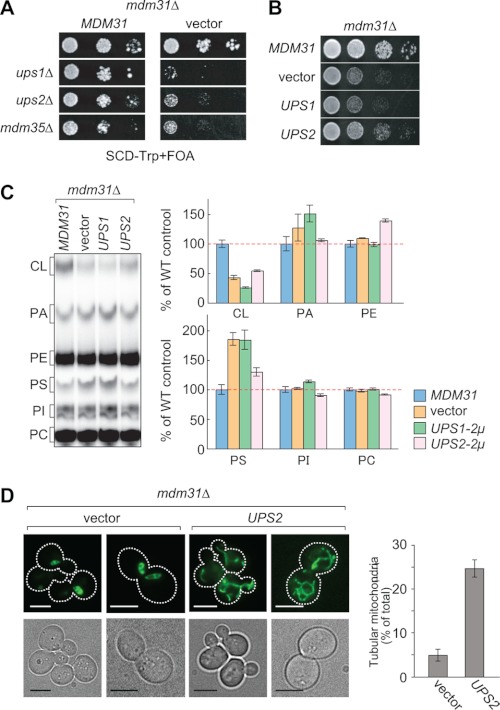
Overexpression of Mdm31p Rescues ups1Δ Cells
Given the similar phenotypes observed in ups1Δ and ERMES-deleted cells, we sought to understand whether Mdm31p overexpression also rescues ups1Δ cells. We introduced a multicopy plasmid harboring the MDM31 gene or an empty vector into ups1Δ cells and spotted the transformed cells onto YPD (Fig. 6A). We found that Mdm31p overexpression improved growth in ups1Δ cells (Fig. 6A). Similarly, Mdm31p overexpression also partially restored CL levels in ups1Δ mitochondria, in which CL was decreased to ∼25% compared with mitochondria. Following Mdm31p overexpression, the level of CL was increased significantly by 2-fold (Fig. 6B). Tubule mitochondrial morphology in ups1Δ cells was also partially restored in the presence of Mdm31p (Fig. 6C). As reported previously (7, 34), ∼80% of ups1Δ cells showed abnormal mitochondrial morphology, such as fragmented tubules and aggregated structures. However, when Mdm31p was overexpressed, ∼50% of ups1Δ cells exhibited tubular mitochondria (Fig. 6C). These results indicate that higher levels of Mdm31p can partially restore cell growth, CL, and mitochondrial morphology in ups1Δ cells. To test whether these effects are specific to ERMES-deleted and ups1Δ cells, we overexpressed Mdm31p in other mutants that are defective in CL biosynthesis. We found that Mdm31p overexpression does not rescue growth phenotypes in cells lacking cardiolipin synthase (Crd1p) or phosphatidylglycerophosphatase (Gep4p), indicating that the suppressive effects incurred by Mdm31p overexpression are specific to ERMES-deleted and ups1Δ cells (Fig. 6D).
FIGURE 6.
Mdm31p overexpression ameliorates cell growth, CL amounts, and mitochondrial morphology in ups1Δ cells. A, serial dilutions of ups1Δ cells with single copy plasmids expressing UPS1 (UPS1), empty vector (vector), multicopy plasmids expressing Mdm31p (MDM31), or Ups2p (UPS2) were spotted onto YPD and cultivated at 30 °C for 2 days. B, yeast cells used in A were grown in YPD in the presence of 32Pi. Phospholipids were extracted from crude mitochondrial fractions and separated by TLC. Amounts of each lipid relative to total phospholipids were determined, and those detected in mitochondria from ups1Δ cells containing the Ups1p plasmid were set to 100% (e.g. UPS1). Values are mean ± S.E. (n = 3). PI, phosphatidylinositol. C, ups1Δ cells containing the multicopy plasmid harboring MDM31 (MDM31) or an empty vector (vector) were grown in YPD to log phase. Mitochondria were visualized using matrix-targeted Su9-GFP. Cells containing tubular mitochondria were scored. Values are mean ± S.E. (n = 3). At least 100 cells were examined in each experiment. Bar, 5 μm. D, serial dilutions of crd1Δ and gep4Δ cells carrying the multicopy plasmid harboring MDM31 (MDM31) or UPS2 (UPS2), or an empty vector (vector), were spotted onto YPD and grown for 2 days.
Overexpression of Ups2p Rescues mdm31Δ Cells
The growth defects of ERMES-deleted cells were enhanced by the addition of ups1Δ, although ups2Δ rescued them. However, unlike ERMES-deleted cells, mdm31Δ cells displayed additional growth defects when combined with ups1Δ and ups2Δ (Fig. 7A). Thus, we assessed whether overexpression of Ups1p or Ups2p could rescue the growth defects in mdm31Δ cells. Interestingly, overexpression of Ups2p, but not Ups1p, relieved the growth defects of mdm31Δ cells, suggesting overlapping functions for Mdm31p and Ups2p (Fig. 7B). We then examined the steady state levels of phospholipids and mitochondrial shape in mdm31Δ mitochondria with or without Ups2p overexpression. Our data show that CL levels were decreased by ∼60% in mdm31Δ mitochondria compared with those in wild-type mitochondria (i.e. mdm31Δ mitochondria expressing Mdm31p) (MDM31, Fig. 7C). In addition, similar to ups1Δ and ERMES-deleted mitochondria, mdm31Δ mitochondria contained increased levels of PS (vector, Fig. 7C). Interestingly, we found that Ups2p overexpression partially decreased PS levels and increased PE levels (Fig. 7C). mdm31Δ cells contained spherical mitochondria that often showed a hollow nontubular structure. We found that the tubular morphology of mitochondria was increased by Ups2p overexpression in mdm31Δ cells (Fig. 7D). Suppression of growth defects by Ups2p appeared to be specific to mdm31Δ as Ups2p overexpression failed to rescue the growth defect in crd1Δ and gep4Δ cells (Fig. 6D). Thus, Ups2p can improve cell growth, CL levels, and mitochondrial morphology in mdm31Δ cells.
FIGURE 7.
Ups2p overexpression partially rescues defects in cell growth, CL levels, and mitochondrial morphology in mdm31Δ cells. A, UPS1, UPS2, or MDM35 gene was deleted in mdm31Δ cells carrying the URA3 single copy plasmid harboring the MDM31 gene. The resulting yeast cells were transformed with the TRP1 single copy plasmid harboring MDM31 or a TRP1 empty vector. These cells were spotted onto the 5′-FOA-containing SCD-Trp medium and incubated for 2 days. B, serial dilutions of mdm31Δ cells carrying the single copy plasmid expressing Mdm31p (MDM31), the multicopy plasmids expressing Ups1p (UPS1) or Ups2p (UPS2), or an empty vector (vector) were spotted onto YPD and grown for 2 days. C, yeast cells used in B were grown in YPD in the presence of 32Pi. Phospholipid was extracted from crude mitochondrial fractions and analyzed by TLC. Amounts of each lipid were quantified and normalized to those in mdm31Δ mitochondria expressing Mdm31p. Values are mean ± S.E. (n = 3). PI, phosphatidylinositol. D, mitochondrial morphology in mdm31Δ cells carrying the multicopy plasmid harboring UPS2 (UPS2) or an empty vector (vector) were visualized using Su9-GFP. Cells were cultivated in YPD to log prior to observation. Cells containing tubular mitochondria were scored. Values are means ± S.E. (n = 3). At least 100 cells were examined in each experiment. Bar, 5 μm.
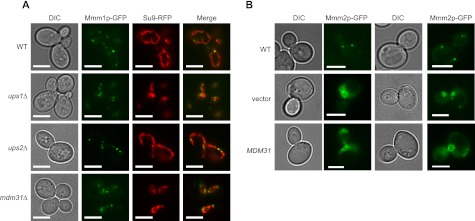
Loss of Ups Proteins and Mdm31p as Well as Mdm31p Overexpression Do Not Affect the ERMES Complex Formation
How are Ups proteins and Mdm31p involved in phospholipid metabolism? A possible function of Ups proteins and Mdm31p is to regulate formations of the ERMES complex. To test this hypothesis, we examined the effects of loss of Ups1p, Ups2p, and Mdm31p in formation of the ER-mitochondria contact sites. To observe the ER-mitochondria contact sites, we GFP-tagged an ER resident subunit of the ERMES protein, Mmm1p, and expressed mitochondrion-targeted RFP (Su9-RFP) in wild-type, ups1Δ, ups2Δ and mdm31Δ cells. As expected, the ERMES complex visualized by Mmm1p-GFP was observed as punctate structures and associated with mitochondria in wild-type cells (Fig. 8A, WT). Similarly, in ups1Δ, ups2Δ, and mdm31Δ cells in which mitochondrial shapes were changed to aggregated or spherical structures, the foci of Mmm1p-GFP formed normally and associated with mitochondria.
FIGURE 8.
Ups1p, Ups2p, and Mdm31p are not required for formation of ER-mitochondria contact sites. A, GFP-tagged Mmm1p (Mmm1p-GFP) and mitochondria-targeted RFP (Su9-RFP) were expressed in wild-type, ups1Δ, ups2Δ, and mdm31Δ cells. B, GFP-tagged Mmm2p (Mmm2p-GFP) was expressed from plasmid, pMY3 (33), in mdm12Δ cells with the multicopy plasmid expressing Mdm31p or an empty vector. Bar, 5 μm; DIC, differential interference contrast.
We also tested whether suppression of ERMES-deleted cells by Mdm31p overexpression is due to restoration of the ERMES complex. We monitored Mmm2p-GFP in mdm12Δ in the presence of absence of Mdm31p overexpression. As reported previously, in the absence of Mdm12p, Mmm2p-GFP defused to entire mitochondria (Fig. 8B, vector). A similar distribution was observed in mdm12Δ cells overexpressing Mdm31p (Fig. 8B, MDM31). These results indicate that Ups1p, Ups2p, and Mdm31p do not control the formation of ER-mitochondria contact sites.
DISCUSSION
In this study, we investigated the roles of the ERMES complex, Ups proteins, and Mdm31p in phospholipid metabolism using gene knock-out and overexpression methods. Individual loss of ERMES subunits, Ups1p, or Mdm31p led to similar phospholipid profiles such as decreased CL levels and increased PS levels. We also found that the ERMES genes (MMM1, MMM2, MDM10, and MDM12), UPS1, UPS2, and MDM31 genetically interact. First, defects in cell growth of ERMES-deleted cells can be rescued by Mdm31p overexpression or loss of Ups2p, which has functions opposite to Ups1p. Second, decreased cell growth in ups1Δ cells was also partially restored by Mdm31p overexpression. Third, additional loss of Ups1p worsens cell growth in ERMES-deleted cells. Fourth, overexpression of Ups2p partially restores growth defects of mdm31Δ cells. Fifth, loss of Mdm31p together with Ups1p or Ups2p causes severe synthetic growth defects. Finally, a previous study has demonstrated synthetic growth defects in ERMES-deleted cells in combination with mdm31Δ (36). These results suggest that these proteins function in related mechanisms for phospholipid metabolism, such as transport of phospholipids between the ER and mitochondria, transport of phospholipids within mitochondria, stability of phospholipids, and regulation of phospholipid biosynthetic enzymes.
Our pulse-chase experiments to analyze conversion of PS to PE and PC delineated at which stage lipid conversion is altered and suggest that Ups1p and Ups2p antagonistically regulate PE to PC conversion. Ups1p promotes PE to PC conversion because conversion rate of PE to PC was slower and PE accumulated in the absence of Ups1p. In contract, Ups2p suppresses PE to PC conversion. Supporting this function, ups2Δ cells showed accelerate PE to PC conversion and reduced amounts of PE. Ups1p and Ups2p may regulate outward trafficking of PE from the IM to the ER or PC biosynthetic enzymes such as Cho2p and Opi3p. Our pulse-chase data are consistent with the observation that ups2Δ mitochondria contain lower levels of PE and slightly increased levels of PC at the steady state (7). Different from the pulse-chase data, we did not observe accumulation of PE in ups1Δ mitochondria at the steady state level. The steady state levels of PE may be compensated by parallel pathways. For example, PE and PC can be synthesized by two distinct mechanisms, the CDP-diacylglycerol and Kennedy pathways. Also, two PE synthases are present in mitochondria and Golgi/vacuole.
In yeast, the ERMES complex is the only machinery reported so far to form ER-OM contact sites. Our data that loss of the ERMES complex can be partially substituted by overexpression of Mdm31p or loss of Ups2p suggest that there may be multiple mechanisms for ER-OM and OM-IM contact sites. In mammals, it has been suggested that there may be multiple mechanisms for the formation of ER-OM contact sites called the mitochondrion-associated ER membrane. For instance, a mitochondrial fusion protein, Mfn2, tethers mitochondria and the ER (53). In addition, many proteins are enriched at the mitochondrion-associated ER membrane (54). OM-IM contact sites are also formed by multiple mechanisms. The mitochondrial translocases in the OM and IM, the TOM and TIM23 complexes, are physically connected and coordinately function (46–49). In addition, recent studies have shown that a protein complex named MICOS, MINOS, or MitOS spans both the OM and IM and mediates OM-IM contact sites as well as cristae junctions (50–52).
The molecular functions of Mdm31p remain to be determined. Loss of Ups2p does not rescue the growth and CL defects of mdm31Δ cells, which show similar phenotypes to those in ups1Δ and ERMES-deleted cells. Therefore, Mdm31p does not appear to antagonize Ups2p like Ups1p and ERMES proteins. On the contrary, our genetic analyses suggest that Mdm31p and Ups2p have overlapping functions. Mdm31p and Ups2p may negatively regulate conversion of PE to PC. This potential role could explain the decreased levels of PE observed in ups2Δ and mdm31Δ cells (7, 35). Ups2p and Mdm31p may also help maintain their abundance of PE and/or CL in the IM because overexpression of Ups2p and Mdm31p increases PE and CL levels under some conditions (i.e. Ups2p overexpression in mdm31Δ cells and Mdm31p overexpression in ERMES-deleted or ups1Δ cells) (Figs. 4D, 6B, and 7C).
Defects in cell growth and mitochondrial morphology in cells lacking the ERMES complex, Ups1p, or Mdm31p do not simply result from a decrease in CL levels. Supporting this notion, mitochondria display tubular morphology in crd1Δ cells, which completely lack CL (55). These cells also do not display severe impairments in cell growth (55). Therefore, morphological defects may be caused by alterations in the levels of multiple phospholipids or their ratio within mitochondrial membranes. It is also possible that continuous trafficking of phospholipids may be critical for maintaining the dynamic nature of mitochondrial morphology and therefore their function. These models are consistent with a previous study showing that artificially reconnecting the ER and mitochondria using ChiMERA, a hybrid protein that associates with both the ER membrane and OM, can restore tubular mitochondrial morphology in ERMES-deleted cells (24).
Supplementary Material
Acknowledgments
We thank Toshiya Endo for valuable comments on the manuscript and antibodies and Benedikt Westermann for antibodies against Mdm31p and Mdm32p. We are grateful to members of the Sesaki and Iijima laboratories for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM084015 (to M. I.) and GM089853 (to H. S.). This work was also supported by a postdoctoral fellowship for research abroad from the Japan Society for the Promotion of Science (to Y. T.).

This article contains supplemental Table S1.
- OM
- mitochondrial outer membrane
- CL
- cardiolipin
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- IMS
- mitochondrial intermembrane space
- IM
- mitochondrial inner membrane
- ER
- endoplasmic reticulum
- ERMES
- ER-mitochondria encounter structure
- 5′-FOA
- 5′-fluoro-orotic acid
- TEV
- tobacco etch virus
- Cyt
- cytochrome
- RFP
- red fluorescent protein.
REFERENCES
- 1. Claypool S. M. (2009) Cardiolipin, a critical determinant of mitochondrial carrier protein assembly and function. Biochim. Biophys. Acta 1788, 2059–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joshi A. S., Zhou J., Gohil V. M., Chen S., Greenberg M. L. (2009) Cellular functions of cardiolipin in yeast. Biochim. Biophys. Acta 1793, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang F., Ryan M. T., Schlame M., Zhao M., Gu Z., Klingenberg M., Pfanner N., Greenberg M. L. (2000) Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275, 22387–22394 [DOI] [PubMed] [Google Scholar]
- 4. van der Laan M., Meinecke M., Dudek J., Hutu D. P., Lind M., Perschil I., Guiard B., Wagner R., Pfanner N., Rehling P. (2007) Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat. Cell Biol. 9, 1152–1159 [DOI] [PubMed] [Google Scholar]
- 5. Kutik S., Rissler M., Guan X. L., Guiard B., Shui G., Gebert N., Heacock P. N., Rehling P., Dowhan W., Wenk M. R., Pfanner N., Wiedemann N. (2008) The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J. Cell Biol. 183, 1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gebert N., Joshi A. S., Kutik S., Becker T., McKenzie M., Guan X. L., Mooga V. P., Stroud D. A., Kulkarni G., Wenk M. R., Rehling P., Meisinger C., Ryan M. T., Wiedemann N., Greenberg M. L., Pfanner N. (2009) Mitochondrial cardiolipin involved in outer-membrane protein biogenesis. Implications for Barth syndrome. Curr. Biol. 19, 2133–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tamura Y., Endo T., Iijima M., Sesaki H. (2009) Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 185, 1029–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harada Y., Tamura Y., Endo T. (2010) Identification of yeast Art5 as a multicopy suppressor for the mitochondrial translocator maintenance protein Tam41. Biochem. Biophys. Res. Commun. 392, 228–233 [DOI] [PubMed] [Google Scholar]
- 9. Choi S. Y., Huang P., Jenkins G. M., Chan D. C., Schiller J., Frohman M. A. (2006) A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 8, 1255–1262 [DOI] [PubMed] [Google Scholar]
- 10. DeVay R. M., Dominguez-Ramirez L., Lackner L. L., Hoppins S., Stahlberg H., Nunnari J. (2009) Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186, 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rujiviphat J., Meglei G., Rubinstein J. L., McQuibban G. A. (2009) Phospholipid association is essential for dynamin-related protein Mgm1 to function in mitochondrial membrane fusion. J. Biol. Chem. 284, 28682–28686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang H., Gao Q., Peng X., Choi S. Y., Sarma K., Ren H., Morris A. J., Frohman M. A. (2011) piRNA-associated germ line nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial surface lipid signaling. Dev. Cell 20, 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lutter M., Fang M., Luo X., Nishijima M., Xie X., Wang X. (2000) Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2, 754–761 [DOI] [PubMed] [Google Scholar]
- 14. Lucken-Ardjomande S., Montessuit S., Martinou J. C. (2008) Contributions to Bax insertion and oligomerization of lipids of the mitochondrial outer membrane. Cell Death Differ. 15, 929–937 [DOI] [PubMed] [Google Scholar]
- 15. Sani M. A., Dufourc E. J., Gröbner G. (2009) How does the Bax-α1 targeting sequence interact with mitochondrial membranes? The role of cardiolipin. Biochim. Biophys. Acta 1788, 623–631 [DOI] [PubMed] [Google Scholar]
- 16. Zaltsman Y., Shachnai L., Yivgi-Ohana N., Schwarz M., Maryanovich M., Houtkooper R. H., Vaz F. M., De Leonardis F., Fiermonte G., Palmieri F., Gillissen B., Daniel P. T., Jimenez E., Walsh S., Koehler C. M., Roy S. S., Walter L., Hajnóczky G., Gross A. (2010) MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat. Cell Biol. 12, 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Voelker D. R. (1989) Reconstitution of phosphatidylserine import into rat liver mitochondria. J. Biol. Chem. 264, 8019–8025 [PubMed] [Google Scholar]
- 18. Simbeni R., Tangemann K., Schmidt M., Ceolotto C., Paltauf F., Daum G. (1993) Import of phosphatidylserine into isolated yeast mitochondria. Biochim. Biophys. Acta 1145, 1–7 [DOI] [PubMed] [Google Scholar]
- 19. Trotter P. J., Pedretti J., Voelker D. R. (1993) Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268, 21416–21424 [PubMed] [Google Scholar]
- 20. Kodaki T., Yamashita S. (1987) Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J. Biol. Chem. 262, 15428–15435 [PubMed] [Google Scholar]
- 21. Chang S. C., Heacock P. N., Clancey C. J., Dowhan W. (1998) The PEL1 gene (renamed PGS1) encodes the phosphatidylglycerophosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 273, 9829–9836 [DOI] [PubMed] [Google Scholar]
- 22. Osman C., Haag M., Wieland F. T., Brügger B., Langer T. (2010) A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 29, 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang S. C., Heacock P. N., Mileykovskaya E., Voelker D. R., Dowhan W. (1998) Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J. Biol. Chem. 273, 14933–14941 [DOI] [PubMed] [Google Scholar]
- 24. Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., Walter P. (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kornmann B., Osman C., Walter P. (2011) The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. U.S.A. 108, 14151–14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stroud D. A., Oeljeklaus S., Wiese S., Bohnert M., Lewandrowski U., Sickmann A., Guiard B., van der Laan M., Warscheid B., Wiedemann N. (2011) Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J. Mol. Biol. 413, 743–750 [DOI] [PubMed] [Google Scholar]
- 27. Kopec K. O., Alva V., Lupas A. N. (2010) Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics 26, 1927–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Burgess S. M., Delannoy M., Jensen R. E. (1994) MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J. Cell Biol. 126, 1375–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sogo L. F., Yaffe M. P. (1994) Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 126, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berger K. H., Sogo L. F., Yaffe M. P. (1997) Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J. Cell Biol. 136, 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dimmer K. S., Fritz S., Fuchs F., Messerschmitt M., Weinbach N., Neupert W., Westermann B. (2002) Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frederick R. L., Okamoto K., Shaw J. M. (2008) Multiple pathways influence mitochondrial inheritance in budding yeast. Genetics 178, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Youngman M. J., Hobbs A. E., Burgess S. M., Srinivasan M., Jensen R. E. (2004) Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 164, 677–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sesaki H., Dunn C. D., Iijima M., Shepard K. A., Yaffe M. P., Machamer C. E., Jensen R. E. (2006) Ups1p, a conserved intermembrane space protein, regulates mitochondrial shape and alternative topogenesis of Mgm1p. J. Cell Biol. 173, 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., Wieland F. T., Brügger B., Westermann B., Langer T. (2009) The genetic interactome of prohibitins. Coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dimmer K. S., Jakobs S., Vogel F., Altmann K., Westermann B. (2005) Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast. J. Cell Biol. 168, 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C. A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 38. Winston F., Dollard C., Ricupero-Hovasse S. L. (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11, 53–55 [DOI] [PubMed] [Google Scholar]
- 39. Sesaki H., Jensen R. E. (1999) Division versus fusion. Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jensen R. E., Schmidt S., Mark R. J. (1992) Mutations in a 19-amino acid hydrophobic region of the yeast cytochrome c1 presequence prevent sorting to the mitochondrial intermembrane space. Mol. Cell. Biol. 12, 4677–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adams A., Gottschling D., Kaiser C., Stearns T. (1997) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42. Vaden D. L., Gohil V. M., Gu Z., Greenberg M. L. (2005) Separation of yeast phospholipids using one-dimensional thin layer chromatography. Anal. Biochem. 338, 162–164 [DOI] [PubMed] [Google Scholar]
- 43. Claypool S. M., McCaffery J. M., Koehler C. M. (2006) Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J. Cell Biol. 174, 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tamura Y., Iijima M., Sesaki H. (2010) Mdm35p imports Ups proteins into the mitochondrial intermembrane space by functional complex formation. EMBO J. 29, 2875–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pon L., Moll T., Vestweber D., Marshallsay B., Schatz G. (1989) Protein import into mitochondria. ATP-dependent protein translocation activity in a submitochondrial fraction enriched in membrane contact sites and specific proteins. J. Cell Biol. 109, 2603–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tamura Y., Harada Y., Shiota T., Yamano K., Watanabe K., Yokota M., Yamamoto H., Sesaki H., Endo T. (2009) Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shiota T., Mabuchi H., Tanaka-Yamano S., Yamano K., Endo T. (2011) In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc. Natl. Acad. Sci. U.S.A. 108, 15179–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chacinska A., Lind M., Frazier A. E., Dudek J., Meisinger C., Geissler A., Sickmann A., Meyer H. E., Truscott K. N., Guiard B., Pfanner N., Rehling P. (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120, 817–829 [DOI] [PubMed] [Google Scholar]
- 49. Mokranjac D., Popov-Celeketić D., Hell K., Neupert W. (2005) Role of Tim21 in mitochondrial translocation contact sites. J. Biol. Chem. 280, 23437–23440 [DOI] [PubMed] [Google Scholar]
- 50. Hoppins S., Collins S. R., Cassidy-Stone A., Hummel E., Devay R. M., Lackner L. L., Westermann B., Schuldiner M., Weissman J. S., Nunnari J. (2011) A mitochondrial focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195, 323–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. von der Malsburg K., Müller J. M., Bohnert M., Oeljeklaus S., Kwiatkowska P., Becker T., Loniewska-Lwowska A., Wiese S., Rao S., Milenkovic D., Hutu D. P., Zerbes R. M., Schulze-Specking A., Meyer H. E., Martinou J. C., Rospert S., Rehling P., Meisinger C., Veenhuis M., Warscheid B., van der Klei I. J., Pfanner N., Chacinska A., van der Laan M. (2011) Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell 21, 694–707 [DOI] [PubMed] [Google Scholar]
- 52. Harner M., Körner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F., Neupert W. (2011) The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30, 4356–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de Brito O. M., Scorrano L. (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 [DOI] [PubMed] [Google Scholar]
- 54. Fujimoto M., Hayashi T. (2011) New insights into the role of mitochondria-associated endoplasmic reticulum membrane. Int. Rev. Cell Mol. Biol. 292, 73–117 [DOI] [PubMed] [Google Scholar]
- 55. Chen S., Liu D., Finley R. L., Jr., Greenberg M. L. (2010) Loss of mitochondrial DNA in the yeast cardiolipin synthase crd1 mutant leads to up-regulation of the protein kinase Swe1p that regulates the G2/M transition. J. Biol. Chem. 285, 10397–10407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.