Abstract
Real-time PCR in nuclear ribosomal DNA (nrDNA) is becoming a well-established tool for the quantification of arbuscular mycorrhizal (AM) fungi, but this genomic region does not allow the specific amplification of closely related genotypes. The large subunit of mitochondrial DNA (mtDNA) has a higher-resolution power, but mtDNA-based quantification has not been previously explored in AM fungi. We applied real-time PCR assays targeting the large subunit of mtDNA to monitor the DNA dynamics of two isolates of Glomus intraradices sensu lato coexisting in the roots of medic (Medicago sativa). The mtDNA-based quantification was compared to quantification in nrDNA. The ratio of copy numbers determined by the nrDNA- and mtDNA-based assays consistently differed between the two isolates. Within an isolate, copy numbers of the nuclear and the mitochondrial genes were closely correlated. The two quantification approaches revealed similar trends in the dynamics of both isolates, depending on whether they were inoculated alone or together. After 12 weeks of cultivation, competition between the two isolates was observed as a decrease in the mtDNA copy numbers of one of them. The coexistence of two closely related isolates, which cannot be discriminated by nrDNA-based assays, was thus identified as a factor influencing the dynamics of AM fungal DNA in roots. Taken together, the results of this study show that real-time PCR assays targeted to the large subunit of mtDNA may become useful tools for the study of coexisting AM fungi.
INTRODUCTION
Communities of arbuscular mycorrhizal (AM) fungi are an integral component of the soil-plant system, colonizing the roots of the majority of land plants, facilitating plant nutrient uptake, and constituting an important channel of carbon movement from plants to soil (35). The PCR amplification of AM fungal DNA with taxon-specific primers enables the targeted detection of AM fungal taxa within a community and detailed studies of their coexistence involving the screening of large sample sets (21, 37, 47). This approach is promising, particularly if applied in conjunction with quantitative real-time PCR (qPCR) (17), which allows for the determination of the abundance of the fungal taxa in samples. Studies using qPCR with taxon-specific primers demonstrated that qPCR is a more sensitive tool to monitor community changes than commonly used nested endpoint PCR (3, 19, 29), and it corroborated the callings for the quantitative evaluation of AM fungal communities (11, 32).
The qPCR assays that have been developed so far for the quantification of AM fungal DNA in roots or in soil rely, with one exception (12), on nuclear markers in the relatively well-characterized nuclear ribosomal DNA (nrDNA). This region, however, lacks the resolution power to discriminate among closely related species, such as Glomus claroideum and G. etunicatum (8, 44), or even isolates of the same species. It is known that high developmental and functional diversity exists at these lower taxonomic levels in AM fungi (20, 28), but interactions among the closely related genotypes remain unexplored due to the lack of suitable tools for the monitoring of their spread in mixed inoculations. The tools with higher-resolution power to lower taxonomic levels could monitor the establishment of isolates inoculated into nonsterile soil and the impact of the introduction of nonnative AM fungi on the resident community. The search for these tools is encouraged by the inoculum production industry for quality control and the verification of the success of inoculation (13).
Higher-resolution power to species and even to intraspecific level has been demonstrated for the large subunit of mitochondrial ribosomal DNA (mtLSU) of AM fungi. The first sequence analysis of this region in the AM fungal species Glomus intraradices indicated that mitochondrial ribosomal genes lack the intraindividual sequence variability (30) encountered in nuclear ribosomal genes (26, 33). This was also confirmed for the mtLSU of other AM fungal species (41) and other mitochondrial genes (24). Within the AM fungal clade Glomus intraradices sensu lato (6), haplotypes of the mtLSU can be distinguished by the presence, size, and sequence of three introns, as well as by their exon sequence (5). Although a similar degree of intraspecific variability was not recorded in the mtLSU of G. mosseae and related species (41), the variability of the mtLSU region within this clade allows the development of primers to discriminate closely related species.
The mtLSU has been successfully used for the detection of a field-inoculated isolate by endpoint PCR (38), but quantification based on AM fungal mitochondrial DNA (mtDNA) remained unexplored. Gamper et al. (11) suggested that the quantification of mitochondrial genes could estimate the general physiological activity of AM fungi. They assumed that the numbers of mitochondria in the fungal structures better correspond with the fungal metabolic activity than those of nuclei. Mitochondria are typically observed as small thread-like organelles that are evenly distributed throughout the cell in filamentous fungi (48). Higher densities of mitochondria are found in the apical region of growing hyphal tips (25). AM fungal spores harbor many mitochondria (23), and the density of mitochondria even increases in hyphae germinating from AM fungal spores at the activation of presymbiotic hyphal branching (4, 39). However, no information is available on the density or distribution of mitochondria in intraradical fungal structures, such as arbuscules or intraradical hyphae.
The main aim of this study was to explore the quantification of AM fungal mitochondrial DNA and to assess its suitability for monitoring the development of AM fungi in plant roots. We compared the mtLSU-based quantification to the quantification based on the large subunit of nuclear ribosomal DNA (nrLSU), because the quantification of AM fungal DNA using nuclear markers set a standard in the field (12). To obtain a general overview, we applied the two approaches to plant roots of various ages grown under typical experimental conditions. Within this experiment, the DNA dynamics of two isolates of the G. intraradices clade was surveyed to evaluate how these two related genotypes interact when colonizing one root system.
MATERIALS AND METHODS
Fungal material.
Two geographic isolates belonging to the Glomus intraradices clade, named PH5 and Chomutov (CH), were selected for the study. The preliminary characterization of these isolates showed that they harbor different mtLSU haplotypes. Both isolates were obtained via trap cultures from degraded sites in the Czech Republic and were propagated as multispore cultures. The PH5 isolate originated from a Pb-contaminated site, and the fungal material used in this study belonged to the PH5-IS lineage propagated in an inert substrate (31). The CH isolate originated from a freshly leveled coal spoil bank near Chomutov, North Bohemia, and has been kept in culture since 2008.
The phylogenetic analysis of mtLSU exon sequences grouped these isolates together with isolates of the “Glomus irregulare” clade presented by Stockinger et al. (36) (see Fig. S1 in the supplemental material). The PH5 haplotype is identical to BEG140 haplotype B of Sýkorová et al. (38), belonging to the Glomus irregulare group of mtLSU haplotypes. The haplotype of the CH isolate belongs to a well-separated sister clade of this group and can therefore not be unambiguously assigned to G. irregulare. Following the approach of Börstler et al. (6), we refer to both isolates as isolates of G. intraradices sensu lato.
Several cultures were established in a mixture of zeolite and sand (1:1) with medic (Medicago sativa) as the host plant for each isolate, using 200 spores collected from a pooled soil sample from three older multispore cultures. These established cultures were used for the characterization of genetic diversity, as sources of spores, and as inocula.
Characterization of nrLSU.
Total genomic DNA was extracted from 200 to 300 spores per accession (i.e., fungal culture) using the NucleoSpin plant kit (Macherey-Nagel). Partial nrLSU was amplified using the primers 250F (38) and FLR4 (15). The PCRs were performed in a total volume of 20 μl and contained 2 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP), 0.2 μM each primer, 0.5 U of Taq DNA polymerase (Fermentas), 1× Taq buffer with KCl (Fermentas), and about 1 to 5 ng of genomic DNA. An initial denaturation step at 94°C (4 min) was followed by 35 cycles of denaturation (94°C, 30 s), annealing (59°C, 30 s), and extension (72°C, 1.5 min), with a final extension step at 72°C (10 min). Each accession was amplified in three independent reactions. For each fungal isolate, the PCR products obtained from two to three accessions were pooled equimolarly, cloned, and further processed by following the approach of Fehrer et al. (9). The inserts were reamplified using the original PCR conditions and sequenced with the PCR primers (Macrogen, Inc., Seoul, South Korea). The positive bacterial colonies were cultured overnight in liquid LB medium with ampicillin (100 μg ml−1), mixed with sterile glycerol to a 13% glycerol concentration, and stored at −80°C. Five accessions of the CH isolate and three accessions of the PH5 isolate were included in the characterization, and 40 and 27 colonies were sequenced for each isolate, respectively.
Sequence alignments were done using Clustal X (42) and edited manually in Bioedit (16). Representative sequences are deposited in GenBank under the accession numbers JF966362 to JF966370, and their alignment is given in Fig. S2 in the supplemental material.
Characterization of mtLSU haplotypes.
The region was amplified from the same spore extracts as the partial nrLSU by nested PCR using the primers RNL5 and RNL28a (5) in the first step and then RNL29 (5) and the newly designed GImt4510R in the second step. The PCRs were performed in a total volume of 20 μl and contained 0.2 mM each dNTP, 0.5 U of Taq DNA polymerase (Fermentas), 1× Taq buffer with KCl (Fermentas), about 1 to 5 ng of genomic DNA, and reaction-specific concentrations of primers and MgCl2. The cycling conditions were as described above, except the annealing temperatures (Ta) differed. The resulting PCR products were sequenced directly, without cloning (GATC Biotech, Konstanz, Germany), using the original PCR and additional internal primers. One sequence type was obtained per isolate and was deposited in GenBank under the accession numbers JF966360 (PH5) and JF966361 (CH). The sequences were aligned as described above together with mtLSU sequences of G. intraradices sensu lato as well as other species that are representative for all published haplotypes (6, 38, 41).
To prepare standards for the qPCR experiments, these fragments had to be cloned. To facilitate the cloning, the regions discriminating among the two isolates were approximated using the primer combinations GImt1631F/GImt2234R (isolate PH5) and GImt2829F/GImt4510R (isolate CH) in the second amplification step. Fragments generated by the primer pair RNL29/GImt4510R were used for the universal mtLSU assay.
The primer sequences, reaction-specific primer and MgCl2 concentrations, and Ta are specified in Table S1 in the supplemental material, and the location of the newly developed primers is schematically indicated in Fig. S3 in the supplemental material. The cloning and processing of clones is described above, and the inserts were reamplified using the particular PCR primers.
Preparation of plasmid standards for qPCR.
Plasmids were isolated from liquid bacterial cultures in selective LB medium with ampicillin (100 μg ml−1) using a plasmid minikit (Qiagen) according to the manufacturer's recommendations. The plasmids were linearized by restriction digestion using the enzyme KpnI, EcoRV, or SacI (Fermentas), dephosphorylated by FastAP thermosensitive alkaline phosphatase (Fermentas), and purified using the QIAquick purification kit (Qiagen) according to the manufacturers' instructions. They were quantified spectrophotometrically on a BioPhotometer (Eppendorf), serially diluted with 10 mM Tris buffer, pH 8, and stored at −20°C. Several plasmids representing the sequence variants of nrLSU of each fungal isolate were isolated. Mixtures of different plasmids derived from the same isolate were prepared prior to serial dilution.
Design and optimization of the qPCR assays.
We aimed to design primers which would perfectly match the nrLSU sequence types identified from our material, as well as the majority of known nrLSU sequence types deposited in GenBank under the names G. intraradices and G. irregulare, and which would be, at the same time, as specific as possible to this clade. Due to high sequence variability, no primers could be found that would amplify all known nrLSU sequence types within G. intraradices sensu lato and not also match sequences of other members of the Glomeromycota. The primers were therefore designed to fulfill the latter requirement and cannot be expected to amplify all isolates of G. intraradices sensu lato with the same efficiency.
A perfect match with the studied isolates was set as another requirement to avoid quantification biases resulting from mismatches at the priming site (40). A duplex of forward primers GIX6-F351 and GIX6-F351B, differing in two nucleotide positions, in combination with one reverse primer, GIX6-R424, was set up (Table 1). The efficiency of this assay was constant regardless of the ratio of the two sequence types (data not shown).
Table 1.
Parameters of the real-time PCR assays used for the quantification of copy numbers of nrLSU and mtLSU of two isolates of Glomus intraradices sensu lato
| Target region, isolate | Primer names (forward/reverse) | Primer sequences (5′→3′) | Primer concn (μM) | Ta (°C) | Amplicon size (bp) | Amplification efficiencya (SD) |
|---|---|---|---|---|---|---|
| nrLSU, both isolates | GIX6-F351 and GIX6-F351b/GIX6-R424b | TTCGGGGCTACTTGTCTGAT and TTCGGGGCTACCTGTCTGGT/CCATCGACTTTGATAACCGTAA | 0.3 and 0.2/0.5 | 62 | 74 | 94.1 (2.6) |
| mtLSU, both isolates | GI-mtLSU-499F/GI-mtLSU-632R | GAGGGAGTGGCAGTTTCTT/GCATTCTTAGCCCAGCTATG | 0.5/0.5 | 60 | 133 | 99.6 (1.5) |
| mtLSU, isolate CH | GI-CH-mtLSU-2545F/GI-CH-mtLSU-2638R | TGGATTTCTGGTTTTGTAGACG/ATTCTGCTTGTGGTTGTATTCC | 0.4/0.4 | 60 | 94 | 92.3 (2.1) |
| mtLSU, isolate PH5 | GI-PH5-mtLSU-219F/GI-PH5-mtLSU-327R | CAATTTGGCTGTATGCTGGA/GTGGTCGTTGAGGGGTTAAA | 0.4/0.4 | 60 | 119 | 94.0 (3.5) |
Amplification efficiency was calculated from at least five independent 5-fold dilutions of the plasmid template.
The nrLSU assay used a duplex forward primer.
Furthermore, we developed three primer pairs targeted to the mtLSU region (Table 1). One primer pair (termed “universal”) was designed to amplify both isolates for direct comparisons of mtLSU copy numbers (CN) between the two isolates. These primers were targeted to a sequence motif within exon 2 that is conserved in all known haplotypes of G. intraradices but different in the mtLSU of other AM species sequenced so far. Two primer combinations (termed “discriminating”) were designed to amplify only one of the two isolates to quantify their DNA dynamics in coexistence. The first primer pair was located in intron 2, which was present in the PH5 isolate, as well as some other known isolates of G. intraradices sensu lato, but was absent from the CH isolate. The second primer pair was targeted to intron 3, which was present in both isolates, but the priming sites were placed in a 750-bp-long insertion specific to haplotype CH. Although the qPCR assay designed for isolate PH5 is not specific for this isolate and will amplify all mtLSU haplotypes possessing intron 2, these assays discriminate the two studied haplotypes reliably without false-positive results. The location of the assays is schematically shown in Fig. S3 in the supplemental material, and their ability to amplify known G. intraradices sensu lato haplotypes is indicated in Fig. S1 in the supplemental material.
The qPCR using a LightCycler 480 SYBR green 1 master kit (Roche) was performed in 10-μl reaction mixtures on the LightCycler 480 II Real-Time PCR instrument (Roche). The following cycling conditions were used: 5 min at 95°C, followed by 40 cycles of denaturation (95°C, 10 s), annealing (assay-specific Ta, 10 s), and extension (72°C, 15 s). The cycling was finalized by a standard melting curve analysis.
The efficiencies of the qPCR assays were estimated from standard calibration curves based on serial 5-fold dilutions of plasmid standards (0.00032 to 1 pg per reaction). The absolute quantification of the target sequences was performed based on the standard calibration curves using the LightCycler 480 software, version 1.5 (Roche). The resulting concentration is expressed as copy number ng−1 template DNA. Copy numbers were calculated for the amplified fragments using the equation copy number ng−1 DNA = (6.022 × 1023)/(L × 109 × 660), where L is the length in bp of the amplified fragment.
To ensure the specificity of the qPCR assays, cross-amplification tests were carried out with a wide range of templates (plasmid standards, medic DNA extracted from roots and leaves, fungal DNA extracted from spores, and DNA extracted from medic roots colonized by each fungal isolate).
Preparation of fungal material.
Most of the analyzed fungal material originated from a greenhouse experiment with medic (Medicago sativa L. cv. Vlasta) in sterilized soil. It comprised three fungal treatments: (i) inoculated with the isolate PH5 (PH5 treatment), (ii) inoculated with the isolate CH (CH treatment), and (iii) inoculated with both isolates (MIX treatment). A noninoculated treatment (NM) was established as well. Each inoculated treatment comprised 30 replicates, the NM treatment 18 replicates. One-third of the replicate plants each were harvested after 6, 12, and 26 weeks (6 months).
The experiment was established as follows. Seedlings of medic were germinated in autoclaved sand and precultivated for 3 weeks in the same substrate as that used for the experiment to the stage of about three leaves. Pots (volume, 1 liter) were filled with a mixture of garden soil and sand (1:4) and sterilized by gamma irradiation (50 kGy), with the following main chemical parameters: pH (H2O), 7.7; Olsen-P (0.5 M NaHCO3 extractable), 13.11 mg kg−1; N, 0.05%; organic C, 1.15%. The precultivated medic plantlets were transplanted into the pots (1 plant per pot) and inoculated at planting in the fungal treatments. To ensure the similar development of both isolates in the experiment, the mycorrhizal inoculation potential of the inocula was standardized. Substrates from three 6-month-old cultures of each isolate were homogenized and air dried. Five serial dilutions of the inocula with the γ-irradiated experimental substrate (1:10 to 1:105, vol/vol) were planted with medic in five replicates. When the roots of the plants had grown through the whole soil volume of 85 ml (after 5 weeks), the presence and absence of root colonization in the root systems was scored and used to calculate the number of infective propagules by the most-probable-number method (1). Both inocula had the same inoculation potential (57 infective propagules ml−1 substrate). Inoculation thus was performed with air-dried homogenized substrate containing spores, extraradical mycelium, and chopped roots of the host plant that had been stored for about 2 months at room temperature. Each plant received 12 g of the inoculation substrate, which corresponded to about 600 infective propagules. The plants in the MIX treatment received 6 g of the inoculum of each isolate, i.e., the same amount of infective propagules in total as the plants in the PH5 and CH treatments, but only half of each isolate. The inoculum was applied directly below the plantlets. To equalize microbial conditions in the different inoculation treatments, all pots were irrigated with 10 ml of bacterial filtrate, which was obtained by passing a suspension from the two inocula through a filter paper (Whatman no. 1). The experiment was cultivated in a greenhouse with light supplement (12 h at 400 W; metal halide lamps).
At harvest, each root system was carefully washed from the substrate, weighed, and cut to 1-cm segments. A subsample of 200 mg fresh weight was immediately frozen in liquid N and stored at −80°C. Another part was used for the microscopic determination of root colonization after staining with 0.05% trypan blue in lactoglycerol (22). The remaining roots and the shoots were dried at 80°C for 24 h.
Additional fungal material (spores and young root colonization) was prepared to confirm the ratios of mtLSU and nrLSU copy numbers on material from independent cultivation. Spores were collected from one 6-month-old culture of each isolate. Only apparently mature, healthy spores were included and cleaned from extraradical mycelia. Young root colonization was prepared by cultivating pregerminated medic plantlets for 3 weeks in the same conditions as those of the inoculation potential test (see above), with an inoculum level of 1:102 (vol/vol). Root colonization ranged from 15 to 50% after the 3 weeks of cultivation, as determined on six additionally established plants by trypan blue staining (22) and the grid-line intersect method (14). Each material (spores and young root colonization) was collected in five replicates per isolate, and one replicate was represented by about 150 spores or the root system of one medic plant (120 to 200 mg fresh weight). The collected material was immediately frozen in liquid N and stored at −80°C.
Data collection.
Shoot and root dry weights were determined for the experimental plants. Root colonization was evaluated by microscopy (×100 magnification; Olympus BX60) according to Trouvelot et al. (46). The following parameters were calculated using the program Mycocalc (http:/www.dijon.inra.fr/mychintec/Mycocalc-prg/download.html): F, frequency in the root system (percentage of root segments with fungal structures); M, intensity of colonization of the root system; A, arbuscule abundance in the root system; and V, vesicle abundance in the root system. No attempts were made to distinguish vesicles from intraradical spores, because this cannot be done with certainty in trypan blue stained roots, and vesicles can be assumed to contain amounts of nucleic acids similar to those of spores (12).
Genomic DNA was extracted from the deep-frozen samples using a DNeasy plant minikit (Qiagen) according to the manufacturer's instructions. DNA extracts from root samples were quantified spectrophotometrically, and 10 ng of total genomic DNA was used as the template in qPCR. The DNA extracts from spores for which the DNA concentration was beyond the spectrophotometrical detection limit were diluted 1:10.
qPCR was performed as described above. Calibration curves constructed from 5-fold dilutions of plasmid standard were included in each run. The amplification of each dilution of plasmid standard was performed in triplicate, whereas the experimental samples were analyzed in duplicate.
Five replicate root systems per treatment and harvest were analyzed from the experimental material. All target sequences were quantified in all root samples. In the additional material (samples of spores and young root colonization), quantification was performed only with the nrLSU assay and the universal mtLSU assay.
Data analysis and statistics.
Generally, all data from the experiment were analyzed by two-way analysis of variance (ANOVA) with the factors harvest and inoculation treatment. Data obtained with the discriminating assays were analyzed by three-way ANOVA with the factors isolate, coexistence (presence of the other fungus), and harvest. Differences between means were tested by Tukey's test at P < 0.05. ANOVA and Tukey's test were also applied to compare the mtLSU-to-nrLSU ratio in spores and young root colonization.
The relationships between nrLSU and mtLSU copy numbers in the experimental material were determined by liner regression analysis.
When necessary, data sets were either logarithmically [y = ln(x + 1)] or square-root (y = x2) transformed prior to statistical analyses to meet the requirements of normal distribution and homogeneity of variance (as determined by Levene's test). If transformation was performed prior to the regression analysis, both quantitative variables were transformed in the same way.
Analyses were performed using the software SPSS 15.0.
Nucleotide sequence accession numbers.
Sequences determined in the course of this work were deposited in GenBank under accession numbers JF966360 to JF966370.
RESULTS
Relationship between CN of mtLSU and nrLSU.
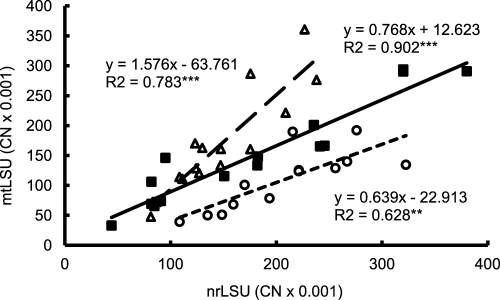
Regression analyses revealed a significant linear relationship between nrLSU and mtLSU copy numbers (CN) (R2 = 0.460; F = 34.9; P < 0.001) in the experimental root material. According to separate regression analyses of the three inoculation treatments, the regression slope was higher in the CH treatment than in the PH5 treatment and was intermediate in the MIX treatment (Fig. 1).
Fig 1.
Relationship between copy numbers (CN) of AM fungal nrLSU and mtLSU in the roots of medic. The mtLSU copy numbers were determined by the universal assay. Each point represents one root sample. The equation of the linear regression, R2, and significance (***, P < 0.001; **, P < 0.01) are given for each inoculation treatment. Open circles with dotted line, isolate PH5; open triangles with dashed line, isolate Chomutov (CH); closed squares with full line, mixed inoculation with both isolates.
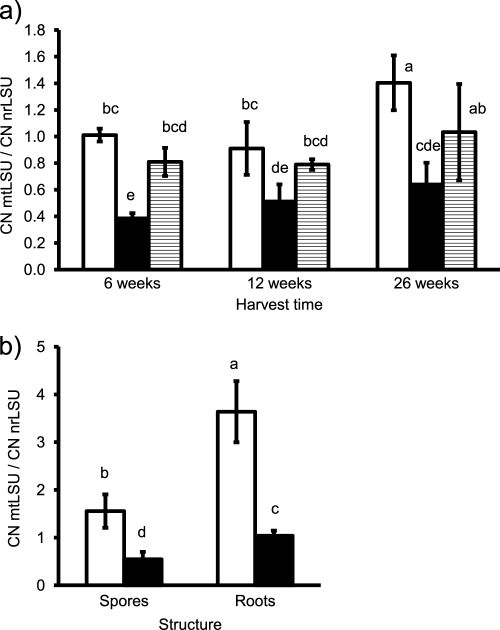
The ratio of mtLSU to nrLSU copy numbers was significantly influenced by inoculation treatment (F = 38.1; P < 0.001) and by harvest date (F = 12.4; P < 0.001), while the interaction of the two factors remained nonsignificant (F = 1.4; P > 0.05). Multiple-comparison test (Fig. 2a) revealed that the ratio was consistently higher in the CH treatment than in the PH treatment. Significant differences between harvests were obtained only for the CH treatment between 12 and 26 weeks of cultivation, while in the other two inoculation treatments, an increasing ratio of mtLSU to nrLSU copy numbers was a nonsignificant trend.
Fig 2.
Ratio of copy numbers (CN) of AM fungal mtLSU to nrLSU in the roots of medic in the experiment with three harvests (a) and in young root colonization and spores (b). The mtLSU copy numbers were determined by the universal assay. Bars represent means of five replicates ± SD. Open bars, isolate Chomutov (CH); black bars, isolate PH5; shaded bars, mixed colonization by both isolates. Differences between means at P = 0.05 are indicated by letters (bars with the same letter are not significantly different).
The higher ratio of mtLSU to nrLSU copy numbers in the CH isolate than in the PH5 isolate was confirmed in the analyzed spores and young root colonization (F = 159.1; P < 0.001) (Fig. 2b). The relationship also significantly differed between the two types of structures (F = 70.9; P < 0.001), with a higher mtLSU-to-nrLSU ratio in the roots than in the spores. The significant interaction of the factors structure and isolate (F = 4.5; P < 0.05) resulted from more pronounced differences between the isolates in the roots than in the spores.
Plant growth and root colonization in the experiment.
Plant shoot and root dry weights steadily increased during the three harvests and were not affected by inoculation (see Table S2 in the supplemental material).
All root colonization parameters significantly increased during the cultivation (Table 2). Only parameter V was significantly affected by inoculation treatment. According to multiple-comparison tests performed for each harvest separately, vesicles were significantly less abundant in the PH treatment than in the other two inoculation treatments after 26 weeks of cultivation, while the abundance of vesicles did not significantly differ among the inoculation treatments after 6 and 12 weeks of cultivation.
Table 2.
Root colonization of medic when inoculated with Glomus intraradices sensu lato isolate PH5, isolate Chomutov, or both isolates together (MIX)
| Harvest time (wk) and inoculation group | Colonization characteristica (%) |
|||
|---|---|---|---|---|
| F | M | A | V | |
| 6 | ||||
| PH5 | 42 (19) | 14 (9) | 10 (7) | 3 (3) |
| CH | 56 (23) c | 24 (13) c | 19 (14) b | 9 (10) b |
| MIX | 55 (23) | 25 (16) | 19 (12) | 8 (9) |
| 12 | ||||
| PH5 | 79 (15) | 32 (13) | 24 (10) | 7 (5) |
| CH | 81 (12) b | 27 (10) b | 20 (8) b | 4 (3) b |
| MIX | 72 (25) | 27 (14) | 22 (12) | 10 (8) |
| 26 | ||||
| PH5 | 100 (0) | 58 (9) | 39 (8) | 15 (7) |
| CH | 98 (3) a | 53 (8) a | 34 (7) a | 29 (8) a |
| MIX | 99 (1) | 57 (14) | 41 (12) | 26 (12) |
Root colonization was evaluated according to Trouvelot et al. (46) after trypan blue staining. F, frequency of root colonization; M, intensity of root colonization; A, abundance of arbuscules; V, abundance of vesicles. Data are means from 9 to 10 replicates (SD are in parentheses). Letters refer to the harvests, with no differentiation between inoculation treatments (for differences among inoculation treatments in V, see Results). Values within each column marked by the same letter are not significantly different (P < 0.05 by Tukey's multiple range test). The significance of effects of factors according to ANOVA for harvest, inoculation, and the combination of the two, respectively, were the following: F, 62.2 (P < 0.001), 0.6 (nonsignificant), and 1.3 (nonsignificant); M, 64.3 (P < 0.001), 0.1 (nonsignificant), and 1.5 (nonsignificant); A, 34.5 (P < 0.001), 1.0 (nonsignificant), and 1.6 (nonsignificant); and V, 43.0 (P < 0.001), 5.5 (P < 0.01), and 2.4 (nonsignificant).
DNA dynamics as determined by the nuclear and mitochondrial marker.
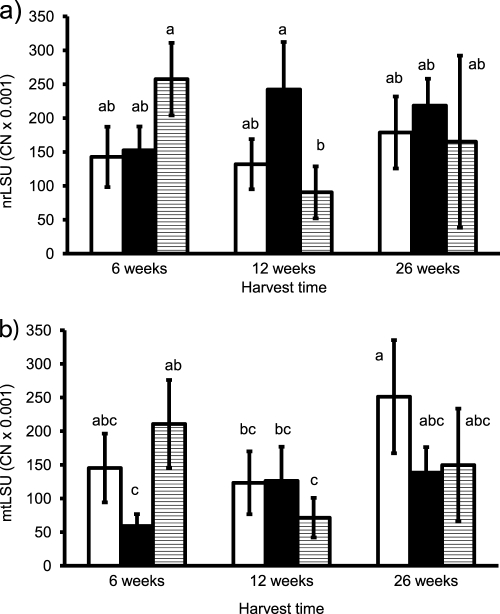
In the experimental material, both nrLSU and mtLSU copy numbers were significantly influenced by the interaction of inoculation treatment and harvest date (for nrLSU, F = 6.1 and P < 0.01; for mtLSU, F = 5.6 and P = 0.001). mtLSU copy numbers also significantly differed among the inoculation treatments (F = 4.3; P < 0.05) and harvests (F = 5.8; P < 0.01).
The interaction of the two factors was a result of different temporal dynamics in the three inoculation treatments, which was described by the two markers consistently (Fig. 3a and b). In the treatments with a single inoculum, there was a mostly nonsignificant trend of increasing gene copy numbers between 6 and 12 weeks in the PH5 treatment and between 12 and 26 weeks in the CH treatment. The latter trend was significant in the mtLSU data. In the MIX treatment, in contrast, the gene copy numbers decreased significantly between 6 and 12 weeks of cultivation.
Fig 3.
Copy numbers (CN; per ng template DNA) of AM fungal nrLSU (a) and mtLSU (b) in the roots of medic plants from the experiment with three harvests. The mtLSU copy numbers were determined by the universal assay. Bars represent means of five replicates ± SD. Open bars, isolate Chomutov (CH); black bars, isolate PH5; shaded bars, mixed colonization by both isolates. Differences between means at P = 0.05 are indicated by letters (bars with the same letter are not significantly different).
Interestingly, both mtLSU and nrLSU CN per ng of template DNA were high in the additionally analyzed young root colonization, about 10 times higher in case of the mtLSU and 3 times higher in case of the nrLSU compared to findings from the first harvest of the experiment. In the CH treatment, the average values (± standard deviations [SD]) were 1,678 × 103 CN of mtLSU (±1,048 × 103) and 506 × 103 CN of nrLSU (±358 × 103); in the PH5 treatment, the average values were 476 × 103 CN of mtLSU (±141 × 103) and 458 × 103 CN of nrLSU (±120 × 103). As evident from the high SD, the determined copy numbers were highly variable, probably due to the variable root colonization of the analyzed root systems (see Materials and Methods).
Coexistence of the two isolates in dual inoculation.
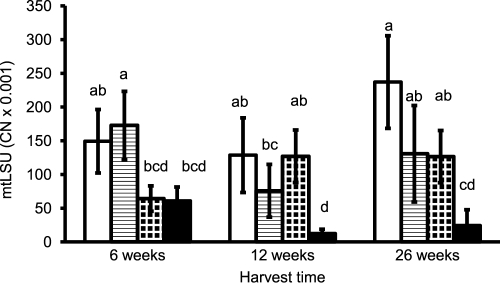
According to three-way ANOVA, mtLSU copy numbers (determined by the discriminating assays) were significantly affected by the factors isolate (F = 43.8; P < 0.001), coexistence with the other isolate (F = 42.0; P < 0.001), and harvest (F = 5.2; P < 0.01). As expected from the ratios of mtLSU to nrLSU copy numbers in the two isolates, overall lower mtLSU levels were obtained for the PH5 isolate than for the CH isolate. mtLSU copy numbers also were lower overall in coexistence than in the single-inoculation treatments. The response to coexistence depended both on isolate (significant interaction of the factors isolate and coexistence; F = 8.4; P < 0.01) and harvest (significant interaction of the factors coexistence and harvest; F = 12.4; P < 0.001).
After 6 weeks, neither of the two isolates was influenced by coexistence with the other (Fig. 4). After 12 and 26 weeks, mtLSU copy numbers of the PH5 isolate were significantly lower in mixed inoculation than when it was growing alone. In the CH isolate, mtLSU copy numbers tended to be lower in the mixed-inoculation treatment as well, but this difference was not significant due to the high variability in the data.
Fig 4.
Copy numbers of AM fungal mtLSU (CN; per ng template DNA) in the roots of medic plants from the experiment with three harvests, as determined by the isolate-discriminating assays. Bars represent means of five replicates ± SD. Open bars, isolate CH growing alone; shaded bars, isolate CH in coexistence with PH5; dotted bars, isolate PH5 growing alone; black bars, isolate PH5 in coexistence with CH. Differences between means at P = 0.05 are indicated by letters (bars with the same letter are not significantly different).
DISCUSSION
This study is, to our knowledge, the first one describing the quantification of AM fungi using mtDNA-based markers. We compared this approach to nrDNA-based quantification, which was previously explored in numerous studies (2, 10, 12, 43). This comparison produced two outcomes: (i) the ratio of mtLSU copy numbers to nrLSU copy numbers differed between the two isolates, and (ii) the ratio of mtLSU to nrLSU copy numbers was rather constant across the root colonization of various ages from the experimental material.
The difference in the mtLSU/nrLSU copy number ratio between the two isolates can be explained by two reasons. First, the two isolates may differ in copy numbers of nrDNA per nucleus or mtDNA per mitochondrion. Variability in nrDNA gene copy numbers has been described among isolates of G. intraradices (7). The mitochondrial ribosomal genes seem to be present in one copy per mitochondrial genome only in G. intraradices (24), but the isolates may differ in the numbers of mitochondrial genomes per mitochondrion, a factor which remains completely unexplored in filamentous fungi. Second, different densities of mitochondria relative to the density of nuclei may play a role. Further studies, including the staining of both organelles and confocal microscopy, should explore this factor in detail.
Gamper et al. (11) suggested that the quantification of AM fungal mitochondrial DNA may better reflect the fungal metabolic activity than the quantification of nrDNA. The high mtLSU/nrLSU ratio of copy numbers encountered in the 3-week-old root colonization, compared to those of spores and older root colonizations from the experiment, are in line with this suggestion. Mitochondria were observed to proliferate in germinating AM fungal hyphae (4, 39) and in actively growing hyphal tips of filamentous fungi in general (25). In the material from the experiment, however, mtLSU and nrLSU copy numbers were closely correlated, and their ratio was rather constant among the three harvests. The last sampling was scheduled 6 months after the establishment of the experiment to capture changes in the colonization structure (e.g., increase in the frequency of vesicles and intraradical spores) and a possible decline in the vitality of the AM fungus in roots indicated by enzymatic staining (34, 45). The evaluation of the experimental material, however, does not indicate that mtDNA contents would provide any additional information to nrDNA contents on the vitality of intraradical structures in established root colonization. This result justifies the search for markers among protein-encoding genes related to certain metabolic processes, as suggested by Gamper et al. (11).
As already outlined by Sanders (32), nrDNA sequence heterogeneity within species and isolates was a problem for the design of AM fungal group-specific primers or primers aimed to discriminate closely related species. We had to combine two forward primers in the nrLSU to match all of the sequence types found within the isolates PH5 and CH. Whereas a bias created by the lower amplification efficiency of some sequence types due to mismatches at the priming sites is acceptable when presence/absence data are collected by conventional endpoint PCR (8, 47) or qPCR (21), such mismatches bias quantitative results obtained by qPCR (40). Considering the demands of qPCR on primer matching, the mtLSU is, due to its sequence homogeneity (30, 41), clearly a superior region for the design of assays. The sequence variability of the region enabled us to design primers discriminating the two closely related isolates, which is generally not possible in nrLSU (8, 44). The lack of haplotype specificity of the PH5 assay indicates that this approach does not allow the specific detection of all known haplotypes. Despite this fact, the application of mtLSU-based markers represents a promising approach, e.g., to the quantitative detection of field-inoculated isolates. The higher-resolution power of this region compared to that of nrDNA increases the chances of distinguishing an introduced isolate from the native background.
The high gene copy numbers at the first harvest and their subsequent decrease in the mixed-inoculation treatment are in accordance with the time course development of gene copy numbers described by Jansa et al. and Thonar (19, 43). In their studies, some of the included Glomus species displayed a peak of nrLSU copy numbers in young root colonization (about 2 to 4 weeks after inoculation). In our experiment, the first harvest was performed 6 weeks after inoculation, and we speculate that in the separate inoculation treatments, the DNA peak occurred before the first harvest. This explanation is supported by the high copy numbers of both nrLSU and mtLSU determined in the 3-week-old roots from independent cultivation. Regardless of whether or not there was a DNA peak in the separate inoculation treatments before the sixth week of cultivation, we show that the coexistence of two isolates in one root system alters the dynamics of AM fungal DNA in the intraradical structures. Accepting the interpretation of the DNA peak as the rapid spread of active mycelium followed by a decrease in vitality (19, 43), this developmental stage was influenced by the coexistence of the two genotypes. AM fungal species colonizing one root system have been shown previously to interact at both the developmental and physiological levels. Coexisting isolates may, in response to each other, redirect their spread in the root zone (3) or alter the expression of some genes (18). Describing such an interaction in closely related isolates, which could not be distinguished by nrLSU-based assays, points to the fact that DNA dynamics of AM fungal isolates, such as those described by Thonar (43), may also depend on the genetic structure of the fungal material.
The isolate-discriminating mtLSU-based markers enabled us to monitor the DNA dynamics of the two closely related isolates in one root system. A direct comparison of the mtLSU levels of the two isolates is biased by the overall lower mtLSU copy numbers in the PH5 isolate than in the CH isolate, the biological basis of which is unclear. The situation is analogous to variable absolute nrLSU copy numbers in different AM fungal species (3, 19, 43), which may be the result of different nuclear densities and/or variation in nrDNA copy numbers per nucleus. Until these differences can be related to genetic or physiological traits, direct comparisons among species or isolates are meaningless. Despite this drawback, the genetic quantification contributes to the understanding of the interactions of AM fungi by describing their dynamics (19, 43) or responses to cultivation factors (3).
In our study, the PH5 isolate reacted to coexistence with the CH isolate by decreasing DNA levels. The interaction of the two isolates thus was competitive, according to previously described interactions in the genus Glomus (27, 43). The depression was established between the 6th and 12th week of cultivation, which suggested that the higher aggressiveness of the CH isolate (according to reference 49) was not the result of its higher initial infectivity. Both isolates colonized roots at a similar rate, as evident from both the microscopic and DNA-based quantification in the separate inoculation treatments. This finding makes the faster occupation of root space by the CH isolate rather improbable. We speculate that the PH5 isolate reacted to coexistence by decreasing the vitality of its intraradical structures, which became evident as a decrease in the levels of mtDNA and, presumably, nrDNA.
In conclusion, our study demonstrated, for the first time, the application of mtDNA-based qPCR assays in the quantification of AM fungi in roots. It provided similar results for their time course development as an nrDNA-based marker. The advantage of mtLSU for the quantification of AM fungi consists mainly in its higher discriminative power to lower taxonomic levels, as demonstrated by the design of assays discriminating two closely related isolates within the G. intraradices sensu lato group. Using these assays, we showed that the DNA dynamics of AM fungi was affected by the coexistence of genotypes which could not be discriminated by nrDNA-based assays. These two closely related genotypes interacted in a competitive manner. Our results thus encourage the application of mtLSU-based assays for the investigation of the coexistence of AM fungal genotypes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Grant Agency of the Czech Republic, grant no. 526/09/0838, the institutional project Z5038910 from the Grant Agency of the Academy of Sciences of the Czech Republic, and the long-term research development project RVO 67985939.
We are grateful to Jana Maršíčková for excellent technical assistance, Karel Müller for advice on the real-time PCR technique, Zuzana Münzbergová for statistical consultations, and Jesse Sadowsky for language revision.
Footnotes
Published ahead of print 9 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alexander M. 1965. Most-probable-number method for microbial populations, p 1467–1472 In Black SA, Evans D, Ensminger LE, White JL, Clark FE. (ed), Methods of soil analysis part 2. Chemical and microbiological properties. American Society of Agronomy, Madison, WI [Google Scholar]
- 2. Alkan N, Gadkar V, Coburn J, Yarden O, Kapulnik Y. 2004. Quantification of the arbuscular mycorrhizal fungus Glomus intraradices in host tissue using real-time polymerase chain reaction. New Phytol. 161:877–885 [DOI] [PubMed] [Google Scholar]
- 3. Alkan N, Gadkar V, Yarden O, Kapulnik Y. 2006. Analysis of quantitative interactions between two species of arbuscular mycorrhizal fungi, Glomus mosseae and G. intraradices, by real-time PCR. Appl. Environ. Microbiol. 72:4192–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Besserer A, et al. 2006. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4:1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Börstler B, Raab PA, Thiéry O, Morton JB, Redecker D. 2008. Genetic diversity of the arbuscular mycorrhizal fungus Glomus intraradices as determined by mitochondrial large subunit rRNA gene sequences is considerably higher than previously expected. New Phytol. 180:452–465 [DOI] [PubMed] [Google Scholar]
- 6. Börstler B, Thiéry O, SýkorovÁ Z, Berner A, Redecker D. 2010. Diversity of mitochondrial large subunit rDNA haplotypes of Glomus intraradices in two agricultural field experiments and two semi-natural grasslands. Mol. Ecol. 19:1497–1511 [DOI] [PubMed] [Google Scholar]
- 7. Corradi N, et al. 2007. Gene copy number polymorphisms in an arbuscular mycorrhizal fungal population. Appl. Environ. Microbiol. 73:366–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Farmer MJ, et al. 2007. Molecular monitoring of field-inoculated AMF to evaluate persistence in sweet potato crops in China. Appl. Soil Ecol. 35:599–609 [Google Scholar]
- 9. Fehrer J, Krak K, Chrtek J. 2009. Intra-individual polymorphism in diploid and apomictic polyploid hawkweeds (Hieracium, Lactuceae, Asteraceae): disentangling phylogenetic signal, reticulation, and noise. BMC Evol. Biol. 9:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Filion M, St-Arnaud M, Jabaji-Hare SH. 2003. Direct quantification of fungal DNA from soil substrate using real-time PCR. J. Microbiol. Methods 53:67–76 [DOI] [PubMed] [Google Scholar]
- 11. Gamper HA, van der Heijden MGA, Kowalchuk GA. 2010. Molecular trait indicators: moving beyond phylogeny in arbuscular mycorrhizal ecology. New Phytol. 185:67–82 [DOI] [PubMed] [Google Scholar]
- 12. Gamper HA, Young JPW, Jones DL, Hodge A. 2008. Real-time PCR and microscopy: are the two methods measuring the same unit of arbuscular mycorrhizal fungal abundance? Fungal Genet. Biol. 45:581–596 [DOI] [PubMed] [Google Scholar]
- 13. Gianinazzi S, Vosatka M. 2004. Inoculum of arbuscular mycorrhizal fungi for production systems: science meets business. Can. J. Bot. 82:1264–1271 [Google Scholar]
- 14. Giovannetti M, Mosse B. 1980. Evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84:489–500 [Google Scholar]
- 15. Gollotte A, van Tuinen D, Atkinson D. 2004. Diversity of arbuscular mycorrhizal fungi colonising roots of the grass species Agrostis capillaris and Lolium perenne in a field experiment. Mycorrhiza 14:111–117 [DOI] [PubMed] [Google Scholar]
- 16. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis suite. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 17. Heid CA, Stevens J, Livak KJ, Williams PM. 1996. Real time quantitative PCR. Genome Res. 6:986–994 [DOI] [PubMed] [Google Scholar]
- 18. Janoušková M, et al. 2009. Development and activity of Glomus intraradices as affected by co-existence with Glomus claroideum in one root system. Mycorrhiza 19:393–402 [DOI] [PubMed] [Google Scholar]
- 19. Jansa J, Smith FA, Smith SE. 2008. Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi? New Phytol. 177:779–789 [DOI] [PubMed] [Google Scholar]
- 20. Koch AM, Croll D, Sanders IR. 2006. Genetic variability in a population of arbuscular mycorrhizal fungi causes variation in plant growth. Ecol. Lett. 9:103–110 [DOI] [PubMed] [Google Scholar]
- 21. König S, et al. 2010. TaqMan Real-Time PCR assays to assess arbuscular mycorrhizal responses to field manipulation of grassland biodiversity: effects of soil characteristics, plant species richness, and functional traits. Appl. Environ. Microbiol. 76:3765–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koske RE, Gemma JN. 1989. A modified procedure for staining roots to detect VA mycorrhizas. Mycol. Res. 92:486–505 [Google Scholar]
- 23. Lang BF, Hijri M. 2009. The complete Glomus intraradices mitochondrial genome sequence–a milestone in mycorrhizal research. New Phytol. 183:4–6 [DOI] [PubMed] [Google Scholar]
- 24. Lee J, Young JPW. 2009. The mitochondrial genome sequence of the arbuscular mycorrhizal fungus Glomus intraradices isolate 494 and implications for the phylogenetic placement of Glomus. New Phytol. 183:200–211 [DOI] [PubMed] [Google Scholar]
- 25. Levina NN, Lew RR. 2006. The role of tip-localized mitochondria in hyphal growth. Fungal Genet. Biol. 43:65–74 [DOI] [PubMed] [Google Scholar]
- 26. Lloyd MacGilp SA, et al. 1996. Diversity of the ribosomal internal transcribed spacers within and among isolates of Glomus mosseae and related mycorrhizal fungi. New Phytol. 133:103–111 [Google Scholar]
- 27. Maherali H, Klironomos JN. 2007. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316:1746–1748 [DOI] [PubMed] [Google Scholar]
- 28. Munkvold L, Kjoller R, Vestberg M, Rosendahl S, Jakobsen I. 2004. High functional diversity within species of arbuscular mycorrhizal fungi. New Phytol. 164:357–364 [DOI] [PubMed] [Google Scholar]
- 29. Pivato B, et al. 2007. Medicago species affect the community composition of arbuscular mycorrhizal fungi associated with roots. New Phytol. 176:197–210 [DOI] [PubMed] [Google Scholar]
- 30. Raab PA, Brennwald A, Redecker D. 2005. Mitochondrial large ribosomal subunit sequences are homogeneous within isolates of Glomus (arbuscular mycorrhizal fungi, Glomeromycota). Mycol. Res. 109:1315–1322 [DOI] [PubMed] [Google Scholar]
- 31. Rydlová J, Vosátka M. 2003. Effect of Glomus intraradices isolated from Pb-contaminated soil on Pb uptake by Agrostis capillaris is changed by its cultivation in a metal-free substrate. Folia Geobot. 38:155–165 [Google Scholar]
- 32. Sanders IR. 2002. Specificity in the arbuscular mycorrhizal symbiosis, p 415–437 In van der Heijden MGA, Sanders IR. (ed), Mycorrhizal ecology. Springer, Berlin, Germany [Google Scholar]
- 33. Sanders IR, Alt M, Groppe K, Boller T, Wiemken A. 1995. Identification of ribosomal DNA polymorphisms among and within spores of the Glomales: application to studies on the genetic diversity of arbuscular mycorrhizal fungal communities. New Phytol. 130:419–427 [Google Scholar]
- 34. Smith SE, Dickson S. 1991. Quantification of active vesicular arbuscular mycorrhizal infection using image-analysis and other techniques. Aust. J. Plant Physiol. 18:637–648 [Google Scholar]
- 35. Smith SE, Read DJ. 2008. Mycorrhizal symbiosis, 3rd ed Academic Press, Amsterdam, The Netherlands [Google Scholar]
- 36. Stockinger H, Walker C, Schüssler A. 2009. ‘Glomus intraradices DAOM197198’, a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 183:1176–1187 [DOI] [PubMed] [Google Scholar]
- 37. Stukenbrock EH, Rosendahl S. 2005. Development and amplification of multiple co-dominant genetic markers from single spores of arbuscular mycorrhizal fungi by nested multiplex PCR. Fungal Genet. Biol. 42:73–80 [DOI] [PubMed] [Google Scholar]
- 38. Sýkorová Z, et al. 2012. Long-term tracing of Rhizophagus irregularis isolate BEG140 inoculated on Phalaris arundinacea in a coal mine spoil bank, using mitochondrial large subunit rDNA markers. Mycorrhiza 22:69–80 [DOI] [PubMed] [Google Scholar]
- 39. Tamasloukht M, et al. 2003. Root factors induce mitochondrial-related gene expression and fungal respiration during the developmental switch from asymbiosis to presymbiosis in the arbuscular mycorrhizal fungus Gigaspora rosea. Plant Physiol. 131:1468–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taris N, Lang RP, Camara MD. 2008. Sequence polymorphism can produce serious artifacts in real-time PCR assays: hard lessons from Pacific oysters. BMC Genomics 9:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thiéry O, Börstler B, Ineichen K, Redecker D. 2010. Evolutionary dynamics of introns and homing endonuclease ORFs in a region of the large subunit of the mitochondrial rRNA in Glomus species (arbuscular mycorrhizal fungi, Glomeromycota). Mol. Phylogenet. Evol. 55:599–610 [DOI] [PubMed] [Google Scholar]
- 42. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thonar C. 2009. Synthetic mycorrhizal communities–establishment and functioning. Ph.D. thesis. ETH, Zürich, Switzerland [Google Scholar]
- 44. Thonar C, Erb A, Jansa J. 2011. Real-time PCR to quantify composition of arbuscular mycorrhizal fungal communities–marker design, verification, calibration and field validation. Mol. Ecol. Resour. 12:219–232 [DOI] [PubMed] [Google Scholar]
- 45. Tisserant B, Gianinazzi-Pearson V, Gianinazzi S, Gollotte A. 1993. In planta histochemical staining of fungal alkaline phosphatase activity for analysis of efficient arbuscular mycorrhizal infections. Mycol. Res. 97:245–250 [Google Scholar]
- 46. Trouvelot A, Kough JL, Gianinazzi-Pearson V. 1986. Mesure du taux de mycorhization VA d'un systeme radiculaire. Recherche de methodes d'estimation ayant une signification fonctionnelle, p 217–221 In Gianinazzi-Pearson V, Gianinazzi S. (ed), Physiological and genetical aspects of mycorrhizae. INRA, Paris, France [Google Scholar]
- 47. Van Tuinen D, Jacquot E, Zhao B, Gollotte A, Gianinazzi-Pearson V. 1998. Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA-targeted nested PCR. Mol. Ecol. 7:879–887 [DOI] [PubMed] [Google Scholar]
- 48. Westermann B, Prokisch H. 2002. Mitochondrial dynamics in filamentous fungi. Fungal Genet. Biol. 36:91–97 [DOI] [PubMed] [Google Scholar]
- 49. Wilson JM. 1984. Competition for infection between vesicular arbuscular mycorrhizal fungi. New Phytol. 97:427–435 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.