Abstract
Comparative genomic hybridization was used to compare genetic diversity of five strains of Leptospira (Leptospira interrogans serovars Bratislava, Canicola, and Hebdomadis and Leptospira kirschneri serovars Cynopteri and Grippotyphosa). The array was designed based on two available sequenced Leptospira reference genomes, those of L. interrogans serovar Copenhageni and L. interrogans serovar Lai. A comparison of genetic contents showed that L. interrogans serovar Bratislava was closest to the reference genomes while L. kirschneri serovar Grippotyphosa had the least similarity to the reference genomes. Cluster analysis indicated that L. interrogans serovars Bratislava and Hebdomadis clustered together first, followed by L. interrogans serovar Canicola, before the two L. kirschneri strains. Confirmed/potential virulence factors identified in previous research were also detected in the tested strains.
INTRODUCTION
Leptospirosis is a zoonotic disease that is found worldwide. It has gained increasing attention because of several recent outbreaks, and it has also become a major public health concern in many developing countries (13, 14, 15, 16, 17, 24, 30). Leptospirosis is caused by a range of leptospires with a broad range of hosts, showing a variety of symptoms including high fever, severe headache, chills, muscle aches, jaundice, and vomiting (7, 10, 25). So far, more than 200 strains and serovars are considered to be pathogenic (2, 17, 18).
Complete genome sequences are currently available for six Leptospira strains, including four pathogenic strains, Leptospira interrogans serovar Lai strain 56601 (23), L. interrogans serovar Copenhageni strain Fiocruz L1-130 (19), Leptospira borgpetersenii serovar Hardjo strains L550 and JB197 (3), and saprophytic Leptospira biflexa serovar Patoc I Paris and Ame strains (20). The availability of these strains has opened the window for scientists to observe and explore the genetic diversity of this species. Based on the comparison of genomes, genetic diversity was observed between saprophytic leptospira (L. biflexa) and pathogens of leptospirosis (20), between different species (L. borgpetersenii and L. interrogans) within the Leptospira genus (3), between different serovars (L. interrogans serovars Copenhageni and Lai), and within species (19). These differences were related to strain evolution events, pathogenesis, and flexibility of survival in various environments. As the leptospires are so diverse, the more genomic information that becomes available, the more the evidence collected would benefit studies on their pathogenesis, adaptation to various environments, and the development of further strategies for diagnosis and vaccine development.
Comparative genomic hybridization (CGH) has proven to be a very powerful tool for high-throughput genomic comparisons between pathogenic and nonpathogenic species and genomic screening of many bacterial species (5, 8, 9, 12, 21, 24, 30). The available genomic sequences of L. interrogans serovars Lai and Copenhageni were used to design a tiling array. In this study, genomic DNA from five strains, three from L. interrogans and two from L. kirschneri (Table 1), were applied to the array, providing a foundation for high-throughput genome screening of leptospire strains. To our knowledge, this is the first report of applying CGH to comparative genomics study on Leptospira species.
Table 1.
Leptospira strains included in this studya
| Species | Serovar | Strain | Source | Country | Sequence type |
|---|---|---|---|---|---|
| L. interrogans | Bratislava | Jes Bratislava | Hedgehog | Czechoslovakia | 24 |
| Hebdomadis | Hebdomadis | Human via guinea pig | Japan | 36 | |
| Canicola | Hon Utrecht IV | Dog | Netherlands | 37 | |
| L. kirschneri | Grippotyphosa | Moskva V | Human | USSR | 62 |
| Cynopteri | 3522C | Bat | Indonesia | 70 |
Data are from Thaipadungpanit et al. (27).
MATERIALS AND METHODS
Strain selection.
Five Leptospira reference strains, three from L. interrogans and two from L. kirschneri, were chosen as representatives in this study. These strains shown in Table 1 were isolated from different hosts and countries and belonged to different sequence types, as determined using multilocus sequence typing (MLST) (27).
Organism growth and DNA isolation.
Leptospires were grown in Ellinghausen-McCullough-Johnson-Harris (EMJH) base medium (Difco, Sparks, MD) supplemented with 1% bovine serum albumin (BSA), 1.25% Tween 80, and 2% rabbit serum at 30°C under aerobic conditions. Genomic DNA extraction was performed as previously described from approximately 100-ml cultures of leptospires grown to late exponential phase (11).
Design of experiment, microarray, and probes.
In this study, L. interrogans serovars Copenhageni and Lai were selected as reference genomes. Probes for the microarray were designed based on the reference genome sequences. Each of the reference strains has two chromosomes. They are L. interrogans Lai strain 56601 chromosome (chr) I (accession number AE010300), L. interrogans Lai strain 56601 chr II (AE010301), L. interrogans Copenhageni strain Fiocruz L1-130 chr I (AE016823), and L. interrogans Copenhageni strain Fiocruz L1-130 chr II (AE016824). The whole genome sequences for these two strains were extracted from sequence files. Each genome was split into fragments 500 bp in length, and these fragments were put through the Oxford Gene Technology's probe design pipeline with up to three probes designed to each fragment. The number of probes was then automatically reduced and optimized in silico to generate the final probe set to fit onto a 4-by-44,000 Agilent array with the aim of conserving probe coverage of at least one probe per gene.
DNA labeling, array hybridization, and data analysis.
DNAs for each sample were quantified before labeling, and quality was also checked. The readings of A260/A280 were more than 1.80 for all the samples. DNA (1.1 μg) was fragmented by sonication with a pulse of 1 s and pause of 1 s for 30 s at 20% amplitude. A total of 21 μl (1 μg) of the sonicated DNA solution was then labeled using Klenow-based incorporation of Cy dye-labeled dCTP. Twenty microliters of 20× random primers was added to each of the samples, and this step was followed by incubation at 95°C for 5 min. The samples were then placed on ice for 5 min when the labeling Master Mix was added to each sample. The Master Mix included 5 μl of 10× deoxynucleoside triphosphate (dNTP) mix, 3 μl of Cy3-dCTP (experiment) or Cy5-dCTP (reference), and 1 μl of Exo-Klenow fragment. The samples were incubated for 2 h at 37°C and then purified using Qiagen QIAquick cleanup columns. The yield and efficiency of dye incorporation (specific activity) of purified labeled DNA were then determined.
The labeled samples were vacuum concentrated and prepared for hybridization. The samples were incubated for 3 min at 94°C and 30 min at 37°C and then applied to the slides and hybridized for 24 h at 65°C and at 20 rpm. Slides were scanned using an Agilent scanner at 100% scan power. For slides and dyes for which the scanning appeared to be saturated at 100%, a lower scanning power was used for the intensity data (10%).
Data analysis.
The data were extracted using Agilent feature extraction 9.5.3. Channels were selected from all slides that had a median intensity across all features of greater than 500 at 100% scan power. Foreground and background intensities for each of these arrays were taken from the featured extracted data and paired across all combinations as if they were the two colors on a normal array. Applied background subtraction and dye normalization (Loess) were performed in exactly the same manner as would be done for a normal two-color array. The fold changes by genomic location for visual identification of patterns of segmentation were plotted.
Principal-component analysis (PCA) was conducted using SYSTAT, version 10, software (SPSS, Inc., Chicago, IL). PCA was employed to evaluate the relationships among strains tested based on microarray hybridization signal intensities.
RESULTS AND DISCUSSION
The raw microarray data of five Leptospira strains were processed with extraction of background, removal of outliers, and normalization using a global Loess method. Probes for the tiling array were designed to cover the entire genome. Therefore, in the tiling array, each gene was designed into 1 to 27 probes based on the length of the gene. A total of 43,235 probes for the L. interrogans serovar Copenhageni and Lai genomes were designed. Five strains (from L. interrogans serovars Bratislava, Canicola, and Hebdomadis and L. kirschneri serovars Grippotyphosa and Cynopteri) from two Leptospira species (Table 1) were applied to the tiling array. The three serovars (L. interrogans serovars Bratislava, Canicola, and Hebdomadis) from the same species as the reference genome detected much higher percentages of the probes (96.23%, 95.45%, and 94.28%, respectively) than the two L. kirschneri serovars Grippotyphosa and Cynopteri (64.54% and 67.84%, respectively). In addition, the genomes of the five strains tested had higher similarity to the reference genome of L. interrogans serovar Copenhageni than to that of L. interrogans serovar Lai (data not shown).
Determination of genes present, absent, and partially present was conducted. If all probes representing one gene had hybridization intensities equal to or greater than 500, then this gene was considered to be present in the tested strains; likewise, if the signal intensities of all probes representing one gene were all less than 500, this gene was considered to be absent; if the signal intensity of one or more than one of the probes representing one gene was less than 500, this gene was considered partially present (or partially absent).
Analysis based on gene function categories.
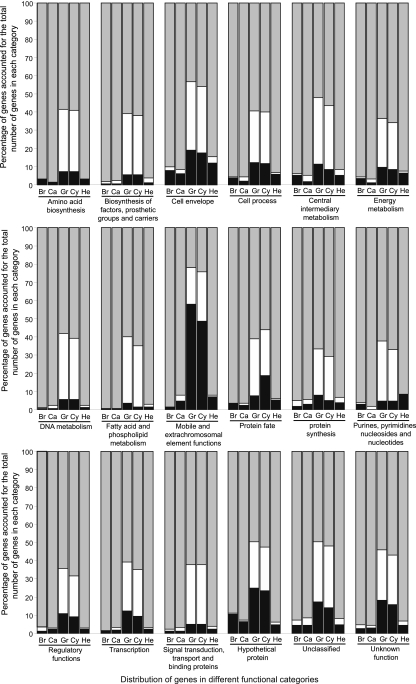
Based on the definition above, the percentage of present, absent, and partially present genes in each functional category (based on published annotation) for each strain was determined (Fig. 1). The known functional diversity showed a broad conservation in L. interrogans serovars Bratislava, Canicola, and Hebdomadis, with 90 to 99.29% of reference genes detected in each functional category, while the highest percentages of reference genes detected in L. kirschneri serovars Grippotyphosa and Cynopteri were 66.99% and 70.83%, respectively, for protein synthesis; the lowest percentages were 21.88% and 24.22%, respectively, for the mobile and extrachromosomal element functions (MEEF). Transposases were predominant in the MEEF category, and a new insertion (IS) element, ISlin1, was identified in L. interrogans serovar Copenhageni; IS1500, IS1501, and IS1533 were discovered previously (19). IS1500 was detected in all tested strains. Other IS elements were present in L. interrogans serovars Bratislava, Canicola, and Hebdomadis, and they were either absent or partially present in L. kirschneri serovar Grippotyphosa; only 4 of 31 ISlin1 elements were present in L. kirschneri serovar Cynopteri. Transposases contributed to creating genetic diversity within species and adaptability to changing living conditions. This suggested that the two L. kirschneri serovars Grippotyphosa and Cynopteri might have less genetic diversity than the three serovars from L. interrogans.
Fig 1.
Distribution of genes in different functional categories for the five tested strains. The y axis shows the percentage of genes relative to the total number of genes in each category. Gray, present genes; white, partially present genes; black, absent genes. Br, L. interrogans serovar Bratislava; Ca, L. interrogans serovar Canicola; He, L. interrogans serovar Hebdomadis; Gr, L. kirschneri serovar Grippotyphosa; Cy, L. kirschneri serovar Cynopteri.
Comparison of tested strains to reference genomes.
Compared with the reference genomes, the percentage of genes present in the tested serovars varied from 51.23% (L. kirschneri serovar Grippotyphosa) to 95% (L. interrogans serovar Bratislava), whereas the percentages of partially similar genes ranged from 1.70% (L. interrogans serovar Canicola) to 27.90% (L. kirschneri serovar Grippotyphosa), and the percentages of absent genes ranged from 3.82% (L. interrogans serovar Bratislava) to 20.87% (L. kirschneri serovar Grippotyphosa) (Table 2). This result suggests that L. interrogans serovar Bratislava is the closest to the reference genome of L. interrogans serovar Copenhageni and that L. interrogans serovar Lai and L. kirschneri serovar Grippotyphosa have the least similarity to L. interrogans serovar Copenhageni.
Table 2.
Percentage of genes detected in five serovars based on the reference genomes
| Gene status | Genome similarity (% of genes)a |
||||
|---|---|---|---|---|---|
|
L. interrogans serovar |
L. kirschneri serovar |
||||
| Bratislava | Canicola | Hebdomadis | Grippotyphosa | Cynopteri | |
| Present | 95 | 93.97 | 92.83 | 51.23 | 54.09 |
| Partially present | 1.18 | 1.70 | 1.81 | 27.90 | 27 |
| Absent | 3.82 | 4.33 | 5.35 | 20.87 | 18.91 |
The percentage was determined relative to that of the reference genome.
Comparison among tested strains.
A total of 3,957 genes were detected in all five tested strains. Of these genes, 54.18% belonged to unclassified, hypothetical, or unknown functions or were unassigned any function; the rest of the genes were predominantly housekeeping genes involved in transport and binding, regulatory functions, transcription, purines, pyrimidines, nucleosides and nucleotides, protein synthesis, protein fate, energy metabolism, central intermediary metabolism, DNA metabolism, cellular processes, cell envelope, amino acid biosynthesis and biosynthesis of cofactors, and prosthetic groups, most of which were consistent with the core leptospiral genes resulting from the comparison of genomes between the saprophyte L. biflexa and pathogenic species L. interrogans and L. borgpetersenii (3, 19, 20, 23). Eighty-four genes were not detectable in all five strains, and these genes did not have functions assigned. The number of genes unique to each tested strain (except for the L. kirschneri serovar Grippotyphosa strain) was 120, 78, 30, and 4 for L. interrogans serovars Bratislava, Canicola, and Hebdomadis and L. kirschneri serovar Cynopteri strains, respectively. Genes unique to L. interrogans and L. kirschneri were also observed. A total of 993 genes were detected in the three strains of L. interrogans while only five genes without assigned functions were detected in the two strains of L. kirschneri. Of the 993 genes, only 183 had known functions, and these were dominated by genes involved in MEEF (47 transposase) and cell envelope (44 lipoproteins and membrane proteins), which are involved in nutrition and signal transduction. In addition, three genes responsible for fruiting body development for long-term survival (28) were shown only in the L. interrogans species. This observation suggests that L. interrogans might better adapt to multiple environments (3, 19, 20) than the L. kirschneri species.
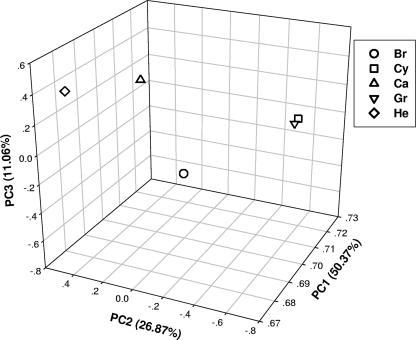
PCA and clustering analysis.
PCA based on signal intensities was used to group and separate strains with similar or dissimilar genetic properties. The results showed that the two L. kirschneri serovars Grippotyphosa and Cynopteri were closely grouped, separated from the three L. interrogans serovars (Fig. 2). Clustering analysis based on microarray signal intensities showed that the cluster formed into two groups, I and II (data not shown). In group I, L. kirschneri serovars Grippotyphosa and Cynopteri were closely clustered together and then clustered with group II from L. interrogans; L. interrogans serovars Bratislava and Canicola grouped first, followed by L. interrogans serovar Hebdomadis. The PCA and clustering results combined showed that strains within species were genetically closer than those from across species. In addition, there are controversies in previous publications about L. kirschneri serovar Grippotyphosa taxonomy. Yasuda et al. (32) assigned L. kirschneri serovar Grippotyphosa to L. interrogans while Ramadass et al. (22) assigned it to L. kirschneri. Our result based on tiling arrays, which covered most of the genome, supported that L. kirschneri serovar Grippotyphosa should be assigned to L. kirschneri instead of L. interrogans, consistent with the most recent result (22) based on DNA hybridization.
Fig 2.
PCA analysis for all five serovars based on microarray hybridization intensity. L. kirschneri serovars Grippotyphosa and Cynopteri were closely grouped together, separated from the three L. interrogans serovars Bratislava, Canicola, and Hebdomadis.
Pathogenic factors. (i) Confirmed or potential factors.
Genes identified to be responsible for pathogenesis were also observed in the tested strains (Table 3). ligA, ligB, and ligC previously reported to be probably involved in host-pathogen interactions (19) were present or partially present in the tested strains except those of L. kirschneri; ligB and ligC were present in three L. interrogans strains while ligB was partially present in L. kirschneri strains; ligA was present in one L. kirschneri serovar Cynopteri strain and partially present in all other tested strains. This is consistent with previous results of McBride et al., who used L. interrogans serovar Canicola strain Kito. The L. interrogans serovar Canicola used in this study had 90.3%, 96.7%, and 98.5% DNA sequence identity of ligA, ligB, and ligC genes, respectively, with those from L. interrogans serovar Copenhageni while L. kirschneri serovar Grippotyphosa had 91.4%, 93.2%, and 90.5%, respectively, sequence identity.
Table 3.
Summary of previously confirmed/potential virulence factors detected in the five tested serovars
| ORF | Name or identification | No. of probes on the array | Presence in L. interrogans serovara |
Presence in L. kirschneri serovara |
Protein | |||
|---|---|---|---|---|---|---|---|---|
| Bratislava | Canicola | Hebdomadis | Grippotyphosa | Cynopteri | ||||
| LIC13101 | 15 | P | U | P | U | U | Outer membrane protein with alpha-integrin-like repeat domains | |
| LIC10021 | 7 | P | P | P | P | P | Outer membrane protein with alpha-integrin-like repeat domains | |
| LIC12259 | 9 | P | P | P | U | U | Cytoplasmic membrane protein | |
| LA1499 | 13 | P | U | U | U | U | FG-GAP repeat domain protein | |
| LA3881 | 9 | P | P | P | U | U | Transposase, putative | |
| LIC13414 | 6 | P | P | P | P | P | LipL45-like protein | |
| LIC10123 | 4 | P | P | P | P | P | LipL45-like protein | |
| LIC20114 | 7 | P | P | P | P | P | LipL45 homologue | |
| LIC11643 | 4 | P | P | P | P | P | LipL45 | |
| LIC13060 | lipL36 | 12 | P | A | P | U | U | LipL36 |
| LIC10011 | lipL21 | 6 | P | P | P | P | P | LipL21 |
| LIC12690 | Lp95 | 9 | P | P | A | A | A | Putative lipoprotein |
| LIC10314 | lsa63 | 9 | P | P | P | P | P | Conserved hypothetical protein |
| LIC10465 | ligA | 23 | U | U | U | U | U | Ig-like repeat domain protein 1 |
| LIC10464 | ligB | 21 | P | P | P | U | U | Ig-like repeat domain protein 3 |
| —b | ligC | 16 | P | P | P | A | U | |
| LIC12575 | 5 | P | P | P | P | P | TolC-like protein | |
| LIC13050 | 4 | P | P | P | P | P | Putative glycosyl hydrolase | |
| LIC10191 | Loa22 | 4 | P | P | P | P | P | OmpA family lipoprotein |
| LIC12032 | 8 | P | P | P | P | P | Catalase | |
| LIC13166 | 3 | P | P | P | P | P | Putative coagulase | |
P, present; U, partially present; A, absent.
Not identified.
Three integrin alpha-like proteins (LIC12259, LIC10021, an LIC13101) from L. interrogans serovar Copenhageni and three (LA1499, LA0022, and LA3881) from L. interrogans serovar Lai were identified as candidates of leptospiral adhesins (19). Except for LA0022 missing on the microarray, others were present or partially present in all tested strains (Table 3).
Eshghi et al. (6) compared global proteome analyses on L. interrogans serovar Copenhageni grown under conventional in vivo conditions and growth mimicking in vivo conditions. Four novel proteins (LIC12575, LIC13050, LIC12032, and LIC13166) and related virulent factors were identified, which were present in all five tested strains (Table 3). The lipoproteins LipL21, LipL45, and LipL36 were unique in pathogenic Leptospira based on the sequenced strains (20). LipL21 and LipL45 were present in all tested strains; however, LipL36 varied among the serovars as it was present in L. interrogans serovars Bratislava and Hebdomadis, partially present in L. kirschneri serovars Grippotyphosa and Cynopteri, but absent in L. interrogans serovar Canicola (Table 3).
Two proteins, Lsa63 (LIC10314) and Lp95 (LIC12690), were observed in all strains tested; they were confirmed to bind laminin and collagen (29) and extracellular matrix components (1), respectively, which were related to invasion of the hosts.
(ii) Possible pathogenic factors.
The genomic comparison between saprophyte L. biflexa and the pathogens of leptospirosis (L. borgpetersenii and L. interrogans) showed that 1,431 genes were unique to the pathogens. These genes may be playing a role in pathogenesis since there were no orthologous genes in L. biflexa (20). The array used in this study contained 1,083 of 1,431 genes, and only 323 genes had assigned functions. A clustering analysis for the 323 genes based on genes present, partially present, and absent in the tested strains was performed; for this, present was replaced by 1, partially present was assigned a value of 0.5, and absent was assigned a value of 0. Five clusters were formed (see Fig. S1 in the supplemental material). In cluster I (see Fig. S1a), the present genes varied among strains, probably related to the survival in the environment for different strains; in clusters II and III (see Fig. S1b and c), genes were present in all the strains tested or partially present in the L. kirschneri serovars Grippotyphosa and Cynopteri, in which sphingomyelinase, phage-related protein, leucine-rich repeat protein, and methylase/methyltransferase were reported to be related to pathogenesis; in clusters IV and V (see Fig. S1d), genes were either present only in L. interrogans serovars Bratislava, Canicola, and Hebdomadis (cluster IV) or present only in L. interrogans serovar Canicola (cluster V). The pathogenic roles of most of these genes, even with assigned functions, were not clear; however, the data based on CGH microarray provided basic genomic information that can become the references for further study on Leptospira.
A tiling array was used in this study, which was designed to cover the whole genome. The advantage of a tiling array compared to an expression array is that the tiling includes not only open reading frames (ORFs) but also intragenic DNA fragments, potentially providing more information. In addition, on the tiling array, a gene can be designed with more probes according to the gene size, thereby allowing identification of genes which were present, partially present, and absent in the tested strains confidently. We proposed the concept partially present in this study so that the gene variation during strain evolution could be identified. Furthermore, genes reported to be involved in pathogenesis were observed in all the five strains. However, a limitation of the application of an array based on the reference genome is that the unique genes, which existed in the tested strains instead of in the reference genomes, cannot be detected because they were not on the array. Our results also showed that the tiling CGH array could clearly distinguish species and identified the differences of genetic content for each strain. Thus, the tiling CGH array designed for this study is appropriate to conduct high-throughput genome screens for Leptospira.
Supplementary Material
ACKNOWLEDGMENTS
We thank Duangjai Suwanchareon at the National Institute of Animal Health, Department of Livestock and Development, Ministry of Agriculture and Cooperatives, Bangkok, Thailand, for providing Leptospira strains.
This work was supported by National Institutes of Health grant SO6GM0816-37.
Footnotes
Published ahead of print 17 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Atzingen MV, et al. 2009. Lp95, a novel leptospiral protein that binds extracellular matrix components and activates e-selectin on endothelial cells. J. Infect. 59:264–276 [DOI] [PubMed] [Google Scholar]
- 2. Bharti AR, et al. 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 3:757–771 [DOI] [PubMed] [Google Scholar]
- 3. Bulach DM, et al. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. U. S. A. 103:14560–14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reference deleted.
- 5. Dziejman M, et al. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. U. S. A. 99:1556–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eshghi APA, Cullen Cowen L, Zuerner RL, Cameron CE. 2009. Global proteome analysis of Leptospira interrogans. J. Proteome Res. 8:4564–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Faine S. 1999. Leptospira and leptospirosis. MediSci, Melbourne, Australia [Google Scholar]
- 8. Fitzgerald JR, Musser JM. 2001. Evolutionary genomics of pathogenic bacteria. Trends Microbiol. 9:547–553 [DOI] [PubMed] [Google Scholar]
- 9. Fukiya S, Mizoguchi H, Tobe T, Mori H. 2004. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 186:3911–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gouveia EL, et al. 2008. Leptospirosis-associated severe pulmonary hemorrhagic syndrome, Salvador, Brazil. Emerg. Infect. Dis. 14:505–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grimberg J, Maguire S, Belluscio L. 1989. A simple method for the preparation of plasmid and chromosomal E. coli DNA. Nucleic Acids Res. 17:8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huyghe A, Francois P, Schrenzel J. 2009. Characterization of microbial pathogens by DNA microarrays. Infect. Genet. Evol. 9:987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson MA, et al. 2004. Environmental exposure and leptospirosis, Peru. Emerg. Infect. Dis. 10:1016–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karande S, et al. 2003. An observational study to detect leptospirosis in Mumbai, India, 2000. Arch. Dis. Child. 88:1070–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ko AI, Galvao MR, Ribeiro CM, Johnson WD, Riley LW. 1999. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet 354:820–825 [DOI] [PubMed] [Google Scholar]
- 16. LaRocque RC, et al. 2005. Leptospirosis during dengue outbreak, Bangladesh. Emerg. Infect. Dis. 11:766–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levett PN. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McBride AJA, et al. 2009. Genetic diversity of the Leptospiral immunoglobulin-like (Lig) genes in pathogenic Leptospira spp. Infect. Genet. Evol. 9:196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nascimento AL, et al. 2004. Genome features of Leptospira interrogans serovar Copenhageni. Braz. J. Med. Biol. Res. 37:459–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Picardeau M, et al. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3:e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pritchard L, et al. 2009. Microarray comparative genomic hybridisation analysis incorporating genomic organisation, and application to enterobacterial plant pathogens. PLoS Comput. Biol. 5:e1000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramadass P, Jarvis BD, Corner RJ, Penny D, Marshall RB. 1992. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int. J. Syst. Bacteriol. 42:215–219 [DOI] [PubMed] [Google Scholar]
- 23. Ren SX, et al. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888–893 [DOI] [PubMed] [Google Scholar]
- 24. Salama N, et al. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. U. S. A. 97:14668–14673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sarkar U, et al. 2002. Population-based case-control investigation of risk factors for leptospirosis during an urban epidemic. Am. J. Trop. Med. Hyg. 66:605–610 [DOI] [PubMed] [Google Scholar]
- 26. Reference deleted.
- 27. Thaipadungpanit J, et al. 2007. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl. Trop. Dis. 1:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thony-Meyer L, Kaiser D. 1993. devRS, an auto regulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J. Bacteriol. 175:7450–7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vieira ML, et al. 2010. Lsa63, a newly identified surface protein of Leptospira interrogans binds laminin and collagen IV. J. Infect. 60:52–64 [DOI] [PubMed] [Google Scholar]
- 30. Wolfgang MC, et al. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:8484–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reference deleted.
- 32. Yasuda PH, et al. 1987. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int. J. Syst. Evol. Microbiol. 37:407–415 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.