Abstract
Survival of the food-borne pathogen Listeria monocytogenes in acidic environments (e.g., in the human stomach) is vital to its transmission. Refrigerated, ready-to-eat foods have been sources of listeriosis outbreaks. The purpose of this study was to determine whether growth at a low temperature (i.e., 7°C) affects L. monocytogenes survival or gene transcription after exposure to a simulated gastric environment (i.e., acid shock at 37°C). L. monocytogenes cells grown at 7°C were less resistant to artificial gastric fluid (AGF) or acidified brain heart infusion broth (ABHI) than bacteria grown at higher temperatures (i.e., 30°C or 37°C). For L. monocytogenes grown at 7°C, stationary-phase cells were more resistant to ABHI than log-phase cells, indicating that both temperature and growth phase affect acid survival. Microarray transcriptomic analysis revealed that the number and functional categories of genes differentially expressed after acid shock differed according to both growth temperature and growth phase. The acid response of L. monocytogenes grown to log phase at 37°C involved stress-related transcriptional regulators (i.e., σB, σH, CtsR, and HrcA), some of which have been implicated in adaptation to the intracellular environment. In contrast, for bacteria grown at 7°C to stationary phase, acid exposure did not result in differential expression of the stress regulons examined. However, two large operons encoding bacteriophage-like proteins were induced, suggesting lysogenic prophage induction. The adaptive transcriptional response observed in 37°C-grown cells was largely absent in 7°C-grown cells, suggesting that temperatures commonly encountered during food storage and distribution affect the ability of L. monocytogenes to survive gastric passage and ultimately cause disease.
INTRODUCTION
Listeria monocytogenes is a food-borne pathogen that has the potential to cause symptoms ranging from gastroenteritis to severe neurological disorders and spontaneous abortions. Postprocessing contamination of a food product with L. monocytogenes is of particular concern for refrigerated ready-to-eat (RTE) products that support L. monocytogenes growth, as this organism can grow even at temperatures below 7°C (64). A 1999 survey (2a) showed that the average temperature of 27% of food products in U.S. home refrigerators was above 5°C. In the same study, the average temperature of RTE deli meats (from deli counters and prepackaged) in the U.S. was reported as >7°C. Temperatures during distribution and home storage of refrigerated RTE foods are thus often higher than 4°C, which is the recommended maximum refrigeration temperature (60). Therefore, to simulate temperature conditions that can be experienced by L. monocytogenes in refrigerated RTE foods, we selected an experimental temperature of 7°C.
Once consumed, L. monocytogenes must survive gastric passage to reach the intestinal epithelial cell barrier, where it can invade and subsequently spread systemically. Human stomach pH ranges from 1 to 3 (55) but can be higher (over 6.0) after food consumption (51). Adaptation of L. monocytogenes to acidic environments involves mechanisms that maintain intracellular pH homeostasis by directing H+ ions out of the cell (e.g., FoF1 ATPases or cation/H+ antiporters) and by consumption of internal H+ through decarboxylation reactions (e.g., glutamate and lysine decarboxylases), generation of ammonium ions (e.g., amino acid deiminases), and macromolecule repair by heat shock proteins (52). A number of transcriptional regulators contribute to acid stress response in L. monocytogenes (23, 42, 52, 67). The negative transcriptional regulators HrcA (heat regulation at CIRCE) and CtsR (class three stress gene repressor), which regulate class I and class III heat shock genes, respectively, have been implicated in acid stress adaptation in L. monocytogenes and other organisms (23, 42). CtsR-regulated genes in L. monocytogenes (38) include clpP and clpE, which both show increased transcript levels after acid shock (42); genes encoding Clp chaperone proteases (e.g., clpC, clpE, and clpP) help rid the bacterial cell of aggregated proteins resulting from environmental stress exposure (33). σH is an alternative sigma factor that regulates the transcription of genes encoding chaperonins, including groEL, clpB, and dnaK in Corynebacterium glutamicum (16). σH also is expressed at higher levels in L. monocytogenes after acid stress (45), indicating that it may be involved in acid response in L. monocytogenes and other Gram-positive bacteria. The alternative sigma factor σB regulates stress response genes in L. monocytogenes (30) and is specifically involved in stationary-phase acid survival (17, 67), as well as regulation of gadD3 (lmo2434) and at least three other genes that contribute to acid resistance (1). L. monocytogenes σB also plays a critical role in a number of stress-related regulatory networks, including transcription of genes encoding other regulatory proteins (e.g., hrcA and prfA) (10, 27, 28, 43). Further, the σB and σH regulons show considerable overlap (10). Therefore, multiple transcriptional regulators appear to be involved in adaptation to acid stress.
Adaptation of L. monocytogenes to certain environmental conditions appears to affect its subsequent response to acidic stress. For example, sublethal exposure to ethanol or acid increases acid resistance of L. monocytogenes (35). In addition, growth rate and growth phase also affect acid resistance in L. monocytogenes (44, 53). Preliminary data also indicate that adaptation to a low temperature (i.e., 10°C) may reduce L. monocytogenes survival at pH 2.5 compared to growth at 30°C (44). These observations suggest that environmental conditions experienced prior to human consumption may affect L. monocytogenes ' ability to survive passage through the acidic gastric environment. The specific goal of this study was to compare the responses to low-pH exposure for L. monocytogenes grown at 7°C and at 37°C. Survival was monitored over time for L. monocytogenes 10403S grown to log or stationary phase at 7°C, 30°C, or 37°C and then exposed to pH 3.5 at 37°C either in artificial gastric fluid or in acidified brain heart infusion broth (BHI). Full-genome microarrays were used to determine changes in gene transcription following exposure to acidified BHI for 5 and 15 min for L. monocytogenes that had been grown to log or stationary phase at 7°C or 37°C.
MATERIALS AND METHODS
Stains and growth conditions.
L. monocytogenes strains 10403S (FSL X1-001; serotype 1/2a; lineage II), FSL J1-194 (serotype 1/2b; lineage I), and Mack (FSL F6-367; serotype 1/2a lineage II) were streaked from frozen stocks (stored at −80°C) onto BHI agar and stored at 4°C for working stocks. For acid survival and microarray studies, one colony from the working stock was inoculated into 5 ml BHI and grown at 37°C overnight (12 to 18 h). The overnight culture was diluted 1:100 in 5 ml BHI and grown with aeration (i.e., shaking at 220 rpm) at 37°C to an optical density at 600 nm (OD600) of 0.4. A 1:100 dilution of this culture was made to 50 ml of BHI in a 500-ml side-arm Nephelo flask (Belco, Vineland, NJ), and this culture was grown at the appropriate temperature with aeration to log phase, defined as an OD600 of 0.4 for 37°C and 7°C, or to early stationary phase, defined as an OD600 of 1.0, followed by an additional incubation for either 3 h (for 30 and 37°C) or 120 h (for 7°C).
Artificial gastric fluid survival.
To assess the effect of growth temperature on L. monocytogenes survival in artificial gastric fluid, 10403S and J1-194 were grown to stationary phase at 7°C, 30°C, or 37°C in BHI. Three independent biological replicates of the artificial gastric fluid survival assay were conducted as described by Garner et al. (21) with slight modifications. Briefly, a 1-ml aliquot of culture was added to 4 ml of 1.25× artificial gastric fluid for a final concentration of 1× (8.3 g/liter Proteose peptone, 3.5 g d-glucose, 2.05 g NaCl, 0.6 g KH2PO4, 0.147 g CaCl2 · 2H2O, 0.37 g KCl, adjusted to pH 2.5 with HCl) and for a final pH of approximately 3.5. Though we acknowledge that metabolic compounds in the 1-ml aliquot of culture could affect response to subsequent acid stress, we have found that centrifugation induces the activity of alternative sigma factor SigB, which is involved in activating the acid response in L. monocytogenes (9). Washing of bacterial cells thus would induce the SigB regulon, which is why the experiment was designed to not include a wash step. Acid-exposed cultures were subsequently incubated at 37°C for 0, 0.5, 1, or 2 h with bacterial enumerations at each time point performed by plating appropriate dilutions on BHI with a spiral plater (Advanced Instruments, Norwood, MA). Separate analyses of variance (ANOVA) of bacterial log reduction values with the factors “temperature” and “strain” were performed for each time point.
Acidified BHI survival.
To assess the effect of growth temperature on the ability of L. monocytogenes to survive exposure to acidified BHI over time, 10403S was grown to log phase or early stationary phase at 7°C or 37°C in BHI buffered with 100 mM morpholinepropanesulfonic acid (MOPS) (adjusted to pH 7.2 with NaOH). At the appropriate growth stage, 5 ml of culture was adjusted to pH 3.5 with 12 N HCl. Acid-exposed cultures were subsequently incubated at 37°C, and viable bacteria were enumerated at different time points by spiral plating appropriate dilutions on BHI. Three independent biological replicates of this experiment were performed. Data were analyzed using ANOVA of bacterial log reduction values.
RNA extraction and microarray.
The effect of growth temperature on the L. monocytogenes transcriptional response to acid shock was evaluated using strain 10403S; this strain was chosen for these microarray experiments because it has the same genetic lineage (II), serotype (1/2a), and ribotype (DUP-1039C) as EGD-e, for which the array was designed (46), and because comprehensive regulon data are available for this strain. Briefly, L. monocytogenes 10403S was grown to log or stationary phase at 7°C or 37°C, and a 5-ml aliquot of each culture was adjusted to a final pH of 3.5 by the addition of 12 N HCl, followed by incubation at 37°C without aeration for 5 or 15 min. This time frame was chosen because prior studies indicated that key events in acid stress response occur within 15 min of acid treatment (42) and because log-phase cultures showed considerable loss in viability after 15 min of acid treatment. Samples (4.5 ml) were taken from untreated, 5-min acid-treated, and 15-min acid-treated cultures. RNA was stabilized by adding 0.5 ml of a 10% acidified phenol in ethanol solution to each sample for a final concentration of 1% phenol (5). Bacterial cells were then pelleted by centrifugation at 1,800 × g for 10 min, and RNA extraction was performed using TRI reagent (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol with slight modifications. Briefly, cell pellets were resuspended in 5 ml TRI reagent, combined with 3 cm3 of 0.1-mm zirconium beads and homogenized in a Mini-Beadbeater-8 (BioSpec Products Inc., Bartlesville, OK) for 4 min. After centrifugation at 3,200 × g for 10 min, supernatants were removed, and 1-bromo-3-chloropropane (0.5 ml) was added to each supernatant. Samples were then allowed to phase separate at room temperature for 10 min, followed by centrifugation at 10,000 × g for 10 min. The aqueous phase was removed, and nucleic acids were precipitated with isopropanol. After washes in 70% and 100% ethanol, nucleic acid pellets were resuspended in nuclease-free water and stored at −80°C. DNase treatment was subsequently carried out as previously reported (46). Following DNase treatment, RNA was purified using an RNeasy minElute cleanup kit (Qiagen, Germantown, Maryland), resuspended in nuclease-free water, and stored at −80°C.
Microarray design, cDNA labeling, competitive hybridization, and scanning.
The microarray used here (Cornell University Food Safety Laboratory Listeria 6K oligonucleotide array, v1.0; Gene Expression Omnibus accession no. GPL5029) has been described in detail (46). Briefly, the array consists of 70-mer oligonucleotides representing 2,857 open reading frames (ORFs) from L. monocytogenes EGD-e; only 45 probes on the array have an EGD-e/10403S cross-hybridization identity of <90 (46). DNA labeling, cleanup, hybridization, and slide scanning were performed as described previously (46), except that in this study, cDNA was synthesized from 6 μg of total RNA. To determine baseline differences in gene transcription between growth temperatures for L. monocytogenes grown to log or stationary phase, competitive hybridizations were performed between total cDNA from non-acid-treated cells grown at 7°C and 37°C. To determine changes in gene transcription after acid shock, separate competitive hybridizations were performed between cDNA from untreated cells (grown at 7°C or 37°C to log or stationary phase) and cDNA from corresponding log- or stationary-phase cells acid treated for either 5 min or 15 min. Four biological replications were completed for the entire experiment. Dye swapping was carried out to account for dye incorporation bias; for each competitive hybridization experiment, two replicate samples from a given treatment were labeled with Cy3, and the other two replicate samples were labeled with Cy5.
Statistical analysis of microarray data.
Microarray results were analyzed as described by Raengpradub et al. (46). Briefly, TIFF images from array scans were analyzed using GenePix Pro 6.0 software. The LIMMA (version 2.16.2) package for R (version 2.8) was used to determine differentially expressed genes. Data were background corrected using “normexp” and normalized using “printtiploess.” To identify genes that were differentially expressed after acid addition, for each temperature and growth phase condition, the modelMatrix design for comparing more than one condition to a reference sample was employed with the untreated sample as the reference sample. The duplicateCorrelation function was used to account for duplicate spots, and a linear model was fitted to the normalized data using lmfit, followed by empirical Bayes smoothing using the eBayes function to calculate moderated t statistics and B statistics. P values were adjusted by controlling for false discovery rate using the Benjamini-Hochberg method (3).
For microarray comparisons to determine differentially expressed genes between untreated cultures grown at 37°C and 7°C, genes were considered differentially expressed if the absolute value of the fold change (FC) between the two temperatures was ≥2.0 with an adjusted P value of ≤0.05. These criteria had been used previously in determining the growth-phase-dependent L. monocytogenes 4°C regulon (7), though in the previous study, cultures were grown at 4°C or 37°C without aeration, whereas in this study, L. monocytogenes cells were cultured at 7°C or 37°C with aeration. Because we exposed the cultures to acid for relatively short times (i.e., 5 or 15 min), we used a less conservative definition of differential expression for the acid shock experiments to increase the sensitivity for identifying acid-induced changes in gene transcription. Specifically, for acid shock experiments, genes were considered differentially expressed if the absolute value of the fold change after acid shock was ≥1.5 with an adjusted P value of ≤0.05.
Gene set enrichment analysis (GSEA).
Rather than confirming changes in gene transcription for a relatively small subset of genes, GSEA was used to identify differences in the transcript levels of gene sets (i.e., sets of genes having the same biological function or sets of genes in the same regulon) between two conditions (e.g., before and after acid shock). GSEA (58) was performed as described previously (7). Briefly, a ranked list of each probe's M value (log2 of fold change between growth temperature or after acid shock) obtained from the LIMMA analyses was compared to lists of gene names categorized by biological function (based on The Institute for Genomic Research Comprehensive Microbial Resource [http://cmr.tigr.org]) or lists of genes that are part of the regulons for selected transcriptional regulators. Briefly, the regulons for σB (7), PrfA (7), σH (10), and σL (10) were defined as genes that had been identified as positively regulated by these proteins in previous microarray studies (7, 10). The regulons for HrcA (27), CtsR (28), and CodY (7) had been defined as genes negatively regulated by these repressors in previous microarray studies (7, 27, 28). For each growth temperature-growth phase combination (e.g., cells grown to log phase at 7°C), separate GSEA analyses were performed to analyze changes in transcript levels after 5 or 15 min of acid shock. Gene sets that were significantly (P ≤ 0.05) enriched for genes with positive changes at a given temperature or after acid shock were reported as showing positive enrichment, whereas gene sets that were significantly enriched for genes with negative changes were reported as showing negative enrichment.
Bacteriophage enumeration.
To enumerate plaque-forming substances in L. monocytogenes supernatants, 5-ml aliquots of 10403S, grown at 7°C to stationary phase, were exposed, as described above, to acidified BHI for 15 min at 37°C, neutralized to pH 7.0 to 7.2 with NaOH, and placed at 7°C or 37°C for 2 or 4 h. Untreated control samples (i.e., no acid shock) were also incubated at 7°C or 37°C for 2 or 4 h. At these time points, L. monocytogenes cells were enumerated by plating the appropriate dilution on BHI, and 0.5 ml of the culture was filtered through a 0.22-μm syringe filter. For identification and quantification of plaque-forming substances (e.g., phages and monocins), 10-fold serial dilutions of the filtrate were prepared, and 100 μl of the appropriate dilution was mixed with 300 μl of an overnight (12 to 18 h at 37°C) culture of L. monocytogenes Mack diluted 1:10 in Luria-Bertani–MOPS (LB-MOPS) broth, followed by the addition of 4 ml LB-MOPS agar (10 g/liter tryptone, 5 g/liter yeast extract, 10.5 g/liter MOPS free acid plus glucose [1 g/liter] and salts [1.47 g/liter CaCl2, 1.02 g/liter MgCl2]). The entire mixture was poured over a plate of LB-MOPS+Glu+salts agar (1.5%). Plates were incubated upright at room temperature (approximately 25°C) for 24 to 48 h. Plaques were enumerated, and results were recorded as PFU/ml. We chose L. monocytogenes Mack as the host strain because it is a host for L. monocytogenes phage A118 (25), which showed induction in our microarray experiments.
To determine differences in phage titer between incubation temperatures and between acid-treated and non-acid-treated cultures, matched-pairs analyses, which take into account differences in initial phage titer for each replicate, were performed for each time after acid shock.
Transmission electron microscopy.
High-titer phage stocks were prepared after two rounds of plaque purification (19). A drop of the 0.2-μm-filtered phage stock (3.5 × 109 PFU/ml) was placed on a carbon adhesive tab and allowed to dry for a minute. One drop of staining solution (2% uranyl acetate; pH 4.2) was transferred onto the carbon film containing the phage sample. Excess stain was removed with filter paper, and the grid was allowed to air dry before imaging with an FEI Tecnai 12 microscope (electron microscopy and optical facility at the Cornell Center for Materials Research, Ithaca, NY).
Microarray accession number.
Raw and analyzed microarray data are available at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE22672.
RESULTS
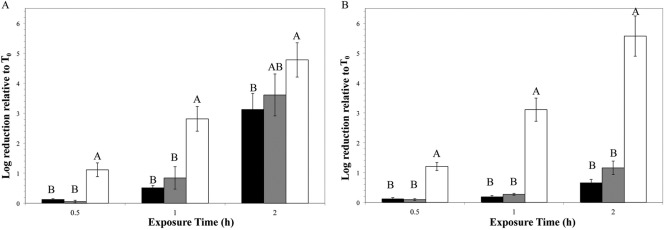
Growth at 7°C increases susceptibility to artificial gastric fluid compared to growth at 30°C or 37°C.
L. monocytogenes 10403S grown at 7°C showed significantly lower survival after 0.5 and 1 h of acid exposure than bacteria grown at 30°C or 37°C (P < 0.05; Tukey's honestly significant difference [HSD]) (Fig. 1A). After 2 h exposure, 10430S grown at 7°C showed 4.78-log reduction, compared to 3.61- and 3.12-log reductions for 10403S grown at 30°C and 37°C, respectively, and at this time point the effect of growth temperature on survival was statistically significant (P = 0.039; ANOVA) (Fig. 1A). Strain J1-194 grown at 7°C also showed significantly reduced survival after 0.5, 1, and 2 h exposure to artificial gastric fluid compared to J1-194 grown at 30°C or 37°C (P < 0.05; Tukey HSD) (Fig. 1b). Overall, these results show that L. monocytogenes cells grown at 7°C are more susceptible to artificial gastric fluid than bacteria grown at higher temperatures.
Fig 1.
L. monocytogenes survival after challenge with artificial gastric fluid. L. monocytogenes 10403S (A) and J1-194 (B) were grown to stationary phase at 37°C (black), 30°C (gray), or 7°C (white) in BHI before being exposed to artificial gastric fluid (pH approximately 3.5) at 37°C for various amounts of time. Log reduction was calculated as log10 CFU/ml before treatment minus log10 CFU/ml after treatment. Capital letters represent statistical groupings of growth temperatures within exposure time (ANOVA; post hoc Tukey HSD; P ≤ 0.05). Bars show the averages of three biological replicates; error bars represent standard deviations.
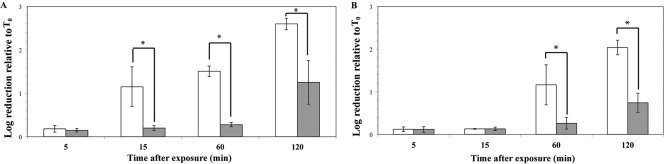
L. monocytogenes cells grown at 37°C are more resistant to acidified BHI than bacteria grown at 7°C.
Strain 10403S was cultured to log or stationary phase at 7°C or 37°C in MOPS-buffered BHI, followed by exposure to pH 3.5 at 37°C (referred to as acid shock below). The average pHs of 7°C cultures were 7.11 (standard deviation [SD] = 0.01) and 6.92 (SD = 0.14) for log- and stationary-phase cells, respectively, and the average pHs for 37°C cultures were 7.08 (SD = 0.06) and 6.89 (SD = 0.15) for log- and stationary-phase cells, respectively. An overall ANOVA showed a significant (P = 0.0059) effect of temperature on survival of bacteria grown to log phase. After 15, 60, and 120 min of acid shock, survival of 10403S grown to log phase at 7°C was significantly lower than that of 10403S grown to log phase at 37°C (P < 0.05; t test) (Fig. 2A). These results indicate that cells grown to log phase at 7°C are more susceptible to acid shock in BHI than cells grown to log phase at 37°C, though the small difference in final culture pH between log- and stationary phase could contribute to increased acid resistance of stationary-phase cells. However, the pH difference between log- and stationary-phase cultures was small, and for cultures grown at 37°C, the pH values for log- and stationary-phase cultures showed overlapping standard deviations.
Fig 2.
L. monocytogenes survival in acidified BHI. L. monocytogenes 10403S was grown to log phase (A) or stationary phase (B) at 37°C (gray bars) and 7°C (white bars) before the cultures were adjusted to pH 3.5 and incubated at 37°C for various amounts of time. Log reduction is log10 CFU/ml before treatment minus log10 CFU/ml after treatment. Time points showing statistically lower log reduction (i.e., higher survival) in 37°C-grown cells are marked with an asterisk. Bars show the averages of three biological replicates; error bars represent standard deviations.
For L. monocytogenes 10403S grown to stationary phase and exposed to acid, the effect of temperature on survival was borderline significant (P = 0.0511; ANOVA). However, after 60 and 120 min of acid shock, L. monocytogenes 10403S grown at 7°C showed numerically lower survival than 10403S grown at 37°C (Fig. 2B). Even though L. monocytogenes cells grown to stationary phase at 7°C showed numerically higher mean survival after 15 min of acid shock than bacteria grown to log phase at 7°C prior to acid shock (1.81 log reduction versus 0.13 log reduction, respectively), the effect of growth phase on survival was not significant (P = 0.2407).
Some acid resistance genes are downregulated at 7°C before acid shock.
Initial analyses identified 355 genes with significantly higher transcript levels (LIMMA; change ≥ 2.0-fold; adjusted P value ≤ 0.05) at 7°C compared to 37°C (see Table S1 in the supplemental material for a complete list of these genes). A total of 192 and 146 genes, respectively, showed higher transcript levels only in the comparison between log-phase cells and only in the comparison between stationary-phase cells grown at the two temperatures. In addition, there were 17 genes that showed significantly higher transcript levels at 7°C in both log- and stationary-phase cells. A total of 298 genes showed lower transcript levels at 7°C than at 37°C (see Table S2 in the supplemental material for a complete list of these genes), including 174 and 100 genes that showed lower transcript levels only in log-phase cells and only in stationary-phase cells, respectively. groEL, which is part of the HrcA regulon, is one of the genes that showed lower transcript levels in stationary-phase cells grown at 7°C; this is relevant, as the GroEL protein is expressed at higher levels after acid exposure (45). A total of 24 genes showed lower transcript levels at 7°C in both log and stationary phase; among these genes are both genes in the gadT2-gadD2 operon (lmo2362-2363), which encode a glutamate transporter and decarboxylase.
GSEA was used to identify predefined functional categories and stress regulons that differed in transcript levels between log or stationary-phase L. monocytogenes grown at 7 and 37°C. For L. monocytogenes grown to log phase, five role categories showed positive enrichment at 7°C compared to 37°C (see Table S3 in the supplemental material), including three categories of transport and binding proteins and the categories “Viral functions: general” and “cellular processes chemotaxis and motility” (see Table S3), while 8 categories showed negative enrichment. For L. monocytogenes grown to stationary phase, only one category (“energy metabolism ATP-proton motive force interconversion”) showed positive enrichment (i.e., higher transcript levels at 7°C than 37°C). For L. monocytogenes grown to log phase, one regulon (i.e., the σH regulon) showed positive enrichment while four regulons (CodY, HrcA, σB, σL) showed negative enrichment at 7°C compared to 37°C (see Table S3). For L. monocytogenes grown to stationary phase, GSEA of the different regulons revealed patterns distinct from those observed for log-phase cells. While the σL regulon showed negative enrichment (i.e., lower transcript levels at 7°C) in both log- and stationary-phase cells, the CodY-regulon showed positive enrichment in stationary-phase cells and the CtsR and σH regulons showed negative enrichment in stationary-phase cells.
Chan et al. (7) previously probed differences in gene transcription levels between L. monocytogenes grown without aeration at 4 and 37°C. Using the same criteria to define differential transcription (i.e., change ≥ 2.0-fold; adjusted P value ≤ 0.05), 209 and 163 genes showed higher transcript levels at 7°C than at 37°C in log and stationary phases, respectively, whereas Chan et al. (7) found 110 and 237 genes transcribed at higher levels at 4°C than at 37°C in log and stationary phases, respectively. Like Chan et al. (7), we found lower transcript levels of PrfA-regulated virulence genes (i.e., plcA, hlyA, actA, and plcB) at 7°C (see Table S2 in the supplemental material), enrichment of CodY-dependent genes at 7°C in stationary phase (see Table S3 in the supplemental material), and enrichment of σL-dependent genes at 37°C in stationary phase (see Table S3). Also consistent with the work of Chan et al. (7), we observed, for L. monocytogenes cells grown to log phase at 7°C, higher transcript levels of lmo450, which encodes an RNA helicase, bacteriophage genes (e.g., lmo0123), and motility genes (e.g., lmo0684) as well as lower transcript levels of groEL, which encodes a heat shock protein, than in cells grown to log phase at 37°C (see Tables S1 and S2 in the supplemental material).
The early transcriptional response to acid shock differs between L. monocytogenes grown at 7 and 37°C.
For L. monocytogenes grown to log phase at 7 and 37°C, a total of 16 and 64 genes, respectively, showed higher transcript levels after acid shock (i.e., after 5 and/or 15 min of acid shock) (Table 1; also, see Tables S4 and S6 in the supplemental material), including 2 and 10 operons, respectively. We identified no genes that showed higher transcript levels after acid shock in L. monocytogenes grown to log phase at 7 and 37°C (Table 1). Also, for bacteria grown to log phase at 7 and 37°C, a total of 13 and 86 genes, respectively, showed lower transcript levels after acid shock (Table 1; also, see Tables S4 and S6 in the supplemental material), including 1 and 11 operons, respectively. Among these genes, nine showed lower transcript levels after acid shock of L. monocytogenes grown to log phase at 7°C and at 37°C (Table 1).
Table 1.
Genes differentially expressed after acid treatment of Listeria monocytogenes grown under various conditionsa
| Growth temp(s) and culture phase(s) prior to acid treatment | Upregulated genes |
Downregulated genes |
||
|---|---|---|---|---|
| No. | Examples | No. | Examples | |
| 7°C log phase only | 13 | lmo0066, lmo0320, lmo1151 | 6 | lmo0299, lmo0735, lmo1114 |
| 7°C stationary phase only | 28 | lmaA, lmaB, lmaD | 6 | lmo0049, lmo0728, lmo2445 |
| 37°C log phase only | 63 | clpC, grpE, clpP | 68 | lmo0680, lmo1406, minD |
| 37°C stationary phase only | 13 | rplU, lmo2362, rplP | 38 | lmo0770, murC, ndk |
| 7°C log phase + 7°C stationary phase | 3 | lmo2290, lmo2291, lmo2292 | 1 | lmo2210 |
| 37°C log phase + 37°C stationary phase | 0 | 5 | lmo0998, lmo1597, lmmo1965 | |
| 7°C log phase + 37°C log phase | 0 | 7 | lmo952, lmo1749, lmo2408 | |
| 7°C stationary phase + 37°C stationary phase | 1 | lmo2293 | 2 | lmo1049, atpI |
| 7°C log phase + 37°C stationary phase | 0 | 0 | ||
| 7°C stationary phase + 37°C log phase | 1 | lmo0903 | 1 | lmo1926 |
| 7°C log phase + 7°C stationary phase + 37°C log phase | 0 | 1 | lmo0883 | |
| 7°C log phase + 7°C stationary phase + 37°C stationary phase | 0 | 0 | ||
| 37°C log phase + 37°C stationary phase + 7°C log phase | 0 | 1 | lmo1639 | |
| 37°C log phase + 37°C stationary phase + 7°C stationary phase | 0 | 3 | lmo0217, lmo1828, deoD | |
| All conditions tested | 0 | 0 | ||
| Total | 122 | 139 | ||
Genes showing a change in expression with an absolute value of ≥1.5-fold and an adjusted P value of ≤0.05 (LIMMA) after acid treatment for 5 and/or 15 min (acid treatment was BHI-MOPS adjusted to pH 3.5 with HCl followed by incubation at 37°C).
For L. monocytogenes grown to stationary phase at 7 and 37°C, a total of 33 and 13 genes, respectively, showed higher transcript levels after acid shock (Table 1; also, see Tables S5 and S7 in the supplemental material), including 5 and 2 operons, respectively. The two operons that showed higher transcript levels after acid shock of stationary-phase cells grown at 37°C encode ribosomal proteins (Table 2), consistent with a previous study that also found higher transcript levels, after organic acid stress, of genes encoding ribosomal proteins (57). Only one gene (lmo2293) showed higher transcript levels after acid shock of cells grown to stationary phase at 7°C and 37°C (Table 1). Also for bacteria grown to stationary phase at 7 and 37°C, a total of 13 and 48 genes, respectively, showed lower transcript levels after acid shock (Table 1; also, see Tables S5 and S7), including 3 operons that showed lower transcript levels after acid shock of stationary-phase cells grown at 37°C (Table 2). Only five genes showed lower transcript levels after acid shock of cells grown to stationary phase at 7°C and cells grown to stationary phase at 37°C (Table 1). These data further support the idea that there is very limited overlap between acid shock responses of L. monocytogenes grown at 7 and 37°C.
Table 2.
L. monocytogenes 10430S operons differentially transcribed after acid treatment of cells grown at 37°C to log or stationary phasea
| Locus tag (gene name) | Gene functionb | Fold change after acid treatment of cells inc: |
Putative binding site(s)d | |||
|---|---|---|---|---|---|---|
| Log phase |
Stationary phase |
|||||
| 5 min | 15 min | 5 min | 15 min | |||
| lmo0113 | Similar to protein gp35 from bacteriophage A118 | −1.81* | ||||
| lmo0114 | Similar to putative repressor C1 from lactococcal bacteriophage Tuc2009 | −1.57* | ||||
| lmo0136 | Similar to oligopeptide ABC transporter, permease protein | 2.27** | 2.10** | |||
| lmo0137 | Similar to oligopeptide ABC transporter, permease protein | 1.64** | 1.54* | B | ||
| lmo0180 | Similar to sugar ABC transporter, permease protein | 1.71* | 1.80* | |||
| lmo0181 | Similar to sugar ABC transporter, sugar-binding protein | 1.75* | 1.87** | h | ||
| lmo0230 | Similar to Bacillus subtilis YacH protein | 1.53* | 1.56* | B, i | ||
| lmo0231 | Similar to arginine kinase | 1.60** | ||||
| lmo0232 (clpC) | Endopeptidase Clp ATP-binding chain C | 1.59** | 1.67** | |||
| lmo0680 | Similar to flagellum-associated protein FlhA | −1.59** | −1.87*** | |||
| lmo0681 | Similar to flagellar biosynthesis protein FlhF | −1.54* | −1.84** | |||
| lmo0683 | Similar to chemotactic methyltransferase CheR | −2.16** | −2.68** | |||
| lmo0806 | Similar to transcription regulator | 2.03* | i | |||
| lmo0808 | Similar to spermidine/putrescine ABC transporter, permease protein | 1.87* | 2.08* | |||
| lmo0809 | Similar to spermidine/putrescine ABC transporter, permease protein | 1.90** | 2.07** | |||
| lmo0810 | Similar to spermidine/putrescine-binding protein | 2.27** | 2.25*** | |||
| lmo0811 | Similar to carbonic anhydrase | 2.55** | 2.78** | |||
| lmo0847 | Similar to glutamine ABC transporter (binding and transport protein) | 2.27** | 2.29** | |||
| lmo0848 | Similar to amino acid ABC transporter, ATP-binding protein | 1.82** | 1.91** | |||
| lmo1041 | Similar to molybdate ABC transporter binding protein | −1.66* | ||||
| lmo1046 | Molybdenum cofactor biosynthesis protein C | −1.58* | ||||
| lmo1089 (tagD) | Highly similar to glycerol-3-phosphate cytidylyltransferase, CDP-glycerol pyrophosphorylase (teichoic acid biosynthesis protein D) | −1.86*** | −2.03*** | |||
| lmo1090 | Similar to glycosyltransferases | −1.51** | −1.64*** | b, h | ||
| lmo1406 (pflB) | Pyruvate-formate lyase | −4.03* | −4.30* | |||
| lmo1407 (pflC) | Pyruvate-formate lyase activating enzyme | −2.86* | −2.72* | |||
| lmo1474 (grpE) | Heat shock protein GrpE | 1.84** | 1.71** | |||
| lmo1475 (hrcA) | Transcription repressor of class I heat shock gene HrcA | 1.83** | 1.76** | B, I, III | ||
| lmo1541 | Similar to unknown protein | 1.60* | 1.57** | |||
| lmo1542 (rplU) | Ribosomal protein L21 | 1.51* | 1.69*** | |||
| lmo1556 (hemC) | Highly similar to porphobilinogen deaminases (hydroxymethylbilane synthase) | −1.59* | h | |||
| lmo1557 (hemA) | Highly similar to glutamyl-tRNA reductase | −1.72*** | −1.87*** | |||
| lmo1589 (argB) | Highly similar to N-acetylglutamate 5-phosphotransferase | −4.37* | b | |||
| lmo1590 (argJ) | Highly similar to ornithine acetyltransferase and amino acid acetyltransferases | −6.76* | −4.84* | |||
| lmo1591 (argC) | Similar to N-acetylglutamate gamma-semialdehyde dehydrogenases | −25.44* | −11.04* | |||
| lmo1833 (pyrD) | Highly similar to dihydroorotase dehydrogenase | 1.57** | h | |||
| lmo1835 (pyrAB) | Highly similar to carbamoyl-phosphate synthetase (catalytic subunit) | 1.83* | 1.91** | |||
| lmo1929 (ndk) | Similar to nucleoside diphosphate kinase | −1.73*** | ||||
| lmo1932 | Heptaprenyl diphosphate synthase component I | −1.51* | ||||
| lmo2040 (ftsL) | Similar to cell division protein FtsL | −1.51*** | ||||
| lmo2041 | Similar to unknown proteins | −1.68*** | ||||
| lmo2114 | Similar to ABC transporter (ATP-binding protein) | 2.28** | 2.25** | |||
| lmo2115 | Similar to ABC transporter (permease) | 2.23** | 2.16** | |||
| lmo2260 | Similar to unknown proteins | 1.87* | 1.78* | |||
| lmo2261 | Similar to unknown proteins | 1.63* | ||||
| lmo2293 | Protein gp10 (bacteriophage A118) | −2.34* | ||||
| lmo2295 | Protein gp8 (bacteriophage A118) | −2.88* | −2.39* | |||
| lmo2296 | Similar to coat protein (bacteriophage SPP1) | −2.32* | ||||
| lmo2362 | Similar to amino acid antiporter (acid resistance) | −2.97** | −2.80** | |||
| lmo2363 | Similar to glutamate decarboxylase | −2.59* | −2.28* | |||
| lmo2408 | Similar to repressor protein | −1.61*** | −1.94*** | |||
| lmo2409 | Unknown | −3.28** | −4.21*** | h | ||
| lmo2484 | Similar to B. subtilis YvlD protein | 2.11* | B | |||
| lmo2487 | Similar to B. subtilis YvlB protein | 1.56* | 1.74** | |||
| lmo2625 (rplP) | Ribosomal protein L16 | 1.69* | ||||
| lmo2630 (rplW) | Ribosomal protein L23 | 1.50* | ||||
| lmo2632 (rplC) | Ribosomal protein L3 | 1.50* | ||||
| lmo2633 (rpsJ) | Ribosomal protein S10 | 1.64* | ||||
Operons with at least two genes showing a fold change with an absolute value of ≥1.5 after acid treatment and an adjusted P value of ≤0.05 after acid treatment. Gene names are from ListiList (http://genolist.pasteur.fr/ListiList), and operon predictions are from ListiList and reference 59. Acid treatment was done at 37°C in BHI adjusted to pH 3.5 with HCl.
Based on annotation provided by TIGR (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi).
Adjusted P values are indicated as follows: ***, <0.001; **, <0.01; *, ≤0.05.
Putative binding sites for σB (B, b), σH (H, h), σL (L, l), CtsR (III, iii), and HrcA (I, i) are based on the hidden Markov model results reported by Hu et al. (27, 28), Raengpradub et al. (46), and Chaturongakul et al. (10). Hits were considered significant if they had an E value of ≤0.02 and were ≤300 bp upstream of the respective coding sequence. Uppercase denotes genes for which regulation has been confirmed by experimental studies (i.e., differential transcription in an isogenic deletion mutant versus the wild type).
Comparisons of genes that showed up- or downregulation after acid shock of both log- and stationary-phase L. monocytogenes grown at the same temperature revealed three genes (lmo2290 to lmo2292) that showed higher transcript levels after acid shock treatment of both log- and stationary-phase cells grown at 7°C (Table 1) and two genes (lmo0883 and lmo2210) that showed lower transcript levels after acid shock treatment of both log- and stationary-phase cells grown at 7°C (Table 1). While there were no genes that showed higher transcript levels after acid shock treatment of both log- and stationary-phase cells grown at 37°C, eight genes showed lower transcript levels after acid shock treatment of both log- and stationary-phase cells grown at 37°C (Table 1). Overall, our data found very limited overlap between genes differentially transcribed after acid shock of bacteria grown under different conditions (i.e., growth phase or temperature) (Table 1).
Few previously reported acid stress response genes were induced after acid shock.
We found that very few genes that were previously reported as playing a role in acid stress response were induced after acid shock. Among a total of 42 previously reported acid response genes representing four categories of acid stress response mechanisms (i.e., decarboxylases, deiminases, macromolecular repair, and ATP synthases) as well as other σB-dependent acid stress genes [i.e., lmo0796, lmo0913, and lmo2391 (1)] (see Table S8 in the supplemental material), only three genes (clpE, clpP, and grpE, all encoding proteins involved in macromolecular repair) showed induction after acid stress. All three of these genes were induced after 5 and 15 min of acid shock exposure of log-phase cells grown at 37°C but were not induced in bacteria grown under any other conditions (see Table S8 in the supplemental material).
Functional categories that show differential gene set enrichment after acid shock differ considerably in L. monocytogenes grown to different growth phases prior to acid shock.
GSEA showed limited overlap between functional gene categories that showed positive or negative enrichment after acid shock treatment of log- and stationary-phase cells. Among the 18 functional categories that showed positive or negative enrichment after 5 or 15 min of acid shock treatment of L. monocytogenes grown at either 7 or 37°C, only three categories (i.e., “protein synthesis: ribosomal proteins: synthesis and modification,” “transport and binding proteins: carbohydrates, organic alcohols, and acids,” and “viral functions: general”) showed positive enrichment after acid shock of both log- and stationary-phase cells (Table 3).
Table 3.
Listeria monocytogenes gene groups showing positive or negative enrichment after acid treatment for 5 or 15 min at 37°C
| Gene category and protein or protein group | No. of genes in group | Enrichment in bacteria grown toa: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Log phase |
Stationary phase |
||||||||
| 7°C |
37°C |
7°C |
37°C |
||||||
| 5 min | 15 min | 5 min | 15 min | 5 min | 15 min | 5 min | 15 min | ||
| Regulons | |||||||||
| CtsR | 18 | +*** | +*** | ||||||
| HrcA | 20 | +** | −** | −* | |||||
| σB | 166 | +*** | +*** | +*** | +*** | −*** | −*** | ||
| σH | 95 | +*** | +*** | −*** | −*** | ||||
| σL | 31 | +* | |||||||
| PrfA | 9 | +* | +*** | ||||||
| CodY | 84 | −* | −* | ||||||
| Functional categoriesb | |||||||||
| Purines, pyrimidines, nucleosides, and nucleotides: pyrimidine ribonucleotide biosynthesis | 18 | + | +* | +* | |||||
| Amino acid biosynthesis: aspartate family | 22 | +* | |||||||
| Amino acid biosynthesis: glutamate family | 14 | −*** | −*** | ||||||
| Cellular processes: chemotaxis and motility | 25 | +* | |||||||
| Cellular processes: cell division | 24 | −** | −* | ||||||
| Cellular processes: adaptations to atypical conditions | 19 | −* | |||||||
| Cell envelope: other | 37 | −** | |||||||
| Energy metabolism: fermentation | 33 | −*** | −** | ||||||
| Protein fate: protein folding and stabilization | 14 | +*** | +** | ||||||
| Amino acid biosynthesis: aromatic amino acid family | 70 | +** | +*** | ||||||
| Energy metabolism: pentose pyruvate dehydrogenase | 65 | +*** | +*** | +*** | |||||
| Energy metabolism: biosynthesis and degredation of polysaccharides | 16 | +* | |||||||
| Energy metabolism: sugars | 44 | +** | |||||||
| Protein synthesis: ribosomal proteins: synthesis and modification | 59 | +** | +*** | +*** | +*** | ||||
| Regulatory functions: other | 180 | −*** | −*** | ||||||
| Transport and binding proteins: carbohydrates, organic alcohols, and acids | 110 | +*** | +** | +** | +*** | ||||
| Transport and binding proteins: cations | 58 | +* | |||||||
| Viral functions: general | 47 | +** | +** | +*** | +*** | +** | |||
Reported changes are based on gene set enrichment analysis. +, genes comprising a group were significantly enriched for higher transcript levels after acid stress relative to the nonstressed control in the same growth phase (i.e., positive enrichment); −, significant enrichment for lower transcript levels after acid stress (i.e., negative enrichment). ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Functional categories from The Institute for Genomic Research Comprehensive Microbial Resource (http://cmr.tigr.org). Only functional categories containing ≥10 genes are reported; 74 total functional categories were tested for enrichment, including 39 that contained ≥10 genes.
For L. monocytogenes grown to log phase, GSEA revealed eight and two gene categories that showed positive and negative enrichment, respectively, after acid shock treatment. Among these 10 categories, only one (i.e., “purines, pyrimidines, nucleosides, and nucleotides: pyrimidine ribonucleotide biosynthesis”) showed significant (positive) enrichment after acid shock of both L. monocytogenes grown at 7 and 37°C (Table 3). Examples of upregulated genes involved in pyrimidine ribonucleotide biosynthesis include pyrD and pyrAB (both encode enzymes involved in pyrimidine biosynthesis), which showed significantly higher transcript levels in cells grown to log phase at 37°C and then exposed to acid shock for 15 min (adjusted P value < 0.01, LIMMA [see Table S4 in the supplemental material]). The gene category “protein fate: protein folding and stabilization” (Table 3) showed positive enrichment after acid shock of log-phase bacteria grown at 37°C; this category includes the heat shock gene grpE, which showed significant upregulation after 15 min of acid shock of cells grown to log phase at 37°C (adjusted P value < 0.01 [see Table S4]). The two gene categories that showed negative enrichment after acid shock treatment of log-phase bacteria were “energy metabolism fermentation” and “amino acid biosynthesis: glutamate family” (Table 3). Examples of genes in the glutamate family category that showed significantly lower transcript levels after 5 and 15 min of acid shock are lmo1589 (argB), lmo1590 (argJ), and lmo1591 (argC); the three enzymes encoded by these genes are involved in conversion of glutamate to n-acetyl-ornithine. Downregulation of these genes is thus expected to increase the pool of intercellular glutamate, which is the substrate for the glutamate decarboxylase pathway, which contributes to acid response and resistance in a variety of organisms (2, 47), including L. monocytogenes (12, 13, 29).
For L. monocytogenes grown to stationary phase prior to acid shock treatment, GSEA revealed seven and four gene categories that showed positive and negative enrichment, respectively. Five categories showed significant (positive) enrichment after acid shock of both L. monocytogenes grown at 7 and 37°C (Table 3). For four of these categories, bacteria grown at 7°C showed significant enrichment after 15 min of acid shock treatment, while bacteria grown at 37°C showed significant enrichment after 5 min of acid shock treatment, indicating more rapid induction of acid stress response in L. monocytogenes grown at 37°C. Bacteria grown to stationary phase at 7°C showed positive enrichment of the gene category “viral functions: general” after 5 min and 15 min of acid shock treatment (Table 3), with bacteria grown at 37°C showing positive enrichment after 15 min of acid shock. Specifically, two large groups of bacteriophage genes showed significantly higher transcript levels after acid shock (see Table S6 in the supplemental material), including the lmo0115-lmo0129 operon, which includes lmaA-lmaD (22, 54), and four operons comprised of genes encoding proteins from a temperate bacteriophage similar to L. monocytogenes phage A118 (lmo2278-lmo2279, lmo2282 to lmo2292, lmo2293 to lmo2299, lmo2303 to lmo2326 [see Table S7 in the supplemental material]).
A number of stress regulons show positive enrichment after acid shock of log-phase L. monocytogenes but not after acid shock treatment of stationary-phase cells.
Regardless of growth temperature, log-phase cells showed highly significant positive enrichment for σB-regulated genes after acid shock (Table 3). For example, the σB-dependent oligopeptide transporter genes lmo0136 and lmo0137 were both induced (FC > 1.5; adjusted P value ≤ 0.05, LIMMA) after 5 and 15 min of acid shock of L. monocytogenes grown to log phase at 37°C (Table 2). Also consistent with our observed enrichment of σB-dependent genes after acid shock, genes involved in chemotaxis (i.e., cheR) and flagellar biosynthesis genes (lmo0676, lmo0680, lmo0681, and lmo0683), which had been shown to be indirectly negatively regulated by σB (46, 59), were downregulated after acid shock of bacteria grown to log phase at 37°C (see Table S4 in the supplemental material). L. monocytogenes grown to log phase at 37°C also showed positive enrichment of the σH and the CtsR regulons (Table 3). The CtsR-regulated genes clpC, clpE, lmo1138, and clpP all showed induction after 5 and 15 min of acid shock treatment of log-phase cells grown at 37°C (adjusted P < 0.05; FC > 1.5, LIMMA [see Table S4]). On the other hand, the regulons for the repressors CodY and HrcA showed a negative enrichment after 5 and 15 min of acid shock treatment of L. monocytogenes grown to log phase at 37°C. Interestingly, the HrcA regulon also showed positive enrichment after 15 min of acid shock treatment of bacteria grown to log phase at 7°C (Table 3). Consistent with the negative enrichment for the HrcA regulon and the positive enrichment of the σB regulon after acid shock of bacteria grown to log phase at 37°C, hrcA and grpE, which are in the same operon, showed significantly higher transcript levels after 5 and 15 min of acid shock of bacteria grown to log phase at 37°C (Table 2). As hrcA transcription is upregulated by σB (10) [even though the hrcA-grpE-dnaK operon is also repressed by HrcA (27)], increased activity of σB after acid shock treatment may contribute to upregulation of hrcA and hence downregulation of the HrcA regulon.
Among the regulons included in the GSEA, only σL and PrfA showed positive enrichment after acid shock treatment of stationary-phase L. monocytogenes (Table 3); both regulons showed positive enrichment after acid shock treatment of bacteria grown at 7°C. The PrfA regulon also showed positive enrichment after acid shock of bacteria grown to log phase at 7°C. GSEA showed negative enrichments for the σB and σH regulons after acid shock treatment of L. monocytogenes grown to stationary phase at 37°C (Table 3), possibly reflecting that these regulons are already highly transcribed in stationary-phase cells [consistent with the fact that σB is induced in stationary phase (31)].
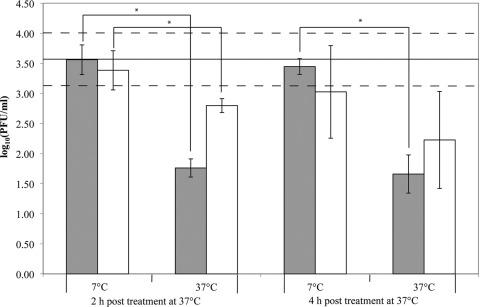
Prophage induction occurs in stationary-phase L. monocytogenes grown at 7°C.
As our transcriptomic analyses indicated a possible prophage induction after acid shock of L. monocytogenes grown to stationary phase at 7°C, we evaluated bacteriophage titers in L. monocytogenes 10403S grown to stationary phase at different temperatures and then exposed to acid shock. Initial experiments showed phage titers below the detection limit (1.3 log PFU/ml) for L. monocytogenes grown to stationary phase at 37°C, while bacteria grown to stationary phase at 7°C showed detectable phage titers. For further experiments, L. monocytogenes was grown to stationary phase at 7°C and then exposed to pH 3.5 for 15 min at 37°C, followed by neutralization by the addition of NaOH and determination of PFU counts after 2 and 4 h of incubation at either 7°C or 37°C. At time zero (i.e., before acid shock), 10403S had an average titer of 3.59 log PFU/ml (Fig. 3). Cultures incubated for 2 and 4 h at 7°C after acid shock showed average phage titers of 3.56 log and 3.45 log PFU/ml, respectively; control cultures not exposed to acid showed similar titers (Fig. 3). For cultures incubated at 37°C for 2 h and 4 h after acid shock treatment, average titers were 1.76 and 1.66 log PFU/ml, respectively, and phage titers for non-acid-shocked control cultures were significantly higher (Fig. 3) (P = 0.0139, matched-pairs analysis). With one exception (4 h with no acid shock; P = 0.0647, matched-pairs analysis), L. monocytogenes incubated at 7°C had significantly higher titers than cells incubated at 37°C (P < 0.05, matched-pairs analysis) (Fig. 3), regardless of acid shock, consistent with our microarray results showing that transcription levels of multiple bacteriophage genes were higher at 7°C than at 37°C (see Table S1 in the supplemental material).
Fig 3.
Plaque-forming substances present in the supernatants of L. monocytogenes 10403S grown to stationary phase at 7°C and then exposed to acid shock (pH 3.5) for 15 min (gray bars) or not acid shocked (white bars) and incubated at 7°C or 37°C for 2 or 4 h. The solid line at 3.59 log10(PFU/ml) represents the mean count at time zero (i.e., stationary-phase culture with no acid stress or subsequent incubation). Dashed lines represent the standard deviations of the counts at time zero. Instances where cultures incubated at 7°C showed significantly higher log10(PFU/ml) than cultures incubated at 37°C are indicated with an asterisk (P < 0.05, matched-pairs analysis). Bars show the averages of four biological replicates; error bars represent standard deviations.
Transmission electron microscopy of a purified phage prepared from a single plaque obtained from L. monocytogenes culture grown at 7°C to stationary phase showed phage morphology similar to that of L. monocytogenes phage A118 [i.e., an isomeric capsid with a long, noncontractile tail (68)].
DISCUSSION
This study provides clear evidence that growth temperature and growth phase of L. monocytogenes affect its survival and transcriptional response to a sudden downshift in pH. Specifically, we found that (i) L. monocytogenes cells grown at ≥30°C are more resistant to acid shock than cells grown at 7°C, (ii) L. monocytogenes log- and stationary-phase cells show distinct transcriptional responses to acid shock, (iii) L. monocytogenes cells grown to log phase at 37°C show an acid response involving induction of multiple regulators that may prime the cells for intracellular survival, and (iv) acid shock treatment of L. monocytogenes grown to stationary phase at 7°C induces rapid upregulation of prophage genes in the absence of any clear other transcriptional responses.
L. monocytogenes grown at ≥30°C are more resistant to acid shock than cells grown at 7°C.
Our finding that L. monocytogenes cells grown at 7°C are more susceptible to inorganic acid shock than cells grown at 37°C is consistent with other studies that indicated that bacterial food-borne pathogens such as Salmonella enterica serovar Typhimurium and E. coli O157:H7 grown at low temperatures mount a less effective acid tolerance response (ATR) and are less resistant to acid challenge than cells grown at higher temperatures (53). One study also showed that L. monocytogenes grown at 10°C had a significantly shorter D value when exposed to pH 2.5 (at 30°C and at 10°C) than cells grown at 30°C (44). Overall, these data suggest that food-borne pathogens, including L. monocytogenes, grown at refrigeration temperatures may survive less well in the low-pH gastric environment than bacteria grown at higher temperatures. As Patchett et al. (44) showed that L. monocytogenes cells growing at lower rates were more resistant to acid shock than cells growing at higher rates and as both stationary-phase and log-phase cells grown at 7°C showed reduced acid resistance in the experiments reported here, growth temperature, rather than reduced growth rate at low temperature, appears to affect the acid resistance phenotype of L. monocytogenes. Hence, growth conditions, including growth temperature, may significantly affect the outcomes of experiments that involve low pH exposure, including possibly oral infection experiments and acidic food challenge experiments.
The transcriptomic data reported here also provide insights into the mechanisms that may be responsible for the reduced acid resistance of L. monocytogenes grown at lower temperatures. For example, L. monocytogenes cells grown at 7°C were found to have lower transcript levels than cells grown at 37°C of some key regulons that have been implicated in acid stress response, including negative enrichment for the σL regulon in both stationary and log phases as well as negative enrichment for the σB regulon in log-phase and the σH regulon in stationary-phase cells grown at 7°C. L. monocytogenes σL has been shown to contribute to osmotic stress (41) and resistance to bacteriocins (48), but no prior publications have reported contributions of σL to acid resistance in L. monocytogenes. While some previous studies of Mycobacterium tuberculosis have suggested that σH is involved in stress response (37), including acid stress (49), no reduced stress resistance phenotypes have been observed in L. monocytogenes sigH-null mutants (10). Most importantly, the σB regulon was found here to be downregulated in cells grown to log phase at 7°C. In addition, the gadT2-gadD2 operon, which encodes a γ-aminobutyric acid (GABA) transporter and associated glutamate decarboxylase, which have been shown to contribute to the survival of L. monocytogenes LO28 in low-pH environments (12, 13), showed lower transcript levels at 7°C than at 37°C in both log- and stationary-phase cells. As σB has previously been shown to play a critical role in the acid stress response in L. monocytogenes (8, 66, 67), downregulation of the σB regulon, along with downregulation of key acid resistance genes in bacteria grown at 7°C, may at least partially explain the reduced acid resistance of L. monocytogenes grown at this temperature.
L. monocytogenes log- and stationary-phase cells show distinct responses to acid shock.
A number of studies, including the one reported here, have shown that stationary and log-phase L. monocytogenes cells differ in their acid resistance phenotypes, with stationary-phase cells typically being more resistant to acid stress (14, 40, 53). The data reported here indicate that log- and stationary-phase cells show distinct differences in their transcriptomic responses to acid shock. Most importantly log-phase cells showed higher transcript levels of key stress response regulons after acid shock, including rapid induction of the σB regulon after acid shock of log-phase cells grown at 7 and 37°C; cells grown to log phase at 37°C also showed rapid induction of the CtsR and σH regulon. Thus, while bacteria grown to log phase at 7°C were more sensitive to acid stress, they were still able to mount a rapid σB-dependent transcriptional response to acid shock. Reduced acid resistance of bacteria grown at 7°C may thus at least partially reflect lower transcript levels of genes encoding key acid response proteins prior to acid shock rather than a reduced ability to mount a rapid σB-dependent transcriptional response. Rapid induction of a transcriptional response to acid shock is consistent with previous reports that showed induction of the σB regulon after 15 min of salt stress (42) and after as little as 3 min of heat shock (61).
By comparison, the transcriptomic response to acid shock of L. monocytogenes grown to stationary phase showed no evidence for induction of stress response regulons (except for induction of the σL regulon after acid shock of stationary-phase cells grown at 7°C); bacteria grown to stationary phase at 37°C actually showed downregulation of σB and σH regulons after acid shock. Previous studies have shown that σB activity and transcription of the σB regulon are at higher levels in stationary-phase than in log-phase L. monocytogenes (31) and that L. monocytogenes cells develop increased acid resistance with entry into stationary phase (14). Our findings, thus, support a model where stationary-phase cells show high transcript levels for the σB regulon, including for σB-dependent acid response genes [e.g., gadD3 (30)], likely reflecting relatively high levels of acid response proteins, and are thus highly resistant to acid exposure (67). As their considerable acid resistance is associated with already high transcript levels of acid resistance genes, stationary-phase cells do not show induction of typical stress response regulons linked to acid resistance (e.g., the σB regulon) but rather can show reduced transcript levels of some of these regulons, possibly reflecting an adaptation to transcription of specific acid response genes or a general metabolic rearrangement of transcription rather than induction of general stress response regulons. Alternatively, growth arrest after acid shock may lead to more rapid declines in transcript levels for genes that are part of these general stress response regulons, as these genes may have short RNA half-lives (26).
L. monocytogenes cells grown to log phase at 37°C show an acid response involving multiple transcriptional regulators, which may prime the cells for intracellular survival.
While mechanisms by which L. monocytogenes adapts to acidic environments include increased expression of FoF1 ATPases pumps, decarboxylation reactions, or deiminases (52), we found that few genes encoding proteins with these H+ removal systems were induced after acid shock. For example, while a number of studies have shown that L. monocytogenes σB is important for acid stress survival (18) and transcriptional regulation of genes that are involved in acid survival (1, 30, 46), including the glutamate decarboxylase (GAD) gene gadD3 (30), none of these genes were transcribed at significantly higher levels after acid shock. Potential explanations for these observations include functional overlaps among the GAD systems found in L. monocytogenes (13, 15) and regulation of some gad genes (e.g., gadT2-gadD2 operon) by multiple transcriptional regulators (27).
Overall, L. monocytogenes grown at 37°C to log phase showed rapid induction of three stress response regulons (i.e., the σB, σH, and CtsR regulons) after acid shock treatment, including upregulation of some genes with obvious or potential roles in acid response. For example, we not only found evidence for derepression, after acid shock, of the CtsR regulon, which is comprised of type III stress genes, but also found increased transcript levels for clpP and clpE, which both belong to the CtsR regulon (38). While CtsR has been shown to be involved in heat stress survival in L. monocytogenes (28), Hu et al. (28) found no evidence that a ΔctsR deletion affects L. monocytogenes resistance to pH 2.5 acid stress. However, genes encoding class III stress proteins are transcribed at higher levels after acid shock in other Gram-positive bacteria, including Streptococcus pneumoniae (36) and Lactobacillus reuteri (65), supporting some role for CtsR in acid resistance. We also found significantly higher transcript levels for grpE after acid shock treatment of log-phase cells grown at 37°C (compared to non-acid-shocked log-phase cells grown at 37°C); grpE encodes a cochaperone for the heat shock protein DnaK (24) and appears to be regulated by both HrcA and σB (27). As these three upregulated genes (i.e., clpP, clpE, and grpE) are all involved in protein repair mechanisms and as genes in the role category “protein fate: folding and stabilization” also showed upregulation after acid shock, it appears that induction of protein repair mechanisms represents an important transcriptional response to acid shock. This is consistent with previous studies showing the contribution of chaperone proteases to acid response in Streptococcus mutans (34) and Staphylococcus aureus (6).
While reduced acid resistance of a mutant with a deletion of the HrcA-regulated gene dnaK (23) suggests a role for HrcA-regulated class I heat shock genes in L. monocytogenes acid stress response, Hu et al. (27) found no evidence that a ΔhrcA deletion affects L. monocytogenes resistance to pH 2.5 stress, consistent with our observations that the HrcA regulon, overall, showed reduced transcript levels after acid shock. Interestingly, however, the HrcA-regulated hrcA-grpE operon showed increased transcript levels after acid shock, likely due to increased transcription of this operon from an upstream σB-dependent promoter (27), as supported by the observation that the σB regulon was rapidly induced after acid shock. As HrcA is a negative regulator, increased hrcA transcription will facilitate repression of the HrcA regulon. Our data thus indicate that acid stress response in log-phase cells grown at 37°C involves both σB-dependent upregulation of a large gene set (i.e., the σB regulon) and σB-dependent downregulation of other genes (through HrcA). Interestingly, our data showed downregulation of transcript levels for the gadT2-gadD2 operon after acid shock treatment of bacteria grown to log phase at 37°C. While one initial study, using gel-based nonquantitative reverse transcription PCR (RT-PCR) (66), proposed that these genes are positively regulated by σB, subsequent microarray studies indicated negative regulation of these genes by σB (46), which is consistent with our findings that these genes are downregulated after acid shock of cells grown to log phase at 37°C even though the σB regulon is upregulated. While other studies have reported transcriptomic data that suggest (indirect) positive regulation of gadT2-gadD2 by the repressors HrcA and CtsR (27, 28), L. monocytogenes ΔhrcA and ΔctsR strains were no more susceptible to acid stress (pH 2.5, HCl) than the parent strain (27, 28). Further studies are clearly needed to clarify regulatory interactions that affect transcription of the gadT2-gadD2 operon, which plays an important role in acid resistance (13).
Additional interactions between regulatory elements also appear to contribute to acid stress response. We specifically found rapid induction of both the σB and σH regulon after acid shock treatment of L. monocytogenes grown to log phase at 37°C, consistent with a significant overlap between the L. monocytogenes σB and σH regulons (10) and proteomics data that showed increased σH levels after acid stress in L. monocytogenes (45). As a previous study (10) showed that a ΔsigH strain was significantly more resistant to pH 2.5 than the parent strain while, as expected, a ΔsigB mutant was more sensitive to acid stress than the parent strain (10), further studies on the σB and σH interactions and regulatory networks will be important to understand the combined roles of these alternative σ factors in L. monocytogenes stress response.
Interestingly, we also found clear evidence that regulons and specific genes that are involved in L. monocytogenes host invasion as well as intracellular survival and multiplication are upregulated after acid shock and particularly after acid shock of cells grown to log phase at 37°C. In addition to upregulation of the PrfA regulon, we also found upregulation of the CtsR-regulated genes clpB and clpP and of pyrimidine ribonucleotide biosynthesis genes, including pyrD and pyrAB (all after acid shock of bacteria grown to log phase at 37°C). clpB and clpP have been shown to contribute to escape from the host phagosome. ClpP was found to be necessary for functional listeriolysin O secretion (20), which is vital to escape from the primary host cell vacuole (56), while a ΔclpC strain was shown to be impaired in escape from the macrophage phagosome (50). Pyrimidine biosynthesis genes may facilitate scavenging of intracellular nutrient sources after phagocytosis, an idea that is supported by a study (32) that found that pyrE, which is cotranscribed with pyrD and pyrAB, was translated at higher levels in the intracellular environment than during growth in BHI. These findings provide potential mechanisms that may contribute to the previously observed increased survival of acid-adapted L. monocytogenes in activated macrophages (11) and suggest that coordinated upregulation of different genes during gastric passage may prime L. monocytogenes for subsequent stages of infection, including intracellular growth and survival.
The acid response of L. monocytogenes grown to stationary phase at 7°C involves upregulation of prophage genes.
Our transcriptomic analyses showed that two groups of genes encoding bacteriophage proteins were specifically induced after acid shock treatment of L. monocytogenes 10403S grown to stationary phase at 7°C. One of these regions represents 15 genes (lmo0115 to lmo0129) that include the lmaDCBA operon. This putative incomplete cryptic prophage has been identified in similar locations in the genomes of L. monocytogenes strains F2365, H7858, and F6858 (39). LmaA has previously been shown to be localized to the bacterial surface (69) and secreted into the medium at 20°C but not at 37°C (54), consistent with the temperature-dependent transcription of this operon observed here. The second prophage region that showed increased transcript levels after acid shock of stationary-phase bacteria grown at 7°C corresponds to a prophage similar to L. monocytogenes bacteriophage A118 (68). While we were able to isolate a bacteriophage with a morphology similar to that of A118 from the supernatant of 10403S cultured at 7°C, we did not observe a reproducible increase in phage titer after acid shock. Further experiments will thus be necessary to characterize the phenotypic consequences of the increased transcription of the A118 prophage genes after acid shock.
Increased levels of bacteriophage gene transcripts after short-term acid shock have been reported in at least one other organism [i.e., Lactobacillus reuteri (65)]. Prophage induction can be linked to the bacterial SOS response (63), and there is evidence that the SOS response is induced after acid shock in L. monocytogenes (62). Induction of prophage gene expression after exposure to lethal acid shock not only may facilitate phage survival but also may facilitate horizontal gene transfer and, in a mixed population, may thus represent an important mechanism for the acquisition of stress resistance genes that facilitate acid stress survival. In addition, increased expression of prophages or incomplete phages (e.g., monocins) could inhibit or kill other strains and may thus provide a competitive advantage. For example, the products of the putative incomplete (cryptic) bacteriophage operon that was induced here after acid shock has been shown to specifically inhibit the growth of other strains of L. monocytogenes (69). Further study is needed to determine whether bacteriophage gene expression is linked to the SOS response in L. monocytogenes and what role, if any, this induction plays in environmental stress survival and persistence.
Conclusion.
Our results show that adaptation to food-relevant temperatures, specifically refrigeration, can impact the phenotypic and transcriptional responses of L. monocytogenes to sudden acid shock. These data contribute to the mounting evidence that growth condition and previous environmental exposure affect how L. monocytogenes responds to stress, consistent with similar reports on other organisms, including yeast (4). Therefore, complete characterization of the stress response of a food-borne pathogen should include examination of multiple strains and multiple pregrowth conditions, including temperature and growth phase.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health award RO1-AI052151-01A1 (to K.J.B.). R.A.I. was supported by USDA National Needs Fellowship grant 2005-38420-15776.
Footnotes
Published ahead of print 23 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abram F, et al. 2008. Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74:6848–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Álvarez-Ordóñez A, Fernandez A, Bernardo A, López M. 2010. Arginine and lysine decarboxylases and the acid tolerance response of Salmonella Typhimurium. Int. J. Food Microbiol. 136: 278–282 [DOI] [PubMed] [Google Scholar]
- 2a. Audits International/FDA 1999. 1999 U.S. food temperature evaluation. U.S. Food and Drug Administration, Washington, DC. http://foodrisk.org/default/assets/File/Audits-FDA_temp_study.pdf
- 3. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Stat. Method.) 1:289–300 [Google Scholar]
- 4. Berry DB, et al. 2011. Multiple means to the same end: the genetic basis of acquired stress resistance in yeast. PLoS Genet. 7:e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhagwat AA, et al. 2003. Computational methods and evaluation of RNA stabilization reagents for genome-wide expression studies. J. Microbiol. Methods 55:399–409 [DOI] [PubMed] [Google Scholar]
- 6. Bore E, Langsrud S, Langsrud O, Rode TM, Holck A. 2007. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153:2289–2303 [DOI] [PubMed] [Google Scholar]
- 7. Chan YC, Raengpradub S, Boor KJ, Wiedmann M. 2007. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 73:6484–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaturongakul S, Boor KJ. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaturongakul S, Boor KJ. 2006. Sigma B activation under environmental and energy stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 72:5197–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chaturongakul S, et al. 2011. Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors sigma B, sigma C, sigma H, and sigma L in Listeria monocytogenes. Appl. Environ. Microbiol. 77:187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conte MP, et al. 2002. Effect of acid adaptation on the fate of Listeria monocytogenes in THP-1 human macrophages activated by gamma interferon. Infect. Immun. 70:4369–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cotter PD, Gahan CGM, Hill C. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465–475 [DOI] [PubMed] [Google Scholar]
- 13. Cotter PD, Ryan S, Gahan CGM, Hill C. 2005. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71:2832–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis MJ, Coote PJ, O'Byrne CP. 1996. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology 142:2975–2982 [DOI] [PubMed] [Google Scholar]
- 15. Dykes GA, Moorhead SM. 2000. Survival of osmotic and acid stress by Listeria monocytogenes strains of clinical or meat origin. Int. J. Food Microbiol. 56:161–166 [DOI] [PubMed] [Google Scholar]
- 16. Ehira S, Teramoto H, Inui M, Yukawa H. 2009. Regulation of Corynebacterium glutamicum heat shock response by the extracytoplasmic-function sigma factor SigH and transcriptional regulators HspR and HrcA. J. Bacteriol. 191:2964–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferreira A, O'Byrne CP, Boor KJ. 2001. Role of SigB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferreira A, Sue D, O'Byrne CP, Boor KJ. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferreira V, et al. 2011. Diverse geno- and phenotypes of persistent Listeria monocytogenes isolates from fermented meat sausage production facilities in Portugal. Appl. Environ. Microbiol. 77:2701–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gaillot O, Pellegrini E, Bregenholt S, Nair S, Berche P. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286–1294 [DOI] [PubMed] [Google Scholar]
- 21. Garner MR, James KE, Callahan MC, Wiedmann M, Boor KJ. 2006. Exposure to salt and organic acids increases the ability of Listeria monocytogenes to invade Caco-2 cells but decreases its ability to survive gastric stress. Appl. Environ. Microbiol. 72:5384–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gohmann S, Leimeister-Wachter M, Schiltz E, Goebel W, Chakraborty T. 1990. Characterization of a Listeria monocytogenes-specific protein capable of inducing delayed hypersensitivity in Listeria-immune mice. Mol. Microbiol. 4:1091–1099 [DOI] [PubMed] [Google Scholar]
- 23. Hanawa T, et al. 1999. The Listeria monocytogenes DnaK chaperone is required for stress tolerance and efficient phagocytosis with macrophages. Cell Stress Chaperones 4:118–128 [PMC free article] [PubMed] [Google Scholar]
- 24. Harrison C. 2003. GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones 8:218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hodgson DA. 2000. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 35:312–323 [DOI] [PubMed] [Google Scholar]
- 26. Hu Y, Coates AR. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 181:469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu Y, et al. 2007. Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and SigB in Listeria monocytogenes. Appl. Environ. Microbiol. 73:7981–7991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu Y, et al. 2007. Phenotypic and transcriptomic analyses demonstrate interactions between the transcriptional regulators CtsR and sigma B in Listeria monocytogenes. Appl. Environ. Microbiol. 73:7967–7980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karatzas K-AG, Brennan O, Heavin S, Morrissey J, O'Byrne CP. 2010. Intracellular accumulation of high levels of γ-aminobutyrate by Listeria monocytogenes 10403S in response to low pH: uncoupling of γ-aminobutyrate synthesis from efflux in a chemically defined medium. Appl. Environ. Microbiol. 76:3529–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. 2003. Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185:5722–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kazmierczak MJ, Wiedmann M, Boor KJ. 2006. Contributions of Listeria monocytogenes sigma B and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152:1827–1838 [DOI] [PubMed] [Google Scholar]
- 32. Klarsfeld AD, Goossens PL, Cossart P. 1994. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol. Microbiol. 13:585–597 [DOI] [PubMed] [Google Scholar]
- 33. Kress W, Maglica Z, Weber-Ban E. 2009. Clp chaperone-proteases: structure and function. Res. Microbiol. 160:618–628 [DOI] [PubMed] [Google Scholar]
- 34. Lemos JA, Chen YY, Burne RA. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074–6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lou Y, Yousef AE. 1997. Adaptation to sublethal environmental stresses protects Listeria monocytogenes against lethal preservation factors. Appl. Environ. Microbiol. 63:1252–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin-Galiano AJ, et al. 2005. Transcriptional analysis of the acid tolerance response in Streptococcus pneumoniae. Microbiology 151:3935–3946 [DOI] [PubMed] [Google Scholar]
- 37. Mehra S, Kaushal D. 2009. Functional genomics reveals extended roles of the Mycobacterium tuberculosis stress response factor σH. J. Bacteriol. 191:3965–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nair S, Derré I, Msadek T, Gaillot O, Berche P. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800–811 [DOI] [PubMed] [Google Scholar]
- 39. Nelson KE, et al. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Driscoll B, Gahan CG, Hill C. 1996. Adaptive acid tolerance response in Listeria monocytogenes: isolation of an acid-tolerant mutant which demonstrates increased virulence. Appl. Environ. Microbiol. 62:1693–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okada Y, et al. 2006. The sigma factor RpoN (sigma54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol. Lett. 263:54–60 [DOI] [PubMed] [Google Scholar]
- 42. Olesen I, Vogensen FK, Jespersen L. 2009. Gene transcription and virulence potential of Listeria monocytogenes strains after exposure to acidic and NaCl stress. Foodborne Pathog. Dis. 6:669–680 [DOI] [PubMed] [Google Scholar]
- 43. Ollinger J, Bowen B, Wiedmann M, Boor KJ, Bergholz TM. 2009. Listeria monocytogenes sigma B modulates PrfA-mediated virulence factor expression. Infect. Immun. 77:2113–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patchett RA, Watson N, Fernandez PS, Kroll RG. 1996. The effect of temperature and growth rate on the susceptibility of Listeria monocytogenes to environmental stress conditions. Lett. Appl. Microbiol. 22:121–124 [DOI] [PubMed] [Google Scholar]
- 45. Phan-Thanh L, Mahouin F. 1999. A proteomic approach to study the acid response in Listeria monocytogenes. Electrophoresis 20:2214–2224 [DOI] [PubMed] [Google Scholar]
- 46. Raengpradub S, Wiedmann M, Boor KJ. 2008. Comparative analysis of the sigma B-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74:158–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Richard HT, Foster JW, Laskin AI, Bennett JW, Gadd GM. 2003. Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 52:167–186 [DOI] [PubMed] [Google Scholar]
- 48. Robichon D, et al. 1997. The rpoN (σ54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J. Bacteriol. 179:7591–7594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rohde KH, Abramovitch RB, Russell DG. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2:352–364 [DOI] [PubMed] [Google Scholar]
- 50. Rouquette C, de Chastellier C, Nair S, Berche P. 1998. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol. Microbiol. 27:1235–1245 [DOI] [PubMed] [Google Scholar]
- 51. Russell TL, et al. 1993. Upper gastrointestinal pH in seventy-nine healthy, elderly, North American men and women. Pharm. Res. 10:187–196 [DOI] [PubMed] [Google Scholar]
- 52. Ryan S, Hill C, Gahan CGM. 2008. Acid stress responses in Listeria monocytogenes. Adv. Appl. Microbiol. 65:67–91 [DOI] [PubMed] [Google Scholar]
- 53. Samelis J, Ikeda JS, Sofos JN. 2003. Evaluation of the pH-dependent, stationary-phase acid tolerance in Listeria monocytogenes and Salmonella Typhimurium DT104 induced by culturing in media with 1% glucose: a comparative study with Escherichia coli O157:H7. J. Appl. Microbiol. 95:563–575 [DOI] [PubMed] [Google Scholar]
- 54. Schaferkordt S, Chakraborty T. 1997. Identification, cloning, and characterization of the Lma operon, whose gene products are unique to Listeria monocytogenes. J. Bacteriol. 179:2707–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schubert ML. 2009. Gastric exocrine and endocrine secretion. Curr. Opin. Gastroenterol. 25:529–536 [DOI] [PubMed] [Google Scholar]
- 56. Scortti M, Monzo HJ, Lacharme-Lora L, Lewis DA, Vazquez-Boland JA. 2007. The PrfA virulence regulon. Microb. Infect. 9:1196–1207 [DOI] [PubMed] [Google Scholar]
- 57. Stasiewicz MJ, Wiedmann M, Bergholz TM. 2010. The combination of lactate and diacetate synergistically reduces cold growth in brain heart infusion broth across Listeria monocytogenes lineages. J. Food Prot. 73:631–640 [DOI] [PubMed] [Google Scholar]
- 58. Subramanian A, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Toledo-Arana A, et al. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956 [DOI] [PubMed] [Google Scholar]
- 60. United States Department of Agriculture Food Safety and Inspection Service 2011. Basics for handling food safely. U.S. Department of Agriculture, Washington, DC. http://www.fsis.usda.gov/PDF/Basics_for_Safe_Food_Handling.pdf
- 61. van der Veen S, et al. 2007. The heat-shock response of Listeria monocytogenes comprises genes involved in heat shock, cell division, cell wall synthesis, and the SOS response. Microbiology 153:3593–3607 [DOI] [PubMed] [Google Scholar]
- 62. van der Veen S, et al. 2010. The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology 156:374–384 [DOI] [PubMed] [Google Scholar]
- 63. Waldor MK, Friedman DI. 2005. Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 8:459–465 [DOI] [PubMed] [Google Scholar]
- 64. Walker SJ, Archer P, Banks JG. 1990. Growth of Listeria monocytogenes at refrigeration temperatures. J. Appl. Microbiol. 68:157–162 [DOI] [PubMed] [Google Scholar]
- 65. Wall T, et al. 2007. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl. Environ. Microbiol. 73:3924–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wemekamp-Kamphuis HH, et al. 2004. Identification of sigma factor sigma B-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70:3457–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. 1998. General stress transcription factor sigma B and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zink R, Loessner MJ. 1992. Classification of virulent and temperate bacteriophages of Listeria spp. on the basis of morphology and protein analysis. Appl. Environ. Microbiol. 58:296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zink R, Loessner MJ, Scherer S. 1995. Characterization of cryptic prophages (monocins) in Listeria and sequence analysis of a holin/endolysin gene. Microbiology 141:2577–2584 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.