Background: It has been unclear how E1 activities in ubiquitin-like modifications are regulated.
Results: This study identified a role of SUMO modification of the Cys domain of the SUMO E1.
Conclusion: The identified modification is a mechanism for “storing” a pool of E1 that can be quickly activated in response to environmental stress.
Significance: Similar regulation likely exists across the homologous E1s of ubiquitin-like modifications.
Keywords: Enzyme Catalysis, Enzyme Inactivation, Post-translational Modification, SUMO, Sumoylation, Ubiquitin, Mass Spectrometry, Activating Enzyme, Heat Shock
Abstract
Although it is well established that ubiquitin-like modifications are tightly regulated, it has been unclear how their E1 activities are controlled. In this study, we found that the SAE2 subunit of the small ubiquitin-like modifier (SUMO) E1 is autoSUMOylated at residue Lys-236, and SUMOylation was catalyzed by Ubc9 at several additional Lys residues surrounding the catalytic Cys-173 of SAE2. AutoSUMOylation of SAE2 did not affect SUMO adenylation or formation of E1·SUMO thioester, but did significantly inhibit the transfer of SUMO from E1 to E2 and overall SUMO conjugations to target proteins due to the altered interaction between E1 and E2. Upon heat shock, SUMOylation of SAE2 was reduced, which corresponded with an increase in global SUMOylation, suggesting that SUMOylation of the Cys domain of SAE2 is a mechanism for “storing” a pool of E1 that can be quickly activated in response to environmental changes. This study is the first to show how E1 activity is controlled by post-translational modifications, and similar regulation likely exists across the homologous E1s of ubiquitin-like modifications.
Introduction
Conjugation of ubiquitin-like modifiers (Ubl),2 such as small ubiquitin-like modifier (SUMO) and ubiquitin, is catalyzed by enzymes generally known as E1, E2, and E3 (for recent reviews, see Ref. 1). SUMOylation and ubiquitination can be up- or down-regulated during stress response. For example, heat shock induces a global increase in SUMOylation accompanied by a decrease in the free SUMO2 and SUMO3 pools within 10–30 min (2). This rapid change in SUMOylation activities indicates that it arises from mechanisms other than increased expression of the enzymes, but such mechanisms have not been clearly defined. Regulation of E1 during SUMO or other ubiquitin-like modifications is largely unknown.
The SUMO-activating enzyme E1 (SAE) is a heterodimer of SAE1/SAE2 (also known as Aos1 and Uba2). The catalytic component of the SUMO E1, SAE2, is an ∼70-kDa protein with multiple domains (3). The adenylation domain, consisting of residues 1–158 and 384–438, catalyzes SUMO adenylation. The Cys domain (residues 159–386) contains a key binding surface for E2 and the catalytic Cys-173 that forms an E1·SUMO thioester conjugate (4). The ubiquitin folding domain (residues 442–549) (5) also contains a key E2-binding surface. A nuclear localization signal is located at the unstructured C-terminal region of SAE2 (6–8).
In this study, we found that SAE2 can be autoSUMOylated and SUMOylated by catalysis of Ubc9 at a very conserved surface of its Cys domain. SAE2 SUMOylation did not inhibit SUMO adenylation or formation of the E1·SUMO thioester conjugate, nor did it enhance SUMOylation of SUMO-interacting motif (SIM)-containing substrates. However, E1 SUMOylation inhibited its proper interaction with E2 (also known as Ubc9) for transferring SUMO from E1 to E2 and thus inhibited SUMOylation of target proteins. DeSUMOylation of E1 was observed upon heat shock, and this deSUMOylation correlated with a global increase in SUMOylation. These findings suggest that SUMOylation at the E1 Cys domain is an inhibitory mechanism that allows for a quick response to environmental changes by removing such a modification to increase the pool of active SUMO E1.
EXPERIMENTAL PROCEDURES
Plasmids, Mutations, and Protein Expression
His6-SUMO1, His6-SUMO3, His6-Ubc9, GST or GST-Ubc9, RanGAP1, GST-Sp100, His6-SENP1, His6-SENP2, SAE1, His6-SAE2, and His6-SAE2 Δ575 were cloned, expressed in Escherichia coli, and purified as described previously (9). The mammalian expression plasmid for SAE1 was acquired from the Dr. Edward Yeh laboratory through the Addgene service. The mammalian expression plasmid for SAE2-green fluorescent protein (GFP) was obtained from OriGene. Mutations were conducted using the QuikChange Lightning mutagenesis kit (Agilent Technology) according to the manufacturer's protocol.
SENP Digestion of Mammalian Cell Lysates
Cells were lysed with the Passive Lysis Buffer (Promega) with the addition of 20 mm N-ethylmaleimide (NEM) (Fluka) and Complete protease inhibitor mixture (Roche Applied Science) using a volume ratio of cells:buffer of 1:2. The mixture was incubated on ice for 30 min followed by the addition of 25 mm free cysteine (Sigma). Lysate (5–10 μl) was diluted 10-fold before the addition of 1 μl of a mixture of SENP1 and SENP2, at 0.5 μg/μl of each SUMO-specific protease (SENP), to a sample. Samples were then incubated (37 °C, 1 h) followed by reduction of the total volume to 8 μl using a speed vacuum. Then, 7 μl of 2× Laemmli protein sample buffer containing 360 mm DTT was added followed by incubation (95 °C, 10 min) before loading on a 15-well Bis-Tris 4–8% gel (Invitrogen) for separation followed by Western blotting.
SUMOylation of E1
In vitro SUMOylation of E1 was carried out by incubating a reaction mixture (500 μl) containing 3.6 μm E1, 63 μm SUMO-1, and 5 mm ATP in buffer (5 mm MgCl2, 20 mm Hepes, pH 7.5, 50 mm NaCl) with or without 20 μm Ubc9. Reaction time points were 5–40 min to obtain SUMOylated E1 or 40 h to drive SUMOylation of E1 to near completion in the absence of E2 in a warm room (37 °C) under argon to prevent oxidation. Reactions were quenched by the addition of 10 mm DTT. A tube of the same mixture as described above but without SUMO, along with a separate tube of SUMO, was incubated at the same time as an E1 control to account for any activity loss during incubation. After the SUMOylation was quenched, the free SUMO was then added into the E1 control tube, and the same volume of assay buffer was also added into the SUMOylated E1 tube to equalize the protein concentrations. Aliquots (50 μl/tube) of samples were made and flushed with argon for 5 min before storage at −80 °C.
In Vitro SUMOylation Assays and Thioester Formation
In vitro SUMOylation of H6-RanGAP1, GST-Sp100, H6-IR1M, H6-MIR2, and thioester formation of E1 and E2 were carried out by incubating a reaction mixture (5 μl) containing a 10-fold dilution of the SUMOylated E1 or unmodified E1 control prepared above to a final concentration of 0.36 μm, in addition to 3.6 μm Ubc9, 12 μm SUMO1, 5 mm ATP in assay buffer (5 mm MgCl2, 20 mm Hepes, pH 7.5, 50 mm NaCl) without substrates (for thioester formation) or with the addition of 8 μm GST-Sp100, H6-IR1M, H6-MIR2, or H6-RanGap1 (for substrate SUMOylation assays). Reactions were then quenched with 5 μl of 2× Laemmli sample buffer with or without 360 mm DTT.
ATP:AMP Isotopic Exchange
The isotope exchange assay was conducted as described previously (10, 11). Briefly, a 20-μl reaction mixture containing 50 mm Tris, pH 7.5, 10 mm MgCl2, 0.2 mm DTT, 2 mm PPi, 0.5 mm ATP, 0.05 m AMP, 50 μm [14C]AMP (PerkinElmer Life Sciences), and 4 μm SUMO was incubated (37 °C, 10 min) with 0.5 μm SUMOylated or unmodified E1 before the reaction was quenched with 20 μl of 8 m urea solution. 4 μl of the reaction solution was spotted on a polyethyleneimine cellulose plate, and ATP was separated from AMP by thin layer chromatography using a solvent mixture of 0.5 m LiCl and 1 m formic acid. The radiogram was obtained by exposure to a PhosphorImager plate and a Typhoon scanner and quantified using ImageQuant.
Western Blot
Western blot analysis was conducted as described previously (12). Briefly, gels were transferred onto a PVDF-FL membrane that was activated by methanol and preincubated in TOWBIN buffer (containing 192 mm glycine and 25 mm Tris base) at 4 °C for 30 min before the transfer. The membrane was washed twice with PBS containing 0.1% Tween 20 buffer and then incubated with blocking buffer overnight. SAE2, actin, and SUMOylated proteins were detected with rabbit anti-SAE2 antibody (Abcam), mouse anti-actin antibody (Sigma), mouse anti-SUMO1 antibody (Abgent), or rabbit anti-SUMO-2/3 antibody (Abcam) followed by donkey anti-mouse or anti-rabbit secondary antibody and scanned by an Odyssey imager (LI-COR Biosciences). Quantification was conducted using Odyssey software.
Identification of Modified SAE2 by Mass Spectrometry
GST or GST-Ubc9 (50 μm) fusion proteins were coated on beads (GE Healthcare) using the manufacturer's recommended protocol. Cell lysates from five 15-cm plates of HEK293T cells, prepared as described for SENP1/2 digestion, were diluted 10-fold in PBS buffer containing 20 mm NEM and Complete protease inhibitor mixture (Roche Applied Science). Bead-bound GST (1 ml, 0.5 mg/ml) was added to the lysate and incubated (4 °C, overnight) to remove any proteins that bound to GST or the beads. Beads were then stripped with SDS protein loading buffer and loaded on gels (Fig. 1A, GST). The “precleared” lysate was then incubated (4 °C, overnight) with 1 ml (1 mg/ml as GST-Ub9 is two times heavier than GST) of beads bound with GST-Ubc9 fusion protein. The bound proteins were stripped using SDS protein loading buffer and loaded on gels (Fig. 1A, GST-Ubc9). Control and sample were analyzed by in-gel digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS).
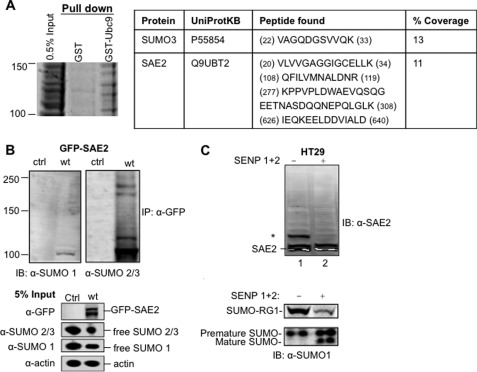
FIGURE 1.
Human SUMO E1 is SUMOylated in cells. A, SAE2 and SUMO3 were identified in a gel section of 100–150-kDa proteins that were specifically pulled down by GST-Ubc9 but not by GST alone. Left, Coomassie Blue-stained SDS-PAGE gel section corresponding to 100–150 kDa. Right, peptide fragments from SAE2 and SUMO3 identified from LC-MS/MS analysis of this section. B, identification of SAE2 SUMOylation in cells transfected with a plasmid expressing the GFP-SAE2 fusion protein. GFP-SAE2 fusion protein overexpressed in HEK293T cells was immunoprecipitated (IP) using beads linked to an anti-GFP antibody and immunoblotted (IB) using anti-SUMO1 (left) or anti-SUMO2/3 (right) antibodies. Ctrl, control. C, detection of SUMOylated endogenous SAE2. Endogenous SUMOylated E1 was detected in whole cell lysates of HT29 cell by Western blot with an anti-SAE2 antibody. Whole cell lysates were treated with a mixture of the SUMO-specific proteases SENP1 and SENP2 (SENP1/2), which significantly reduced the intensity of a higher molecular mass band (asterisk) and increased the intensity of a lower molecular mass band. To ensure that SENP1/2 was active, the same membrane was probed with anti-SUMO1 antibody using a second channel of the Odyssey imager to detect the corresponding reduction in intensities of SUMOylated RanGAP1 (SUMO-RG1) and SUMO1 precursor bands with the appearance of the mature SUMO1 band.
Immunoprecipitation of SUMOylated E1
Anti-GFP antibody (Clontech) was linked with AL20 beads (Applied Biosystems) following the manufacturer's protocol with slight modifications. Anti-GFP antibody (40 μl; 2 mg/ml) in PBS (Clontech) was added to 0.3 mg of AL20 beads. Then, 13 μl of 1 m sodium cyanoborohydride (AL buffer) (Applied Biosystems) was added, the mixture was shaken at room temperature for 5 min, and 23 μl of Na2SO4 was added. Na2SO4 (3 μl) was then added to the mixture 10 times, once every 5 min, after which the mixture was shaken at room temperature overnight. The next day, beads were separated from the solution, which was analyzed by SDS-PAGE to compare the anti-GFP antibody before and after the coating reaction to ensure attachment of the antibody to the beads. The separated beads were submerged in 40 μl of 0.5 m Tris buffer, pH 8.0, and 12 μl of 1 m sodium cyanoborohydride and shaken at 4 °C for 1 h. Beads were then spun down at 12,000 rpm for 10 min and washed five times with 400 μl of 1.5 m NaCl, 50 mm Tris buffer, pH 7.5. Beads were then washed twice with 100 μl of 50 mm Tris buffer, pH 7.5. The finishing product was stored in 100 μl of 50 mm Tris buffer, pH 7.5.
Immunoprecipitation was conducted by incubating cell lysates with the beads at 4 °C overnight followed by five washes with 500 μl of 1.5 m NaCl in 50 mm Tris buffer, pH 7.5. Immunoprecipitation beads were then resuspended in 2× Laemmli protein loading buffer for SDS-PAGE followed by Western blot analysis.
To pull down His6-SUMOylated proteins, 100 μl of Ni2+ beads (Qiagen) was added to 1 ml of cell lysate. Samples were then shaken at room temperature for 1 h and 40 min. The beads were then spun down and washed three times with 2 ml of wash buffer containing 1% SDS and 1 m NaCl in 50 mm Tris buffer, pH 7.6. Bound proteins were eluted by 200 mm imidazole in 2× Laemmli protein loading buffer, separated by SDS-PAGE, and detected by Western blot.
Cell Culture and Transfection
HEK293T, HT-29, and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) (CellGro) supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, 100 units/ml streptomycin, and 0.1 μg/μl puromycin. HeLa cells that stably express His6-tagged SUMO-1 were a generous gift from the Dr. Ronal Hay laboratory (13). For HeLa cells that stably expressed His6-taggged SUMO1, SUMO2, or SUMO3, the three plates of SUMO paralogs were combined before cell splitting and transfection for analysis of SUMOylated proteins using anti-SUMO antibodies.
DNA transfection was performed using Lipofectamine following the manufacturer's protocol (Invitrogen). After 25 h, cells were washed twice with 3 ml of PBS and lysed with 1 ml of radioimmune precipitation assay (RIPA) buffer containing 1% SDS. The lysate was stored at −80 °C.
Mass Spectrometry
SUMOylated SAE2 was excised from the SDS-PAGE gel and subjected to in-gel reduction, alkylation, Glu-C (Roche Applied Science), trypsin (Promega), or a mixture of Glu-C and trypsin. Asp-N (Roche Applied Science) and pepsin (Sigma-Aldrich) were also used, but did not generate many peptide spectra. LC-MS/MS data were acquired using an Eksigent nanoLC-2D equipped with a self-packed C18 column connected to a hybrid linear ion trap (LTQ-FT) mass spectrometer (Thermo Electron). MS/MS spectra were matched to a database of generated FASTA files created by ChopNSpice using the Global Proteome Machine Organization (GPM) X!Tandem database search engine. All identified SUMOylated MS spectra were reconfirmed manually. In the figures, the b and y ion series members are numbered from the N terminus. Capital letters are used for SUMO ion series, and lowercase letters are used for the substrate peptide ion series.
RESULTS
Identification of SAE2 SUMOylation in Cells
In a proteomic study to identify proteins that interact with Ubc9 noncovalently, we carried out a pulldown of whole cell extracts of 293T cells using beads bound with GST-Ubc9. Cell lysate was first precleared with beads bound with GST. The beads were then stripped with SDS protein loading buffer and loaded on gels (Fig. 1A, GST). The precleared lysate was then incubated with beads bound with GST-Ubc9 fusion protein, and bound proteins were stripped with SDS protein loading buffer and loaded on gels (Fig. 1A, GST-Ubc9). Both lanes (GST and GST-Ubc9) of gels were cut into sections, and all sections were analyzed by in-gel digestion and LC-MS/MS. The analysis detected SAE2 within a gel section corresponding to a molecular mass of 100–150 kDa (Fig. 1A), but not in the sample pulled down with GST only. Although identification of SAE2 was not a surprise because it binds tightly to Ubc9 (14, 15), its appearance within the 100–150 kDa range was not consistent with its molecular mass (∼70 kDa). This finding indicates that SAE2 is post-translationally modified, adding 30–80 kDa to the molecular mass. In addition, SUMO3 was also found in the same section of the gel, suggesting SUMOylation of SAE2 (Fig. 1A). To confirm that SAE2 is indeed SUMOylated in vivo, a plasmid that expressed a GFP-SAE2 fusion protein was transfected into 293T cells. The expressed protein was immunoprecipitated with an anti-GFP antibody and then immunoblotted with an anti-SUMO2/3 antibody. Multiple bands were detected by either anti-SUMO1 or anti-SUMO2/3 antibodies in the immunoprecipitated sample, but not in the control (Fig. 1B, upper panel). The GFP-SAE2 fusion protein was detected in the directly loaded sample of transfected cells, but not in the control (Fig. 1B, lower panel). In addition, levels of free SUMO1 and SUMO2/3 proteins were reduced in GFP-SAE2-expressing cells, indicating increased SUMOylation (Fig. 1B, lower panel).
To examine SUMOylation of endogenous E1 in other cell lines, we analyzed endogenous SAE2 in HT29 whole cell lysates. Cells were lysed in high concentrations of a SENP inhibitor, NEM, to inhibit SENP cleavage of SUMO from substrates. Free cysteine was then added to react with accessible NEM followed by treatment with a mixture of SENP1 and SENP2 (SENP1/2) to selectively cleave the attached SUMO from modified proteins (Fig. 1C). Anti-SAE2 antibody revealed bands higher than the molecular mass of SAE2, suggesting modifications of SAE2. Treatment of cell lysates with the SENP1/2 mixture resulted in the disappearance or a reduction in the intensity of the higher molecular mass bands and enhancement of the intensity of the lower molecular mass bands (Fig. 1C). The activity of the added SENP1/2 was confirmed by cleavage of full-length SUMO1 into the mature SUMO1, as well as a reduction in the amount of SUMOylated RanGAP1 after treatment with SENP1/2 (Fig. 1C, bottom). These results indicate that SUMOylation of SAE2 is not restricted to a particular cell line.
Identification of SUMOylation Sites on E1
As reported previously, SUMOylated SAE2 was observed by incubation of E1 with SUMO, ATP, and Mg2+ (16) (supplemental Fig. S1A). In addition, Ubc9 promoted SUMO modification of E1 in the catalytic Cys-93-dependent manner of Ubc9, suggesting that Ubc9-catalyzed E1 SUMOylation is due to interaction with the Ubc9·SUMO thioester, as found previously (16) (supplemental Fig. S1A). This mechanism is likely to be relevant in cells because E1 and E2 are associated in such a manner that their catalytic Cys residues are close enough to form disulfide bonds upon H2O2 treatment of cells (17). Furthermore, the different SUMO paralogues do not have intrinsic differences in modifying the E1 (supplemental Fig. S1B).
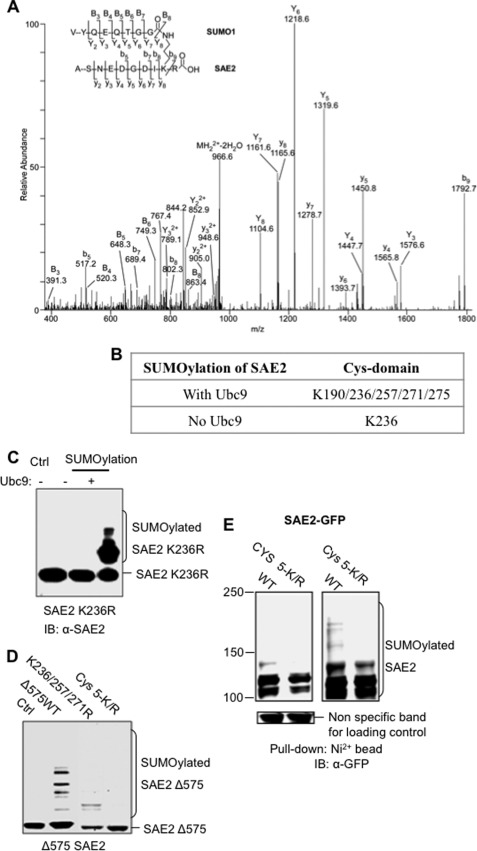
Using the in vitro SUMOylated E1, we identified SAE2 SUMOylation sites using LC-MS/MS. Identification of SAE2 SUMOylation sites was not straightforward, and none of the Lys residues form canonical SUMOylation motifs (for reviews, see Refs. 18 and 19). The standard trypsin digestion approach proved difficult because SUMOylation on a given Lys prevented cleavage at that Lys, resulting in large fragments with an attached peptide from SUMO that were too big to be readily identified. Searching for SUMOylation sites in MS/MS data using conventional approaches was also not feasible because SUMOylation forms T-shaped peptides (Fig. 2A), for which MS/MS fragmentation patterns were not recognized by standard data search engines. To overcome these difficulties, we optimized the sequence coverage by combining digested fragments from treatments with four different enzymes: trypsin, Glu-C, Asp-N, and pepsin, either separately or combined (see “Experimental Procedures”). For the SUMOylated peptide search, we used the recently published ChopNSpice program to generate a custom database for single enzyme digestion, and we manually mimicked ChopNSpice to create a database for double enzyme digestion (20). Using these approaches, we identified multiple SUMOylation sites on the SAE2 Cys domain from multiple bands, as well as from the single main SUMOylated SAE2 band (Fig. 2, A and B).
FIGURE 2.
SUMOylation sites in SAE2 are clustered at the Cys domain. A, a representative SAE2 ion mass spectrum (MS/MS spectrum) showing the identification of the SAE2 Lys-236 SUMOylation site. B, summary of the identified SUMOylation sites when E1 is SUMOylated in the presence and absence of Ubc9. K190/236/257/271/275R, K190R/K236R/K257R/K271R/K275R. C, investigation of Lys-236 SUMOylation. Mutation of Lys-236 to Arg removed SUMOylation in the absence of Ubc9 in vitro. The addition of Ubc9 resulted in multiple SUMOylated bands, corresponding to SAE2 SUMOylation. Ctrl, control; IB, immunoblot. D, validation of the SUMOylation sites in vitro. E1 enzymes in which the SAE2 C terminus was deleted starting from residue 575 (Δ575) and with or without mutations at the SUMOylation sites were catalyzed by Ubc9. Mutations of Lys-236, Lys-257, and Lys-271 to Arg reduced SAE2-Δ575 SUMOylation greatly (K236R/K257R/K271R (designated as K236/257/271R)), and mutation of all SUMOylation sites from Lys to Arg (K190R/K236R/K257R/K271R/K275R (designated as Cys 5-K/R)) eliminated all SUMOylation bands. E, validation of the SUMOylation sites in cells. Plasmids expressing wild-type GFP-SAE2 fusion protein and the Cys 5-K/R were transfected into HeLa cells that stably expressed His6-tagged SUMO1, SUMO2, and SUMO3. Cells were lysed in 1% SDS in radioimmune precipitation buffer, and SUMOylated proteins were pulled down by nitrilotriacetic acid beads. Western blotting using anti-GFP antibodies detected the SUMOylated SAE2 proteins, shown by low intensity (left) and high intensity (right) scans using a LI-COR Odyssey scanner. SUMOylated SAE2 was greatly decreased in SAE2 Cys 5-K/R as compared with WT SAE2. The SUMOylated SAE2 shown in SAE2 Cys 5-K/R likely came from the SAE2 C-terminal region.
Lys-236 was modified with and without Ubc9 catalysis, and four additional SUMOylation sites were found, Lys-190, Lys-257, Lys-271, and Lys-275, upon catalysis by Ubc9 (Fig. 2B). SUMOylation of Lys-236 was confirmed by mutating Lys-236 to an Arg (K236R) in the full-length SAE2. The SAE2 K236R mutant was not SUMOylated in the absence of Ubc9 (Fig. 2C), confirming that Lys-236 is the autoSUMOylation site. In the presence of Ubc9, the SAE2 K236R mutant was SUMOylated at multiple sites (Fig. 2C), suggesting that Ubc9-dependent SUMOylation is independent of Lys-236 SUMOylation.
The SUMOylation sites catalyzed by Ubc9 were confirmed by site-directed mutagenesis in vitro and in vivo. Previous studies have shown that deletion of the unstructured region of SAE2 from residue 575 to the C terminus does not alter the enzymatic activity of SAE2 (3). Therefore, we mutated SUMOylation sites in the truncated SAE2 in in vitro SUMOylation assays. The truncated wild-type (Δ575) and mutant proteins were co-expressed with wild-type SAE1 in E. coli, and the expressed proteins were tested in Ubc9-catalyzed SUMOylation. Although multiple SUMOylation bands were observed for the wild-type Δ575 SAE2, mutation of three SUMOylation sites, Lys-236, Lys-257, and Lys-271, to Arg reduced the SUMOylation of this construct (Fig. 2D). Mutation of all SUMOylation sites in the Cys domain eliminated all SUMOylation bands, confirming that all five Lys residues, Lys-190, Lys-236, Lys-257, Lys-271, and Lys-275, are SUMOylation sites (Fig. 2D). These SUMOylation sites were further validated in cells. Full-length SAE2 instead of the Δ575 mutant was used in the cellular experiments because the C-terminal segment harbors the nuclear localization signal, and its deletion caused mislocalization of the protein (6). HeLa cells that stably expressed His6-tagged SUMO1, SUMO2, and SUMO3 were transfected with GFP fusion SAE2 plasmids with or without mutation of SAE2 Lys-190/Lys-236/Lys-251/Lys-271/Lys-275 to Arg. The SUMOylated proteins were pulled down by nitrilotriacetic acid beads under denaturing conditions. Immunoblotting with an anti-GFP antibody was used to detect SUMOylated SAE2 (Fig. 2E). As expected, the GFP-SAE2 Cys domain mutant had lower SUMOylation levels than did the wild-type GFP-SAE2 (Fig. 2E). The observed SUMOylation bands for the GFP-SAE2 Cys domain mutant are likely due to SUMOylation at the C terminus of SAE2 (data not shown).
Inhibition of SUMOylation by Inhibiting Ubc9 Binding
Although the identified SUMOylation sites are not close to each other in the primary sequence, they are clustered at a binding surface for recruiting E2 for the transfer of SUMO from E1 to E2 (Fig. 3A) (4), suggesting that the modifications likely alter the enzymatic activity of E1. To investigate how SUMOylation of the E1 Cys domain affects the enzymatic activity of E1, we formed SUMO-modified E1 in vitro. SUMO-modified E1 enzyme was made in the absence of Ubc9 to avoid the complication of simultaneous Ubc9 SUMOylation. Unmodified E1 control was incubated under the same conditions at the same time to eliminate other effects, such as potential oxidation during incubation, although the samples were under the inert gas argon. Direct comparison of the two E1 samples allowed the assessment of the effect of SUMO modification on the enzymatic activities of E1. Because E1 loses activity during lengthy purification steps, SUMOylated E1 was not purified to avoid loss of activity due to factors unrelated to SUMOylation.
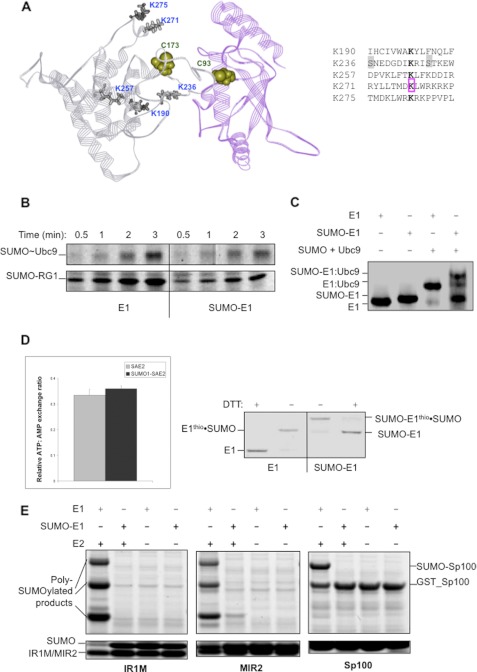
FIGURE 3.
SAE2 SUMOylated sites are located in the Ubc9-binding surface, and SUMOylation inhibits binding to E2. A, SUMOylated lysines at the Cys domain are at the interface for binding Ubc9 and surround the SAE2 catalytic Cys-173 (4). Lys-236 is the closest to Cys-173 of SAE2, and it was found to be SUMOylated in both the presence and the absence of Ubc9, whereas Lys-190, Lys-257, Lys-271, and Lys-275 were SUMOylated in the presence of E2 (Fig. 1B). Sequences surrounding all five SUMOylation sites (bolded K) of SAE2 are shown to the right. Lys-236, which is the autoSUMOylation site, is flanked by consensus phosphorylation sites (highlighted in gray). Lys-271 is an acetylation site (pink box). B, time-dependent formation of Ubc9·SUMO thioester (upper panel) and SUMOylated RanGAP1 (SUMO-RG1, bottom panel) showing that the unmodified E1 (left) is more efficient than the SUMOylated E1 (SUMO-E1) (right). C, native gel image showing that SUMOylated E1 (SUMO-E1) does not bind E2 (Ubc9), like the WT E1. SUMO-modified E1 (SUMO-E1) was obtained in the absence of Ubc9, and it ran slightly differently from the WT E1 control in the absence of Ubc9. The addition of a mixture of SUMO and Ubc9 shifted the band of the WT E1 completely, indicating the formation of a tight complex. However, a majority of the SUMOylated E1 did not form a tight complex upon the addition of the same mixture of SUMO and Ubc9, indicated by the high intensity of free SUMOylated E1 band. The small amount of free WT E1 in the SUMOylated E1 sample due to incomplete SUMOylation formed a tight complex. In addition, a small amount of SUMOylated E1 also formed tight complex that may or may not be the correct interaction mode for the transfer of SUMO from E1 to E2. D, SUMO modifications of E1 did not inhibit SUMO adenylation or formation of the E1·SUMO thioester conjugate. Left panel: AMP:ATP exchange assay showing that unmodified E1 and SUMOylated E1, preformed in the absence of Ubc9, have similar exchange rates. Right panel: unmodified wild-type E1 (left) and SUMOylated E1 (right) can both form thioester conjugates with SUMO (middle two lanes, E1thio·SUMO). Error bars indicate S.D. E, SUMOylated E1 inhibited SUMOylation of SIM-containing substrates and did not bypass E2 in SUMOylation of these substrates. The SIM-containing substrates IR1M, MIR2, and Sp100 had reduced SUMOylation rates when catalyzed by the SUMOylated E1 in comparison with the unmodified E1, in a similar manner as RanGAP1 that does not contain a SIM (B). Both IR1M and MIR2 were poly-SUMOylated, as found previously (33). In addition, SUMOylated E1 did not SUMOylate SIM-containing substrates in the absence of Ubc9.
The obtained SUMOylated E1 contained a single modification at Lys-236, as discussed above. This modified E1 significantly reduced the formation of E2·SUMO thioester as well as SUMO-modified RanGAP1 (Fig. 3B). The formation of the small amount of E2·SUMO thioester and SUMOylated RanGAP1 could have resulted from a small amount of unmodified SAE2 left over from the reaction. To understand how E1 activity was inhibited by SUMOylation, we evaluated the E1-catalyzed SUMO adenylation and thioester formation steps quantitatively using AMP:ATP exchange assays, which depend on both SUMO adenylation and the formation of E1·SUMO thioester conjugate (10, 11). AutoSUMOylated E1 versus unmodified E1 showed similar AMP:ATP exchange rates (Fig. 3D, left panel). In addition, autoSUMOylated E1 was able to form the thioester conjugate to SUMO with similar activity as the unmodified E1 (Fig. 3D, right panel). These data suggest that autoSUMOylation of E1 did not affect the first two steps of SUMO conjugation, and therefore, the inhibition of Ubc9·SUMO thioester formation could only be due to inhibition of the transfer of SUMO from E1 to E2. This is consistent with the SUMOylation sites in the SAE2 Cys domain being at or near the surface of SAE2 that forms a key interaction with Ubc9 for the transfer of SUMO from E1 to E2 (Fig. 3A). Residues Ile-235 and Ile-238 of SAE2 flank the autoSUMOylation site Lys-236, and it has been shown that mutation of these residues to Ala impairs the interaction of E1 with Ubc9 and the transfer of SUMO from E1 to Ubc9 (4). Thus, the results suggest that the bulky SUMO moiety on Lys-236 interferes with the proper interaction with Ubc9 by steric hindrance. Indeed, autoSUMOylated E1 has an altered interaction with Ubc9 and SUMO (Fig. 3C). Also, all SUMO paralogues bind to Ubc9 noncovalently on a surface distal from the catalytic Cys (15, 21), and thus, the SUMO moiety of the modified E1 could also bind to E2 in a manner that prevents the transfer of SUMO from E1 to E2. SUMO modifications on the other sites, Lys-190, Lys-257, Lys-271, and Lys-275, that surround Lys-236 are likely to cause similar inhibitory effects on binding E2 because they are located on or close to the binding surface for E2 (Fig. 3A).
We also investigated whether autoSUMOylated E1 can affect SUMOylation of target proteins that contain SIMs (22). Conversion of thioester bonds to peptide bonds is not unique to ubiquitin-like modifications, but is used by other cellular peptide synthesis processes, such as intein-mediated protein ligation (23) and nonribosomal peptide synthesis (24). Ubiquitin-like modifications are unique in that an additional transthioesterification step is required: transfer of SUMO from the E1·Ubl thioester to the E2·Ubl thioester, which is responsible for substrate recruitment (25). We tested whether autoSUMOylated E1 can bypass E2 to recruit SIM-containing substrates for their SUMOylation. Three SIM-containing substrates were tested, a segment of Sp100, the IR1-M domain, and the M-IR2 domain of RanBP2. All three substrates were SUMOylated efficiently, and the RanBP2 domains also formed poly-SUMO chains using unmodified E1 and E2 (Fig. 3E, lane 1 of the gels on the left and in the middle). However, these modifications did not occur with autoSUMOylated E1 alone and were severely inhibited with autoSUMOylated E1 and wild-type E2 (Fig. 3E). Thus, autoSUMOylated E1 did not enhance SUMOylation of SIM-containing substrates.
Physiological Significance of E1 SUMOylation in Heat Shock Response
To identify the physiological relevance of the finding that SUMOylation of E1 is an inhibitory mechanism, we investigated whether E1 SUMOylation is affected by heat shock, which is known to enhance global SUMOylation of SUMO 2/3 within 10–30 min but not global SUMO1 modifications likely due to the lack of free SUMO1 (2). The increase in substrate modification corresponded to a reduction in free SUMO2 and SUMO3 levels, suggesting that the increase in global SUMOylation is, at least in part, due to an increase in the enzymatic activity of the SUMOylation machinery. However, the mechanism for this regulation is not well understood. We analyzed endogenous SAE2 in HT29 cells in response to heat shock. The cells were subjected to heat shock for 0–30 min at 42 °C followed by recovery for 3 h at 37 °C. Using an antibody against SAE2, Western blot analysis showed that the overall SAE2 level did not appear to change significantly upon heat shock (see Fig. 5). However, the intensity of the band corresponding to the SUMO-modified endogenous SAE2, which was reduced by treatment with SENP1/2 (Fig. 1C and supplemental Fig. S2), was reduced after heat shock for 30 min and then reappeared after recovery from heat shock (Fig. 4A). This finding is consistent with a recent proteomic study of plant cells showing that SUMOylation of the plant homologue of SAE2 is reduced by 50% by heat shock, although this study did not investigate the effect of SAE2 SUMOylation (26). Also, a 50% reduction of SAE2 SUMOylation was reported in the supplemental material for a study of HeLa cell response to heat shock (27). The disappearance and reappearance of SUMO-modified SAE2 were consistent with a corresponding increase and decrease of global SUMO2/3 modifications as well as concomitant changes in free SUMO2 and SUMO3 levels (Fig. 4B). Taken together, these results indicate that deSUMOylation of the SUMO-modified endogenous SAE2 resulted in increased amounts of catalytically active SAE2 that correlated with up-regulation of global SUMO2 and SUMO3 modifications in response to heat shock in cells.
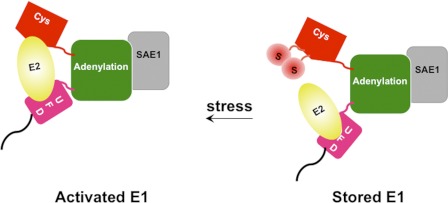
FIGURE 5.
Model for regulation of SUMO E1 enzymatic activity by SUMOylation at the Cys domain. The SAE1 subunit is indicated in light gray, whereas each domain in SAE2 is represented in a different color. Bulky SUMO moieties (circles labeled with S) on Lys-236 and other sites on SAE2 interfere with the proper interaction with Ubc9 by steric hindrance. Also, the SUMO moieties on modified E1 could bind to E2 in a manner that prevents the transfer of SUMO from E1 to E2. DeSUMOylation of E1, upon environmental stress such as heat shock, allows proper interaction of E1-E2 to activate SUMOylation. UFD, ubiquitin fold domain.
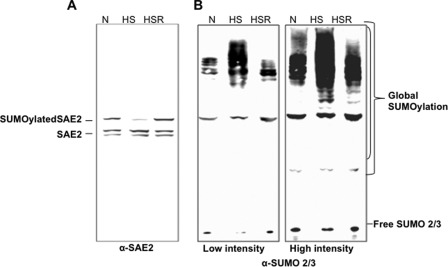
FIGURE 4.
Heat shock induced deSUMOylation of SAE2 that corresponded to up-regulation of global SUMOylation of SUMO2/3. A, HT29 cells without heat shock (normal condition as a control, N), with heat shock for 30 min at 42 °C (HS), or with heat shock followed by recovery at 37 °C for 3 h (HSR) were lysed in the presence of high concentrations of protease inhibitors and NEM (see “Experimental Procedures”). Western blot analysis was conducted using anti-SAE2, anti-actin, and anti-SUMO1 and anti-SUMO2/3 antibodies. The band corresponding to SUMO-modified SAE2 (Fig. 1C) was not present in cells that received heat shock but were not allowed to recover, and was present in cells that recovered after heat shock. B, the disappearance and reappearance of the SUMOylated SAE2 band correlated with the up- and down-regulation of global SUMOylation of SUMO2/3. Both low and high intensity scans of the same membrane are shown.
DISCUSSION
Post-translational Mechanism for Regulating SUMOylation Activity
In this study, we have conducted the first investigation of the effects of an enzymatically catalyzed post-translational modification of SUMO E1 that alters in response to environmental changes (Fig. 5). SUMOylation of E1 inhibits its interaction with E2 for the transfer of SUMO from E1 to E2 and thereby inhibits the SUMOylation reaction. We have shown that E1 is SUMOylated in cells under normal conditions and deSUMOylated upon exposure to heat shock, which in turn correlates with a dramatic increase in global SUMOylation activities. Such a quick and reversible response is consistent with enzymatically catalyzed activation of a pool of inactive stored SUMOylated E1 by deSUMOylation, providing a mechanism for a rapid regulation of the conjugation machinery. It was proposed previously that phosphorylation may play a role in up-regulating substrate SUMOylation during heat shock because heat shock activates kinases that up-regulate phosphorylation of specific target proteins that may account for the enhanced SUMOylation (28, 29). However, there are substrates, such as c-Myb, that have an increased SUMOylation level upon exposure to heat shock, and yet their SUMOylation is independent of phosphorylation state (30). As shown here, heat shock induces deSUMOylation of the pool of SUMOylated E1, leading to its activation and providing a mechanism for how phosphorylation-independent SUMOylation of target proteins can be up-regulated in heat shock response. DeSUMOylation of SAE2 may not be the only mechanism that leads to enhanced SUMOylation during heat shock response. However, the data presented here suggest that SUMO modification of SAE2 Cys domain serves as an on/off switch for E1 activity (Fig. 5).
The Modification Sites Suggest a Conserved Regulatory Mechanism
The identified SUMOylation sites suggest that analogous modifications occur in E1s of other ubiquitin-like modifications. The SUMOylation site Lys-236 is located on an extended flexible loop in the vicinity of the catalytic Cys-173 of SAE2 (Fig. 3A). When Cys-173 is modified with thioester-bonded SUMO, this Lys side chain would be capable of moving to the thioester bond and carrying out nucleophilic attack to form an isopeptide bond with SUMO. The Lys-190, Lys-257, Lys-271, and Lys-275 sites were found to be SUMOylated upon catalysis by Ubc9 (Figs. 2B and 3A), likely due to binding of the Ubc9·SUMO thioester to the Cys domain of SAE2, as shown previously (4, 17). The scattered SUMOylation sites at the Ubc9-binding surface indicate that the interaction does not occur in a single manner. In particular, NMR studies suggest that the interaction between the Cys domain and Ubc9 is dynamic, consistent with multiple Lys modifications catalyzed by Ubc9 (4). Analogous interaction also exists between the ubiquitin E1 and E2 (31). None of these SUMOylation sites consist of the commonly found consensus motif, ΨKX(E/D), or its variants, such as the inverted motif, phosphorylation-dependent SUMOylation, or negatively charged amino acid-dependent SUMO motif (18, 19). These commonly found SUMOylation consensus motifs are Ubc9-binding motifs that bind to the Ubc9·SUMO thioester and lead to SUMO modifications. However, the binding of SAE2 to Ubc9 does not require these consensus motifs (4, 10) because SAE2 contains two Ubc9-binding sites that recruit Ubc9 cooperatively (10). Thus, the SUMOylation sites in the SAE2 Cys domain do not contain the consensus SUMOylation sequence (Fig. 3A, right panel). Because E1-E2 interactions are conserved among ubiquitin-like modifications, analogous modifications may occur in other E1s that catalyze ubiquitination or other ubiquitin-like modifications to regulate their activities.
“Hotspot” for Post-translational Modifications
Sequence comparison of SAE2 from various species shows that SUMOylation occurs at highly a conserved region of the enzyme. One of the lysines that is SUMOylated in the presence of Ubc9, Lys-271, was reported as an acetylation site (Fig. 3A, right, pink box) (32), suggesting that this SUMOylation site is subject to regulation by cross-talk between SUMOylation and acetylation. The Lys-236 site that is autoSUMOylated is flanked by two consensus casein kinase II phosphorylation sites, (S/T)XX(E/D), suggesting that its SUMOylation may be regulated by phosphorylation (Fig. 3A, right, gray highlights). Taken together, the evidence suggests that the SUMOylation sites occupy a highly conserved surface of SAE2 that is regulated by various post-translational modifications. Given the critical functions of the SUMO E1 in regulating global SUMOylation, it is conceivable that its activity is tightly regulated by the cross-talk of these different post-translational modifications.
In summary, our study has shown how an enzymatically catalyzed post-translational modification on E1 can rapidly change, allowing quick response to environmental changes by regulating protein-protein interaction networks, protein trafficking, and degradation mediated by SUMOylation. Future studies are needed to understand how SAE2 is deSUMOylated and potentially modified by other post-translational modifications. Similar regulation may occur of other E1s of ubiquitin-like modifications.
Supplementary Material
Acknowledgments
We thank the City of Hope, Beckman Research Institute Mass Spectrometry Core. We are also grateful to Drs. Ronald Hay and Michael Tatham for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM086171 and R01 GM074748 (to Y. C.) through the NIGMS and National Institutes of Health Grant P30 CA033572 from the NCI.

This article contains supplemental Figs. S1 and S2.
- Ubl
- ubiquitin-like modifier(s)
- SUMO
- small ubiquitin-like modifier
- SAE
- SUMO-activating enzyme E1
- SIM
- SUMO-interacting motif
- NEM
- N-ethylmaleimide
- SENP
- SUMO-specific protease
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Ulrich H. D. (2009) The SUMO system: an overview. Methods Mol. Biol. 497, 3–16 [DOI] [PubMed] [Google Scholar]
- 2. Saitoh H., Hinchey J. (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275, 6252–6258 [DOI] [PubMed] [Google Scholar]
- 3. Lois L. M., Lima C. D. (2005) Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang J., Hu W., Cai S., Lee B., Song J., Chen Y. (2007) The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications. Mol. Cell 27, 228–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walden H., Podgorski M. S., Huang D. T., Miller D. W., Howard R. J., Minor D. L., Jr., Holton J. M., Schulman B. A. (2003) The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell 12, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 6. Dohmen R. J., Stappen R., McGrath J. P., Forrová H., Kolarov J., Goffeau A., Varshavsky A. (1995) An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 270, 18099–18109 [DOI] [PubMed] [Google Scholar]
- 7. Johnson E. S., Schwienhorst I., Dohmen R. J., Blobel G. (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Desterro J. M., Rodriguez M. S., Kemp G. D., Hay R. T. (1999) Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274, 10618–10624 [DOI] [PubMed] [Google Scholar]
- 9. Wang J., Lee B., Cai S., Fukui L., Hu W., Chen Y. (2009) Conformational transition associated with E1-E2 interaction in small ubiquitin-like modifications. J. Biol. Chem. 284, 20340–20348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J., Cai S., Chen Y. (2010) Mechanism of E1-E2 interaction for the inhibition of Ubl adenylation. J. Biol. Chem. 285, 33457–33462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haas A. L., Rose I. A. (1982) The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 257, 10329–10337 [PubMed] [Google Scholar]
- 12. Truong K., Su Y., Song J., Chen Y. (2011) Entropy-driven mechanism of an E3 ligase. Biochemistry 50, 5757–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rodriguez M. S., Desterro J. M., Lain S., Midgley C. A., Lane D. P., Hay R. T. (1999) SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18, 6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bencsath K. P., Podgorski M. S., Pagala V. R., Slaughter C. A., Schulman B. A. (2002) Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 277, 47938–47945 [DOI] [PubMed] [Google Scholar]
- 15. Tatham M. H., Kim S., Yu B., Jaffray E., Song J., Zheng J., Rodriguez M. S., Hay R. T., Chen Y. (2003) Role of an N-terminal site of Ubc9 in SUMO-1, -2, and -3 binding and conjugation. Biochemistry 42, 9959–9969 [DOI] [PubMed] [Google Scholar]
- 16. Subramaniam S., Mealer R. G., Sixt K. M., Barrow R. K., Usiello A., Snyder S. H. (2010) Rhes, a physiologic regulator of sumoylation, enhances cross-sumoylation between the basic sumoylation enzymes E1 and Ubc9. J. Biol. Chem. 285, 20428–20432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bossis G., Melchior F. (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell 21, 349–357 [DOI] [PubMed] [Google Scholar]
- 18. Matic I., Schimmel J., Hendriks I. A., van Santen M. A., van de Rijke F., van Dam H., Gnad F., Mann M., Vertegaal A. C. (2010) Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39, 641–652 [DOI] [PubMed] [Google Scholar]
- 19. Gareau J. R., Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsiao H. H., Meulmeester E., Frank B. T., Melchior F., Urlaub H. (2009) “ChopNSpice,” a mass spectrometric approach that allows identification of endogenous small ubiquitin-like modifier-conjugated peptides. Mol. Cell Proteomics 8, 2664–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Q., Jin C., Liao X., Shen Z., Chen D. J., Chen Y. (1999) The binding interface between an E2 (UBC9) and a ubiquitin homologue (UBL1). J. Biol. Chem. 274, 16979–16987 [DOI] [PubMed] [Google Scholar]
- 22. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Evans T. C., Jr., Xu M. Q. (1999) Intein-mediated protein ligation: harnessing nature's escape artists. Biopolymers 51, 333–342 [DOI] [PubMed] [Google Scholar]
- 24. Marahiel M. A., Stachelhaus T., Mootz H. D. (1997) Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97, 2651–2674 [DOI] [PubMed] [Google Scholar]
- 25. Lin D., Tatham M. H., Yu B., Kim S., Hay R. T., Chen Y. (2002) Identification of a substrate recognition site on Ubc9. J. Biol. Chem. 277, 21740–21748 [DOI] [PubMed] [Google Scholar]
- 26. Miller M. J., Vierstra R. D. (2011) Mass spectrometric identification of SUMO substrates provides insights into heat stress-induced SUMOylation in plants. Plant Signal. Behav. 6, 130–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Golebiowski F., Matic I., Tatham M. H., Cole C., Yin Y., Nakamura A., Cox J., Barton G. J., Mann M., Hay R. T. (2009) System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2, ra24. [DOI] [PubMed] [Google Scholar]
- 28. Hong Y., Rogers R., Matunis M. J., Mayhew C. N., Goodson M. L., Park-Sarge O. K., Sarge K. D. (2001) Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276, 40263–40267 [DOI] [PubMed] [Google Scholar]
- 29. Hietakangas V., Ahlskog J. K., Jakobsson A. M., Hellesuo M., Sahlberg N. M., Holmberg C. I., Mikhailov A., Palvimo J. J., Pirkkala L., Sistonen L. (2003) Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 23, 2953–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sramko M., Markus J., Kabát J., Wolff L., Bies J. (2006) Stress-induced inactivation of the c-Myb transcription factor through conjugation of SUMO-2/3 proteins. J. Biol. Chem. 281, 40065–40075 [DOI] [PubMed] [Google Scholar]
- 31. Huang D. T., Zhuang M., Ayrault O., Schulman B. A. (2008) Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nat. Struct. Mol. Biol. 15, 280–287 [DOI] [PubMed] [Google Scholar]
- 32. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 33. Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.