Abstract
This research was initiated to search for novel antimicrobial compounds produced by food or environmental microorganisms. A new bacterial strain, designated OSY-SE, which produces a unique and potent antimicrobial agent was isolated from soil. The isolate was identified as a Paenibacillus sp. through cultural, biochemical, and genetic analyses. An antimicrobial compound was extracted from Paenibacillus OSY-SE with acetonitrile and purified using liquid chromatography. After analyses by mass spectrometry (MS) and nuclear magnetic resonance (NMR), the antimicrobial compound was determined to be a cyclic lipopeptide consisting of a C15 fatty acyl (FA) chain and 13 amino acids. The deduced sequence is FA-Orn-Val-Thr-Orn-Ser-Val-Lys-Ser-Ile-Pro-Val-Lys-Ile. The carboxyl-terminal Ile is connected to Thr by ester linkage. The new compound, designated paenibacterin, showed antagonistic activities against most Gram-positive and Gram-negative bacteria tested, including Listeria monocytogenes, methicillin-resistant Staphylococcus aureus, Escherichia coli O157:H7, and Salmonella enterica serovar Typhimurium. Paenibacterin is resistant to trypsin, lipase, α-glucosidase, and lysozyme. Its antimicrobial activity was lost after digestion by pronase and polymyxin acylase. Paenibacterin is readily soluble in water and fairly stable to exposure to heat and a wide range of pH values. The new isolate and its antimicrobial agent are being investigated for usefulness in food and medical applications.
INTRODUCTION
The emerging resistance of pathogenic bacteria to antibiotics poses a serious health challenge. Resistance of Staphylococcus aureus to methicillin (38), Pseudomonas aeruginosa and Clostridium difficile to fluoroquinolones (22, 31), and Salmonella serovars to multiple drugs (5) are examples of this challenge. The rate at which new antimicrobial agents are being discovered and approved for use does not match the rate at which the currently used antibiotics lose efficacy. This discrepancy makes it urgent to search for new potent and safe antimicrobial agents. The natural environment remains an important reservoir for microorganisms capable of producing potent antimicrobials (6). Advances in sensitivity testing, material separation, and chemical structure elucidation facilitate the discovery of novel antimicrobials from natural sources.
Interest in Paenibacillus spp. as a source of new antimicrobials has been increasing. These spore-forming species are widely distributed in the environment, and some are involved in plant growth promotion and nitrogen fixing (28, 36, 44, 46). Although Paenibacillus larvae causes disease in larval honeybees, no Paenibacillus species has been reported as a human pathogen (18). Strains of Paenibacillus spp. produce diverse antimicrobial agents, including lantibiotics (21), lipopeptides (34), and macrolide (48). Lipopeptides are nonribosomally synthesized compounds which are active against a wide range of bacteria, fungi, and oomycetes (39). In addition, lipopeptides can act as antiviral and antitumor agents, immunomodulators, or specific toxin and enzyme inhibitors (40).
The goal of this study was to screen food and environmental samples for microorganisms producing antimicrobial agents that are effective against human pathogens. Here we report a new bacterial isolate, designated OSY-SE, which produces an antimicrobial agent that has not been reported previously. The new compound exhibits antimicrobial activities against both Gram-positive and Gram-negative bacteria.
MATERIALS AND METHODS
Abbreviations.
TSBYE, tryptic soy broth supplemented with 0.6% yeast extract; MS, mass spectrometry; MALDI-TOF MS, matrix-assisted laser desorption ionization time-of-flight mass spectrometry; MS/MS, tandem mass spectrometry; 1D and 2D, one and two dimensional; HMBC, heteronuclear multiple-bond correlations; HSQC, heteronuclear single-quantum correlations; NMR, nuclear magnetic resonance; NOE, nuclear Overhauser effect; NOESY, NOE spectroscopy; TOCSY, total correlation spectroscopy; GC-MS, gas chromatography-mass spectrometry; LC-MS/MS, liquid chromatography-tandem mass spectrometry.
Strain screening.
Soil and food samples were collected and screened for microorganisms that produce antimicrobial agents. Soil samples were collected from different locations in Columbus, OH. Food samples were purchased from local food stores (Columbus, OH); these included vegetables (Korean-style fermented vegetables and Chinese-style pickled vegetables), fermented soybeans, and cheeses (Mozzarella, Emmenthaler, Bleu d'Auvergne, Cheddar, and Wisconsin Munster). Briefly, samples were suspended in 0.1% sterile peptone water and homogenized using a blender or a stomacher. For soil samples, suspensions were serially diluted and aliquots (100 μl) were spread-plated on soil-extract agar (19) and dilute nutrient agar (23). Inoculated agar plates were incubated at 25°C for 2 to 8 weeks. Several hundred isolates were screened for the ability to produce antimicrobial agents, as follows. Colonies were transferred to tryptose agar in triplicate and incubated at 30°C for several days. The incubated tryptose agar plates were overlaid with soft-agar medium seeded with Listeria innocua ATCC 33090 or Escherichia coli K-12. The plates were incubated at 37°C overnight and checked for evidence of antimicrobial activities against the indicator bacteria. For food samples, the homogenates were serially diluted and passed through a hydrophobic grid membrane filter with a pore size of 0.45 μm (Iso-grid; Neogen Corporation, Lansing, MI). The membranes were mounted on tryptose agar, and the plates were incubated at 30°C for 48 h. Membranes with colonies were transferred to fresh tryptose agar plates and held at 4°C. The incubated plates, from which the membranes had been removed, were overlaid with soft-agar medium seeded with L. innocua ATCC 33090 or E. coli K-12. The overlaid plates were incubated at 37°C overnight and inspected for the presence of inhibition zones. Isolates which corresponded to these inhibition zones were located on the hydrophobic grid membranes and streaked onto tryptose agar. A soil sample yielded an isolate (OSY-SE) that produces potent antimicrobial agents.
Cultures and media.
Tryptose agar was used for propagation of OSY-SE. For stock preparation, the culture was incubated overnight in tryptic soy broth supplemented with 0.6% yeast extract (TSBYE). Incubated cultures were mixed with glycerol (final concentration, 20%) and stored at −80°C.
Strain identification.
The morphological characteristics of OSY-SE isolate were examined after Gram staining, spore staining with malachite green, and scanning electron microscopy. For scanning electron microscopy examination (27), cells of OSY-SE were incubated in TSBYE overnight, harvested by centrifugation, washed three times with phosphate buffer (0.05 M, pH 7.0), resuspended in fixative (2.5% glutaraldehyde in 0.1 M phosphate buffer with 0.1 M sucrose, pH 7.4) and held at 4°C overnight. The cells were separated from the mixture by centrifugation and resuspended in phosphate buffer. The cell suspension was filtered through a 0.22-μm microbial filter (Millipore Corp., Bedford, MA), and bacteria on the filter were postfixed for 1 h in 1% osmium tetroxide. After dehydration using an ascending series of ethanol solutions, the ethanol was substituted with an ascending series of hexamethyldisilazane in ethanol, and the filter was air-dried. Subsequently, bacteria were coated with a thin layer of gold-palladium using a Cressington 108 sputter coater (Ted Pella Inc., Redding, CA) and examined under a scanning electron microscope (Nova NanoSEM 400; FEI, Hillsboro, OR). The accelerating voltage was 5 kV, and images were collected digitally from the emitted secondary electron signal.
Isolate identity was determined by sequencing its 16S rRNA gene (10). Briefly, genomic DNA of the isolate was extracted using a commercial DNA extraction kit (DNeasy blood and tissue kit; Qiagen, Valencia, CA). Universal primers specific for bacterial 16S rRNA genes (47) were used to amplify the corresponding gene. The targeted DNA sequence was amplified in a thermocycler as follows. After an initial 3-min incubation at 94°C, the reaction mixture was subjected to 30 cycles, each including 1 min at 94°C, 1 min at 52°C, and 2 min at 72°C. The final extension was performed at 72°C for 10 min. Amplified 16S rRNA genes was purified using a commercial DNA extraction kit (QIAquick gel extraction kit; Qiagen, Valencia, CA). Resultant DNA was ligated with a commercial vector (pGEM-T Easy; Promega Corporation, Madison, WI) and electrotransformed into E. coli DH5α cells. Recombinant plasmid was extracted from the overnight culture of E. coli DH5α using a kit (QIAprep spin miniprep; Qiagen, Valencia, CA) and sequenced with an automated DNA analyzer (Applied Biosystems, Foster City, CA). The resultant DNA sequence was compared to known bacterial sequences in the National Center for Biotechnology Information database (NCBI GenBank) using the Basic Local Alignment Search Tool (BLAST) algorithm.
Biochemical tests were conducted to confirm isolate identity, including catalase, oxidase, nitrate reduction, production of acetylmethylcarbinol, dihydroxyacetone, and indole, deamination of phenylalanine, and hydrolysis of starch and casein (17). Two commercial biochemical test kits (API 50CH and API 20E test strips; bioMérieux, Inc., Durham, NC) were also used to characterize the new isolate. The results were recorded after the inoculated kit wells had been incubated at 30°C for 24 and 48 h, and identification was done by referring to the database provided by the kit manufacturer.
Isolation and purification of antimicrobial agents.
The isolate OSY-SE was streaked onto tryptose agar plates and incubated at 37°C for 4 days. The colonies were scraped into a centrifuge tube, mixed with acetonitrile, and agitated at 200 rpm for 30 min. The mixture was then centrifuged at 7,710 × g for 15 min. The supernatant, which contained antimicrobial agents, was collected and evaporated in a chemical hood. The resulting powder was dissolved in 2 ml distilled water followed by filtration (0.22 μm; Millipore, Carrigtwohill, County Cork, Ireland). The solution (crude extract) was applied to a high-performance liquid chromatography (HPLC) system (Hewlett Packard 1050; Agilent Technologies, Palo Alto, CA) for purification. The purification was achieved using a reverse-phase column (Biobasic C18, 250 by 4.6 mm, 5-μm particle size; Thermo Electron Corp., Bellefonte, PA) by linear gradient elution. The mobile phases consisted of acetonitrile (ACN) with 0.1% trifluoroacetic acid (TFA) (mobile phase A) and HPLC-grade water containing 0.1% TFA (mobile phase B). Each run included loading a 40-μl extract to the column and separation by a linear gradient of 0 to 70% ACN over 20 min at a flow rate of 1 ml/min. Elution was monitored using a UV detector set at 220 nm. Fractions from multiple runs were collected and combined for an antimicrobial activity assay. Purified antimicrobial agents were stored at 4°C until analyzed.
Antimicrobial activity determination.
The spot-on-lawn method (21) was used for the bioassay of antimicrobial activity. Bacterial indicators were incubated at 37°C for 24 h, except Pseudomonas putida, Clostridium difficile, and methicillin-resistant Staphylococcus aureus, which were incubated for 48 h (Table 1). The indicator overlay was prepared by pouring 10 ml soft agar (seeded with 10 μl indicator culture) onto tryptose agar as the basal medium in a petri dish. Purified antimicrobials were 2-fold serially diluted, and aliquots (10 μl each) were spotted onto the soft agar. The spotted plates were incubated overnight and inspected for the presence of inhibition zones. Antimicrobial activity was expressed in arbitrary units (AU)/ml; this value is the reciprocal of the highest dilution displaying a zone of inhibition corresponding to 1 ml of the nondiluted antimicrobial preparation.
Table 1.
Relative antimicrobial activity of purified paenibacterin against selected Gram-positive and Gram-negative bacteria
| Straina | Broth mediumd | Antimicrobial activity (AU/ml)e |
|---|---|---|
| Gram-negative bacteria | ||
| Escherichia coli K-12 | LB | 3,200 |
| E. coli O157:H7 EDL 933 | LB | 1,600 |
| E. coli O157:H7 ATCC 43889 | LB | 1,600 |
| Pseudomonas putida ATCC 45491 | TSBYE | 400 |
| Salmonella enterica serovar Typhimurium | TSBYE | 400 |
| S. enterica serovar Typhimurium DT 109 | TSBYE | 400 |
| S. enterica serovar Enteritidis | TSBYE | 800 |
| Yersinia enterocolitica | TSBYE | 1,600 |
| Gram-positive bacteria | ||
| Bacillus cereus ATCC 14579 | TSBYE | 800 |
| B. cereus ATCC 11178 | TSBYE | 200 |
| Clostridium difficile A515b | BHIYE | 200 |
| C. difficile CL148c | BHIYE | 0 |
| Enterococcus faecalis ATCC 29212 | MRS | 0 |
| Listeria monocytogenes Scott A | TSBYE | 800 |
| L. monocytogenes OSY-8578 | TSBYE | 1,600 |
| L. innocua ATCC 33090 | TSBYE | 1,600 |
| Lactobacillus plantarum ATCC 8014 | MRS | 400 |
| L. lactis ATCC 11454 | MRS | 800 |
| Staphylococcus aureus ATCC 6538 | TSBYE | 100 |
| S. aureus (methicillin resistant) | TSBYE | 100 |
Unless otherwise indicated, strains were obtained from the culture collection of The Ohio State University food safety laboratory.
Obtained from J. T. Lejeune, College of Veterinary Medicine, The Ohio State University.
Obtained from W. A. Gebreyes, Department of Veterinary Preventive Medicine, The Ohio State University.
LB, Luria-Bertani medium; TSBYE, tryptic soy broth supplemented with 0.6% yeast extract; MRS, Lactobacillus MRS broth; BHIYE, brain heart infusion supplemented with 5% yeast extract (41).
Reciprocal of the highest dilution displaying a zone of inhibition corresponding to 1 ml of the undiluted antimicrobial preparation.
Sensitivity to heat, pH, and enzymes.
Extracts of OSY-SE were tested for sensitivity to heat and pH change. For the thermal stability test, cell extracts were exposed to 37°C, 55°C (in incubators) or 80°C (in a water bath) for 24 h or autoclaved at 121°C for 5 min. For the pH stability test, the extracts were diluted with 25 mM phosphate buffer (pH 7.0) and adjusted to pH 3.0, 5.0, and 9.0, followed by incubation for 12 h. Treated extracts were neutralized to pH 7.0 before being tested for remaining antimicrobial activity. Purified antimicrobial compounds (∼1 mg/ml) were tested for sensitivity to selected enzymes, including trypsin (type I; 12,705 U/mg), lipase (type I; 9 U/mg), pronase (6.31 U/mg), α-glucosidase (type I; 50 U/mg), lysozyme (46,400 U/mg), and polymyxin acylase (16 U/mg). All enzymes were purchased from Sigma (St. Louis, MO) except polymyxin acylase (Wako Chemicals USA, Inc., Richmond, VA). Polymyxin acylase was prepared at a concentration of 0.2 mg/ml in 50 mM phosphate buffer (pH 8.0), whereas other enzymes were prepared at 1.0 mg/ml in 50 mM phosphate buffer (pH 7.0). Equal volumes of enzyme solution and antimicrobial compound (20 μl each) were mixed and incubated at 37°C for 10 h. A quantitative spot-on-lawn bioassay was used to measure the remaining antimicrobial activities after these treatments.
Alkaline hydrolysis.
A mild alkaline hydrolysis was used to open a potential lactone linkage within the purified peptide (50). The peptide was dissolved in 1 M NaOH and held at 25°C for 12 h. After acidification, the solution was desalted by a peptide desalting trap (Michrom Bioresources Inc., Auburn, CA), and the ring-opened compound was analyzed by MALDI-TOF MS and MS/MS as described below.
MALDI-TOF MS analysis.
MALDI-TOF MS analysis was performed on a mass spectrometer (Bruker Reflex III time-of-flight spectrometer; Bruker Daltonics Inc., Billerica, MA). Briefly, a sample of the purified antimicrobial compound was mixed with a matrix at a ratio of 1:5. The matrix was α-cyano-4-hydroxy cinnamic acid, prepared as a saturated solution in 50% acetonitrile with 0.1% TFA in water. The mixture was then spotted (1 μl) on the target plate and allowed to air dry. The instrument was operated in reflection-positive ion mode at an accelerating voltage of 28 kV. The N2 laser was operated at the minimum threshold level required to generate signal and minimize dissociation.
Quadrupole–time-of-flight MS/MS.
The MS/MS analysis was performed on a Micromass Q-Tof II apparatus (Micromass, Wythenshawe, United Kingdom) equipped with an orthogonal electrospray source (Z-spray) and operated in positive-ion mode. The instrument was calibrated with the angiotensin fragment prior to use. A sample of purified antimicrobial agent, diluted in a mixture of H2O-ACN-HAc (50:50:2.5), was infused into the electrospray source at a 2-μl/min flow rate. To achieve the optimal electrospray, the capillary voltage was set at 3 kV, the source temperature was 100°C, and the cone voltage was 40 V. The first quadrupole, Q1, was set to pass ions between 200 and 2500 m/z. The target ion was isolated and fragmented within the second quadrupole. A voltage of 20 to 40 V was adjusted for the best quality of MS/MS spectra. The fragment ions were then analyzed in the time-of-flight tube (100 to 2000 m/z). Data were acquired in continuum mode until well-averaged data were obtained.
NMR analysis.
The compound was subjected to 1D and 2D NMR analysis using a standard protocol (49) to determine the constituents and their sequential arrangement. The first NMR sample was prepared by dissolving ∼1 mg of the purified antimicrobial agent in 500 μl 90% H2O–10% D2O (referred to here as H2O). This sample was then lyophilized and reconstituted in 500 μl 100% D2O for a parallel NMR data set. The second NMR sample contained ∼5 mg of the pure compound dissolved in 500 μl 99.8% CD3OD (Cambridge Isotope Inc., Andover, MA). Unless stated otherwise, NMR experiments were performed at room temperature on a Bruker DMX-600 spectrometer (Bruker, Karlsruhe, Germany) equipped with a 5-mm (1H, 13C, 15N) triple-resonance probe and three-axis gradients. NMR experiments included 2D 1H-homonuclear COSY, TOCSY (60-ms DIPSI2 mixing time), and NOESY (200-ms mixing time), 2D heteronuclear 1H-13C HSQC, multiplicity-edited 1H-13C HSQC, 1H-13C HSQC-TOCSY (60-ms DIPSI2 mixing time), 1H-13C HSQC-NOESY (200-ms mixing time), 1H-15C HMBC, and 1H-15N HSQC, all using standard Bruker pulse sequences. Water suppression was typically achieved using the 3-9-19 WATERGATE technique (43) for the sample dissolved in H2O or presaturation to suppress residual HDO signal for the sample in D2O or CD3OD. NMR data were processed with NMRPipe (8) and visualized using NMRView (25). Data were typically zero-filled prior to application of window functions followed by Fourier transform. Chemical shifts were referenced externally to sodium 2,2-dimethyl-2-silapentane-5-sulfonate at 0.00 ppm.
GC-MS analysis for confirmation of the acyl moiety.
A mixture of the antimicrobial agent and polymyxin acylase in phosphate buffer (pH 8.0) was incubated at 37° for 24 h, followed by acidification to pH 3.0 and extraction with chloroform (29). The chloroform phase, which contained any released fatty acids, was washed sequentially by saturated sodium chloride solution and distilled water, and the chloroform in the extract was evaporated by a stream of nitrogen gas. The resulting fatty acid was dissolved in a methylating reagent (MethElute; Thermo Scientific, Bellefonte, PA) and applied to a capillary column (DB-23: 30 m by 0.25 mm [inner diameter], 0.25-μm film thickness; Agilent Technologies, Palo Alto, CA) on a gas chromatograph (Trace2000 GC; Thermo-Finnigan, West Palm Beach, FL) coupled to a mass spectrometer (Trace MS; Thermo, West Palm Beach, FL). Pentadecanoic acid (Acros Organics, Morris Plains, NJ) was dissolved in the methylating reagent and analyzed as a reference compound.
LC-MS/MS analysis.
The antimicrobial compound was digested by trypsin (sequencing grade; Promega, Madison, WI) in 100 mM NH4HCO3 buffer (pH 8.0) at 37°C overnight before the reaction was quenched by adding 0.1% TFA. The digests were analyzed by LC-MS/MS for amino acid sequence determination. Capillary liquid chromatography-nanospray tandem mass spectrometry was performed on a mass spectrometer (LTQ Orbitrap; Thermo-Finnigan) equipped with a nanospray source, operated in positive-ion mode (Michrom Bioresources Inc., Auburn, CA). Samples were separated on a capillary column (0.2 by 150 mm, Magic C18AQ, 3 μm, 200 Å; Michrom Bioresources Inc., Auburn, CA) using an HPLC system (UltiMate 3000; LC-Packings, Sunnyvale, CA). Each sample was injected into the trapping column (LC-Packings) and desalted with 50 mM acetic acid for 10 min. The injector port was then switched to the “inject” setting, and the peptides were eluted off the trap onto the column. Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. The flow rate was set at 2 μl/min. Typically, mobile phase B was increased from 2% to 50% in 30 min before being increased again from 50% to 90% in 5 min and then kept at 90% for another 5 min before being decreased quickly to 2% in 1 min. The column was equilibrated at 2% of mobile phase B (or 98% A) for 30 min before the next sample injection. The MS/MS spectrum was acquired with a nanospray source operated with a spray voltage of 2 kV, and a capillary temperature of 175°C was used. The scan sequence of the mass spectrometer was based on the data-dependent TopTen (10 most intense peaks) method. Briefly, the analysis was programmed for a full scan recorded between 300 and 2,000 Da and an MS/MS scan to generate product ion spectra to determine amino acid sequence in consecutive scans of the 10 most abundant peaks in the spectrum. The resolution of the full scan was set at 3 × 104 to achieve high-mass-accuracy MS determinations. The collision-induced dissociation fragmentation energy was set at 35%.
RESULTS
Isolation and identification of an antimicrobial-producing strain.
A large number of isolates, from different sources, were screened for activity against L. innocua ATCC 33090 and E. coli K-12. A soil isolate showed activities against the two indicator bacteria; the isolate was given the strain designation OSY-SE. The new isolate formed irregular and shiny colonies on tryptose agar and exhibited facultatively anaerobic behavior in broth culture. Morphologically, OSY-SE is a rod-shaped, 0.6- by 4.2-μm, Gram-positive, spore-forming bacterium (Fig. 1). The bacterium formed ellipsoidal spores in swollen sporangia. Motile cells could be directly observed under a light microscope. The isolate is positive for catalase, oxidase, and hydrolysis of starch and casein but negative for nitrate reduction, deamination of phenylalanine, and production of acetylmethylcarbinol, dihydroxyacetone, and indole. Genetic analysis indicated that this strain belongs to the genus Paenibacillus. Its 16S rRNA gene sequence shares high similarity with that of Paenibacillus apiarius (99%), Paenibacillus alvei (96%), and Paenibacillus thiaminolyticus (95%). Carbohydrate fermentation analysis (API 50 CH strips and API CHB medium) indicated 96.2% similarity between this strain and P. thiaminolyticus. Another set of biochemical tests (API 20E) showed that the isolate was positive for β-galactosidase, H2S production, and urease and negative for other reactions. The similarity of OSY-SE with P. thiaminolyticus increased to 99.9% when the results of the two sets of biochemical tests were combined. However, OSY-SE was negative for nitrate reduction, indole production, and citrate utilization but positive for urease and H2S production; these reactions are not typical of P. thiaminolyticus (see Table S1 in the supplemental material). Meanwhile, the biochemical traits of OSY-SE make it a mismatch with other Paenibacillus species, e.g., P. apiarius (oxidase and H2S negative and nitrate reduction and citrate utilization positive) and P. alvei (acetylmethylcarbinol, dihydroxyacetone, and indole positive). Therefore, the new bacterial strain was designated Paenibacillus strain OSY-SE, pending further identification.
Fig 1.
Scanning electron microscope examination of Paenibacillus OSY-SE cells.
Isolation and purification of antimicrobial agents produced by Paenibacillus OSY-SE.
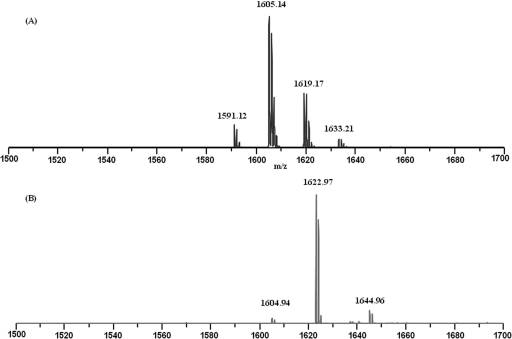
Antimicrobial agents were harvested by extracting Paenibacillus OSY-SE cells with acetonitrile. After evaporation of the organic solvent, the powder containing antimicrobial agents was readily soluble in neutral water. The HPLC fraction corresponding to the peak with a retention time of 17.02 min (see Fig. S1 in the supplemental material) showed antagonistic activities against L. innocua and E. coli, and a single peak was displayed when this fraction was reinjected into the HPLC. MALDI-TOF MS analysis indicated that the fraction contained a major compound with a molecular mass of 1,604 Da, which was designated paenibacterin, and three minor compounds with molecular masses of 1,590, 1,618, and 1,632 Da (Fig. 2A). The purified antimicrobial agent was subjected to MS/MS analysis, and fragmentation patterns of the major and minor compounds were quite similar, which suggested that paenibacterin and the three minor components were homologues (data not shown).
Fig 2.
MALDI-TOF MS analysis of paenibacterin and its linear form produced by alkaline hydrolysis. (A) Spectrogram of paenibacterin (m/z 1605.14) and its three homologues (m/z 1591.12, 1619.17, and 1633.21). (B) Spectrogram of linearized paenibacterin (m/z 1622.97); the ion at m/z 1644.96 corresponds to a sodium adduct.
Antimicrobial spectrum and stability.
Purified paenibacterin was used for the antimicrobial spectrum test. Microorganisms tested for sensitivity to this compound included pathogenic and nonpathogenic, Gram-positive and Gram-negative bacteria (Table 1). Paenibacterin showed activity against the majority of microorganisms tested, but these organisms varied in relative sensitivity to the antimicrobial agent. Paenibacterin exhibited activity against important Gram-negative pathogens, including E. coli O157:H7, Salmonella sp., and Yersinia enterocolitica. The antimicrobial agent was active against several Gram-positive pathogens, including L. monocytogenes and a methicillin-resistant S. aureus strain. Paenibacterin was active against one of two Clostridium difficile strains tested but showed no activity against Enterococcus faecalis ATCC 29212. The crude extract of Paenibacillus OSY-SE was resistant to heat and changes in pH. Most of its antimicrobial activity was retained after holding at 37, 55, and 80°C for 24 h, autoclaving at 121°C for 5 min, and exposure to pH 3.0, 5.0, and 9.0. Paenibacterin was resistant to treatment with trypsin, lipase, α-glucosidase, and lysozyme, but the activity was lost after digestion by pronase or polymyxin acylase. Polymyxin acylase is an enzyme which deacylates lipopeptide (37). Inactivation by polymyxin acylase suggested that paenibacterin is a lipopeptide.
Amino acid sequence of paenibacterin.
Initially, MS/MS analysis failed to sequence the antimicrobial agent due to the lack of fragmentation information, leading to the speculation that the agent could be a cyclic compound. After the open-ring reaction, a peak with m/z at 1622.97 was observed (Fig. 2B). The mass difference between the compound exposed to the open-ring reaction and the intact peptide was 18 Da, suggesting that the compound has a ring structure that can be opened by mild alkaline hydrolysis. Further MS/MS analysis was performed on the open-ring compound by using the Q-Tof. While more fragmentation information was obtained, no conclusive result about the amino acid composition could be achieved. Therefore, we resorted to NMR to elucidate the structure of paenibacterin.
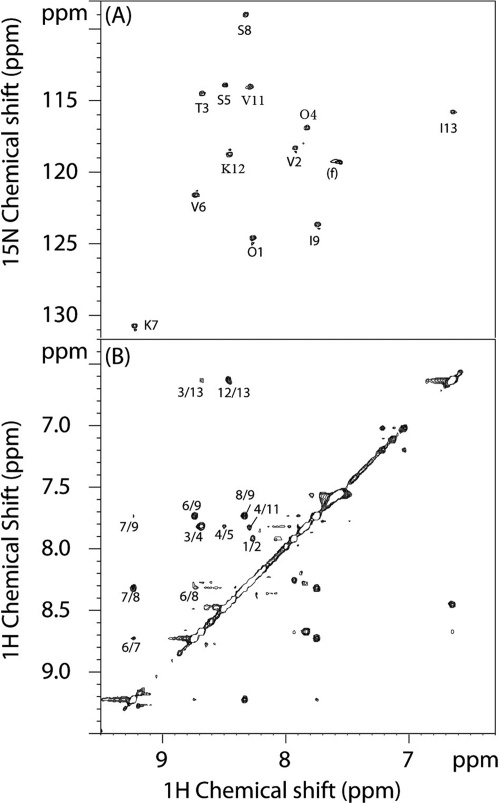
Preliminary analysis of NH amide cross-peaks in 2D 1H-15N HSQC (Fig. 3A) and Cα protons in 2D 1H-13C HSQC (Fig. 4A) indicated the presence of 13 amino acids in the peptidyl fragment, including one proline residue, evidenced by the observation of CH2δ1/δ2. The complete spin system of each amino acid was subsequently established from the COSY and TOCSY spectra. These results, taken together with the results of 2D 1H-13C HSQC and HMBC analyses, led to the identification of 3 Val, 2 Ile, 2 Ser, 1 Thr, 1 Pro, 2 Lys, and 2 Orn, the unnatural amino acid that has been reported previously (1). The sequence of these residues was first deduced by analyzing sequential NOEs such as HN(i)-HN(i + 1), Hα(i)-HN(i + 1) and Hβ(i)-HN(i + 1). The observation of strong NOEs between Pro10 Hδ1,δ2 and Val11 Hα led to their sequential assignment as well as the identification of the trans conformation adopted by Pro10. However, the NOE-based sequential assignment could be equivocal, particularly when the cyclic nature of this peptide moiety is taken into account, as described below. For example, long-range NOEs such as the one between Thr3 HN and Ile13 HN could complicate the analysis without prior knowledge of the linkage location (Fig. 3B). Therefore, a 2D 1H-13C HMBC of very high quality was required for unambiguous sequence-specific assignments on the basis of 1Hα(i)-13C′(i + 1) multiple-bond J-coupling correlations.
Fig 3.
NMR analysis of the peptidyl fragment in the amide region. (A) 2D 1H-15N HSQC recorded for the sample in H2O, showing the 12 main-chain NH amide cross-peaks and a cluster of folded peaks (f) attributable to an Arg, Lys, or Orn side chain NH3+ group; (B) 2D 1H NOESY of the same sample, showing the amide region cross-peaks, with assignments.
Fig 4.
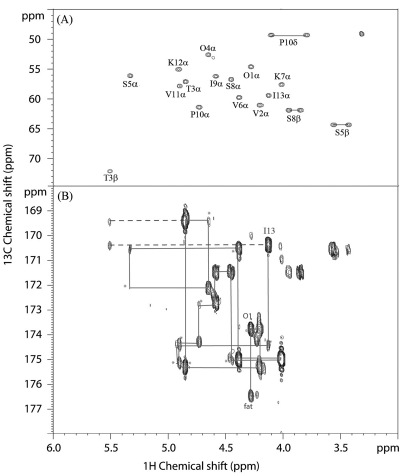
Elucidation of amino acid sequence and linkage of paenibacterin by HMBC. (A) 2D 1H-13C HSQC recorded for the sample dissolved in CD3OD, showing the 13C resonances in the region between 45 and 75 ppm. The CHα assignments of the 13 amino acids, together with Thr3 CH2β, Ser5 CH2β, Ser8 CH2β, and Pro10 CH2δ assignments, are labeled to assist the analysis of cross-peaks in panel B. The unlabeled methyl group at 3.30/49.1 ppm (1H/13C) is attributed to the residual solvent methanol. (B) 2D 1H-13C HMBC acquired for the same sample, showing the connectives associated with Hα protons. Sequential assignment is traced according to intraresidue Hα(i)-C′(i) and C′(i − 1)-Hα(i) (asterisk) multiple-bond J-coupling connectivities. The stretch starts from the connectivity of fatty acid carbonyl carbon (fat) to Orn1 Hα and ends with Lys12 C′ to Ile13 Hα. Also noted by the broken lines are the long-range J couplings of Thr3 Hβ-Thr3 C′ and Thr3 Hβ-Ile13 C′. The latter is the strong evidence for a cyclic peptide with an ester bond formed between the Thr3 hydroxyl group and the Ile13 C-terminal carboxylic group. It is important to note that the tilted and spit HMBC cross peaks are due to 1H-1H coupling (J modulation) (15).
A relatively large sample (∼5 mg) of the purified antimicrobial agent was prepared for this insensitive 2D 1H-13C HMBC analysis. However, severe line broadening was observed when the sample was dissolved in H2O. CD3OD was then used as the alternative NMR solvent, and the experiment was conducted on a Bruker DRX-800 spectrometer equipped with a cryoprobe. Some 2D experiments were also repeated to assist the NMR assignments. As shown in Fig. 4B, almost all of the intraresidue 1Hα(i)-13C′(i) as well as sequential 13C′(i − 1)-1Hα(i) multiple-bond correlations were observed, enabling the unequivocal determination of the peptide sequence as follows: Orn1-Val2-Thr3-Orn4-Ser5-Val6-Lys7-Ser8-Ile9-Pro10-Val11-Lys12-Ile13.
Linkage elucidation.
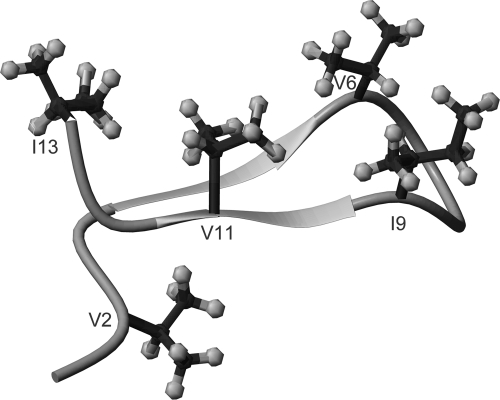
The HMBC spectrum described above also revealed multiple-bond correlations of Thr3 Hβ proton (5.49 ppm) with two carbonyl atoms: Thr3 C′ at 170.8 ppm and Ile13 C′ at 171.8 ppm (Fig. 4B). The latter suggests that Thr3 forms an ester linkage through its hydroxyl group to the C-terminal carboxylic group of Ile13. Consistently, both of Thr3 Hβ and Cβ chemical shifts experience unusual downfield shift similar to threonine shifts reported for other lipopeptides in which cyclization involves a Thr side chain (16, 26). Furthermore, this cyclic nature was supported by the long-range NOEs that have been observed, such as the one between Thr3 Hβ and Ile13 Hδ1. Finally, it appears that the peptide moiety possesses some rigid conformation, most likely adopting a β-hairpin conformation. A tertiary structure of the peptidyl fragment was calculated using CNS software (4) with a total of 162 NMR constraints, including 156 NOE-derived distance constraints (84 intraresidue, 40 sequential, and 32 nonsequential ones) and six χ1 constraints (V2, T3, V8, I9, V11, and K12) extracted from COSY and NOESY data sets, all from the NMR data recorded in aqueous solution. As shown in Fig. 5, the residues Orn4-Val6 and Ile9-Lys12 form an anti-parallel β-sheet, stabilized by hydrogen bonds between Orn4 and Val11 as well as between Val6 and Ile9. It was also noticed that four of the five bulky aliphatic side chain groups (Val6, Ile9, Val11, and Ile13) are on one side of the β-sheet and interact with each other. This structural feature may contribute to the amphipathic nature of paenibacterin.
Fig 5.
Tertiary structure of the peptide moiety of paenibacterin calculated from NMR constraints in aqueous solution. The five bulky aliphatic side chains (V2, V6, I9, V11, and I13) are labeled.
Determination of the acyl moiety.
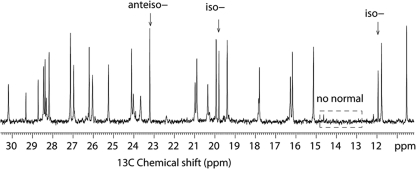
Based on the peptide sequence derived from NMR, there was a discrepancy between the molecular mass of the 13 amino acids and that of the whole compound. This indicated that paenibacterin contains another component, designated R (Fig. 6). MS was then performed on the b2 ion at m/z 339, which confirmed that it comprises R and Orn (data not shown). Therefore, the molecular mass of R was calculated as 225 Da, either from the molecular mass difference between the 13 amino acids and linearized paenibacterin or from the b2 ion, and the formula of R was established as C15H29O. This suggested that paenibacterin is a lipopeptide containing a saturated C15 fatty acid and 13 amino acids. The analysis of the 1D 13C NMR together with 2D 1H-13C HSQC and HMBC suggested that the fatty acid is a mixture of anteiso- and iso-branched forms, as evidenced by the presence of 13C peaks at 11.9 and 23.2 ppm, respectively (Fig. 7) (30). In 1D 13C NMR, the furthest downfield carbonyl carbon resonating at 180.1 ppm was assigned to the first atom of the fatty acid moiety. This C′ atom shows HMBC correlations to the first CH2 group at 2.30/38.2 ppm as well as the second CH2 group at 1.59, 1.56/28.1 ppm. More importantly, it also has an HMBC correlation to Orn1 Hα, indicating that the lipid chain is amidated to the N-terminal amine of Orn1. NOE was also observed between Orn1 HN and the first methylene protons (2.30 ppm) of the fatty acid side chain. A thorough analysis of the NMR data sets, particularly 2D 1H-13C HSQC-TOCSY, HSQC-NOESY, HMBC, and multiplicity-edited HSQC, led to the complete assignments of fatty acid side chains.
Fig 6.
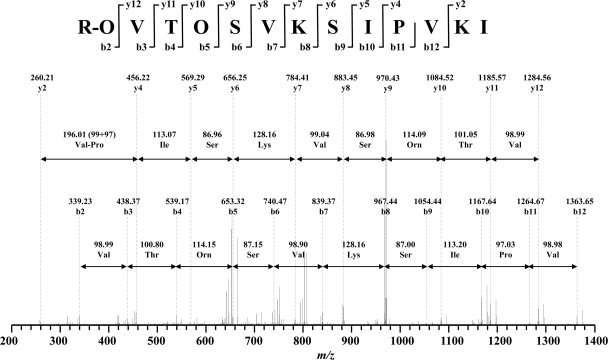
Fragmentation of b and y ion series of linearized paenibacterin, examined by MS/MS.
Fig 7.
1D 13C NMR spectrum, revealing the iso- and anteiso- fatty acyl chains.
Structure confirmation by GC-MS and LC-MS/MS.
GC-MS and LC-MS/MS were performed to verify the acyl moiety and the peptide sequence of paenibacterin, respectively. The fatty acids were successfully released from paenibacterin by polymyxin acylase digestion and analyzed by GC-MS as methyl esters. Three peaks at retention times of 4.87, 5.06, and 5.42 min were identified as methyl esters of iso-, anteiso- and normal-chain C15 fatty acids, respectively, by comparing pentadecanoic acid chromatogram and referring their mass spectra to the Wiley database (see Fig. S2 in the supplemental material). Although the normal-chain fatty acid was not evident in the NMR analysis, it was detected by GC-MS in low abundance. The dominant fatty acid in the sample was the anteiso-chain form, but iso-chain and normally branched forms were also detected. Therefore, the C15 fatty acyl chain of paenibacterin could be normal or an iso- or anteiso- form.
The peptide sequence was confirmed by analyzing tryptic-digested paenibacterin using LC-MS/MS. Paenibacterin was found to be resistant to trypsin, based on an antimicrobial activity test in phosphate buffer (pH 7.0); however, digested products were detected, including VTOSVKSIPVKI, SVKSIPVKI, and SIPVKI (see Fig. S3 in the supplemental material). It was noticed that the linkage between Thr and C-terminal Ile was probably broken during incubation in the NH4HCO3 buffer (pH 8.0) during enzyme digestion, as evidenced by the presence of linearized paenibacterin in the same buffer without trypsin.
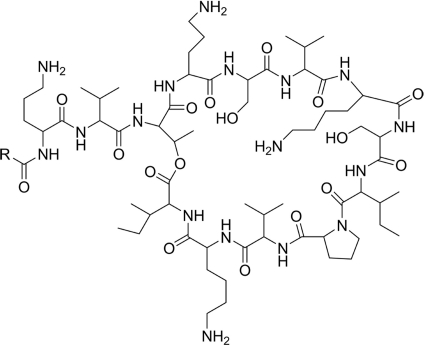
In summary, paenibacterin was identified as a lipopeptide consisting of a C15 fatty acyl chain (normal, iso-, or anteiso- form) and 13 amino acids (Fig. 8). The chemical shift assignments of the peptidyl fragment and the fatty acyl chain in aqueous solution are summarized in Table 2 and Table 3, respectively, while the corresponding assignments in methanol-d4 are provided in Tables S2 and S3 in the supplemental material.
Fig 8.
Molecular structure of paenibacterin; R = CH3(CH2)13 (normal chain), CH3CH(CH3)(CH2)11 (iso- chain), or CH3CH2CH(CH3)(CH2)10 (anteiso- chain).
Table 2.
Chemical shift assignments of peptidyl fragment of paenibacterin in aqueous solution at pH 4.5 and 298.0 K
| Residue | Shift(s) (ppm) |
|||
|---|---|---|---|---|
| 1HN/15N | 1Hα/13Cα | 1Hβ/13Cβ | Others [1H/13C(15N) and C′] | |
| Orn1 | 8.26/124.6 | 4.24/56.5 | 1.80,1.75/30.7 | CH2γ 1.74, 1.67/26.1; CH2δ 3.00/41.6; C′ 177.0 |
| Val2 | 7.92/118.3 | 4.09/62.8 | 2.07/32.8 | CH3γ1,γ2 1.19/22.1, 0.95/21.7; C′ 178.1 |
| Thr3 | 8.67/114.5 | 4.93/58.9 | 5.50/74.4 | CH3γ2 1.14/17.6; C′ 172.0 |
| Orn4 | 7.82/116.9 | 4.62/54.5 | 2.03, 1.76/32.7 | CH2γ 1.58, 1.53/24.6; CH2δ 2.96/41.6; C′ 173.9 |
| Ser5 | 8.49/113.9 | 5.31/57.4 | 3.57, 3.41/65.8 | C′ 172.9 |
| Val6 | 8.73/121.6 | 4.27/61.9 | 1.88/34.9 | CH3γ1,γ2 0.93/21.6, 0.90/20.7; C′ 176.9 |
| Lys7 | 9.22/130.7 | 4.07/58.9 | 1.86/32.1 | CH2γ 1.52, 1.49/25.0; CH2δ 1.70/29.0; CH2ε 2.99/41.7; C′ 178.0 |
| Ser8 | 8.32/109.0 | 4.43/58.5 | 3.90, 3.86/63.1 | C′ 173.7 |
| Ile9 | 7.74/123.6 | 4.68/57.5 | 2.10/39.5 | CH3γ2 0.98/16.6; CH2γ1 1.48, 1.23/28.9; CH3δ1 0.83/11.7; C′ 173.8 |
| Pro10 | 4.70/63.0 | 2.36, 1.96/32.8 | CH2γ 2.16, 1.97/27.5; CH2δ 3.95, 3.77/51.4 | |
| Val11 | 8.28/114.0 | 4.86/59.6 | 2.33/36.0 | CH3γ1,γ2 1.01/22.0, 0.73/19.2; C′ 176.8 |
| Lys12 | 8.45/118.8 | 4.59/56.8 | 2.05, 1.76/31.7 | CH2γ 1.45, 1.42/24.9; CH2δ 1.67/29.1; CH2ε 2.99/41.7; C′ 177.3 |
| Ile13 | 6.65/115.8 | 4.14/61.3 | 1.81/38.0 | CH3γ2 0.79/17.4; CH2γ2 1.26, 1.09/27.5; CH3δ1 0.80/13.3; C′ 174.0 |
Table 3.
Chemical shift assignments of fatty acyl chains of paenibacterin in aqueous solution at pH 4.5 and 298.0 K
| Position | Iso- form | 1H/13C shift (ppm) | Anteiso- form | 1H/13C shift (ppm) |
|---|---|---|---|---|
| 1 | C′ | 180.1 | C′ | 180.1 |
| 2 | CH2 | 2.30/38.2 | CH2 | 2.30/38.2 |
| 3 | CH2 | 1.59, 1.56/28.1 | CH2 | 1.59, 1.56/28.1 |
| 4 | CH2 | 1.26/31.5 | CH2 | 1.26/31.5 |
| 5 | CH2 | ∼1.25/31.5 | CH2 | ∼1.25/31.5 |
| 6 | CH2 | ∼1.25/31.5 | CH2 | ∼1.25/31.5 |
| 7 | CH2 | ∼1.25/31.5 | CH2 | ∼1.25/31.5 |
| 8 | CH2 | ∼1.25/31.5 | CH2 | ∼1.25/31.5 |
| 9 | CH2 | ∼1.25/31.5 | CH2 | ∼1.25/31.5 |
| 10 | CH2 | 1.26/29.2 | CH2 | ∼1.25/31.5 |
| 11 | CH2 | 1.27, 1.08/38.6 | CH2 | 1.26/29.2 |
| 12 | CH2 | 1.29/36.5 | CH | 1.14/41.2 |
| 13 | CH | 1.30, 1.10/31.7 | CH2 | 1.521/29.04 |
| 14 | CH3 | 0.81/13.4 | CH3 | 0.82/24.8 |
| 15 | CH3 | 0.81/21.5 | CH3 | 0.82/24.8 |
DISCUSSION
A new bacterial strain, OSY-SE, was isolated from soil samples during a screening for microorganisms with potent antibacterial activity. The isolate was identified as a Paenibacillus sp. based on morphological, physiological, biochemical, and genetic characteristics. The bacterium produces a new antimicrobial agent, paenibacterin, which is active against several Gram-positive and Gram-negative pathogens. MS analysis of the HPLC-purified antimicrobial agent revealed three minor components that were coeluted with paenibacterin in a single HPLC peak; these are considered paenibacterin homologues, based on MS/MS analyses. However, the low abundance of these homologues prevented our obtaining their structure information, except for the compound with a molecular mass of 1,618 Da. This compound was detected after an open-ring reaction and was found to differ from paenibacterin either in the acyl moiety structure or in the fourth-position amino acid. Similar observations were reported previously for lipopeptides such as fusaricidins, surfactins, fengycins, and iturins (2, 39).
Paenibacterin is readily soluble in water and probably forms micelles at high concentrations in aqueous solution, as observed by NMR. The cyclic nature of paenibacterin was initially assumed due to a lack of MS/MS fragmentation and then confirmed by mild alkaline hydrolysis. However, after the open-ring reaction, chloroform failed to extract the linearized compound as described by Yakimov et al. (50), which indicates that paenibacterin is more hydrophilic than lichenysin A and its analogues. The acyl moiety is considered to be important for the antimicrobial activity in lipopeptides. Deacylation had been reported to inactivate lipopeptide antibiotics, and the peptide moieties regained antimicrobial activity after reacylation (3, 7). Malina and Shai (33) demonstrated that the attachment of aliphatic acid to biologically inactive cationic peptides can produce lipopeptides with antimicrobial activity. Additionally, Majerle et al. (32) reported that development of acyl analogues of the peptide fragment of human lactoferrin enhanced its antibacterial activity. The importance of fatty acid residues for antimicrobial activity was also confirmed in this study, in which paenibacterin lost activity after digestion by polymyxin acylase. Paenibacterin is a cationic lipopeptide containing four basic amino acids, two Orn and two Lys. Positive charges are helpful for interaction of antimicrobials with bacterial cell wall and for promotion of uptake of the antimicrobial agent itself (20). It is inferred that the number of basic amino acids correlates with antimicrobial activity up to a limit that depends on the antimicrobial peptide (14). Besides positive residues, the amphipathic structure contributes to the antimicrobial activity of cationic peptides (24). In spite of the fatty acyl residue, hydrophobic amino acids of paenibacterin are evenly distributed in the peptidyl fragment, and the compound adopted an anti-parallel β-sheet configuration with hydrophobic chains on one side. This structure may help paenibacterin to disrupt the cytoplasmic membrane of target cells. Disruption of the cytoplasmic membrane and leakage of cell components are considered the mechanism of action of many cationic antimicrobial peptides, such as polymyxin, octapeptin, and magainin (9, 11, 35, 42). Polymyxin, which is produced by Paenibacillus polymyxa strains, contains six diaminobutyric acids and has five net positive charges (12). Polymyxin is regarded as the last line of therapy against multi-drug-resistant Gram-negative bacteria such as P. aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae (45). However, polymyxin B has no activity against Gram-positive bacteria and anaerobes, whereas polymyxin E is inactive against Gram-positive and Gram-negative aerobic cocci, Gram-positive aerobic bacilli, and all anaerobes (13, 51). Compared to polymyxin, paenibacterin showed broader antimicrobial activity, including activity against Gram-positive and anaerobic bacteria.
In conclusion, a new strain, Paenibacillus OSY-SE, was isolated and identified. It produces a new antimicrobial agent, designated paenibacterin, which is active against both Gram-positive and Gram-negative pathogens. The new isolate and associated antimicrobials are potentially useful in food or medical applications. The usability of this lipopeptides in food and medical applications is being investigated.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by an endowment from Ginni and Frank Bazler and by a scholarship to Y. Guo from the Chinese Government.
We are grateful to Brian Kemmenoe (Campus Microscopy and Imaging Facility) for assistance with scanning electron microscopy examination, J. T. Lejeune (College of Veterinary Medicine, The Ohio State University) and W. A. Gebreyes (Department of Veterinary Preventive Medicine, The Ohio State University) for providing C. difficile strains, K. Green-Church (Mass Spectrometry and Proteomics Facility, The Ohio State University) for suggestions on GC-MS analysis, and Jocelyn Hach (Mass Spectrometry and Proteomics Facility, The Ohio State University) for performing the GC-MS analysis.
Footnotes
Published ahead of print 24 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ball LJ, Goult CM, Donarski JA, Micklefield J, Ramesh V. 2004. NMR structure determination and calcium binding effects of lipopeptide antibiotic daptomycin. Org. Biomol. Chem. 2:1872–1878 [DOI] [PubMed] [Google Scholar]
- 2. Beatty PH, Jensen SE. 2002. Paenibacillus polymyxa produces fusaricidin-type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Can. J. Microbiol. 48:159–169 [DOI] [PubMed] [Google Scholar]
- 3. Boeck LD, Fukuda DS, Abbott BJ, Debono M. 1989. Deacylation of echinocandin B by Actinoplanes utahensis. J. Antibiot. 42:382–388 [DOI] [PubMed] [Google Scholar]
- 4. Brunger AT, et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905–921 [DOI] [PubMed] [Google Scholar]
- 5. Chen S, et al. 2004. Characterization of multiple-antimicrobial-resistant Salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 70:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clardy J, Fischbach MA, Walsh CT. 2006. New antibiotics from bacterial natural products. Nat. Biotechnol. 24:1541–1550 [DOI] [PubMed] [Google Scholar]
- 7. Debono M, et al. 1988. Enzymatic and chemical modifications of lipopeptide antibiotic A21978C: the synthesis and evaluation of daptomycin (LY146032). J. Antibiot. 41:1093–1105 [DOI] [PubMed] [Google Scholar]
- 8. Delaglio F, et al. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 9. Dixon RA, Chopra I. 1986. Leakage of periplasmic proteins from Escherichia coli mediated by polymyxin B nonapeptide. Antimicrob. Agents Chemother. 29:781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drancourt M, et al. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38:3623–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duwe AK, Rupar CA, Horsman GB, Vas SI. 1986. In vitro cytotoxicity and antibiotic activity of polymyxin B nonapeptide. Antimicrob. Agents Chemother. 30:340–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Evans ME, Feola DJ, Rapp RP. 1999. Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant gram-negative bacteria. Ann. Pharmacother. 33:960–967 [DOI] [PubMed] [Google Scholar]
- 13. Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40:1333–1341 [DOI] [PubMed] [Google Scholar]
- 14. Findlay B, Zhanel GG, Schweizer F. 2010. Cationic amphiphiles, a new generation of antimicrobials inspired by the natural antimicrobial peptide scaffold. Antimicrob. Agents Chemother. 54:4049–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Furihata K, Seto H. 1998. Constant time HMBC (CT-HMBC), a new HMBC technique useful for improving separation of cross peaks. Tetrahedron Lett. 39:7337–7340 [Google Scholar]
- 16. Gerard J, et al. 1997. Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J. Nat. Prod. 60:223–229 [DOI] [PubMed] [Google Scholar]
- 17. Gordon RE, Haynes WC, Pang CH-N. 1973. The genus Bacillus. Agriculture handbook no. 427. United States Department of Agriculture, Washington, DC [Google Scholar]
- 18. Govan VA, Allsopp MH, Davison S. 1999. A PCR detection method for rapid identification of Paenibacillus larvae. Appl. Environ. Microbiol. 65:2243–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamaki T, et al. 2005. Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J. Biosci. Bioeng. 99:485–492 [DOI] [PubMed] [Google Scholar]
- 20. Hancock RE, Chapple DS. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He Z, et al. 2007. Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl. Environ. Microbiol. 73:168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsu DI, Okamoto MP, Murthy R, Wong-Beringer A. 2005. Fluoroquinolone-resistant Pseudomonas aeruginosa: risk factors for acquisition and impact on outcomes. J. Antimicrob. Chemother. 55:535–541 [DOI] [PubMed] [Google Scholar]
- 23. Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jenssen H, Hamill P, Hancock RE. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson BA, Blevins RA. 1994. NMR VIEW: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4:603–614 [DOI] [PubMed] [Google Scholar]
- 26. Kajimura Y, Kaneda M. 1996. Fusaricidin A, a new depsipeptide antibiotic produced by Bacillus polymyxa KT-8. Taxonomy, fermentation, isolation, structure elucidation and biological activity. J. Antibiot. 49:129–135 [DOI] [PubMed] [Google Scholar]
- 27. Kaletunc G, Lee J, Alpas H, Bozoglu F. 2004. Evaluation of structural changes induced by high hydrostatic pressure in Leuconostoc mesenteroides. Appl. Environ. Microbiol. 70:1116–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khianngam S, Akaracharanya A, Tanasupawat S, Lee KC, Lee JS. 2009. Paenibacillus thailandensis sp. nov. and Paenibacillus nanensis sp. nov., xylanase-producing bacteria isolated from soil. Int. J. Syst. Evol. Microbiol. 59:564–568 [DOI] [PubMed] [Google Scholar]
- 29. Kline T, Holub D, Therrien J, Leung T, Ryckman D. 2001. Synthesis and characterization of the colistin peptide polymyxin E1 and related antimicrobial peptides. J. Pept. Res. 57:175–187 [DOI] [PubMed] [Google Scholar]
- 30. Lin SC, Minton MA, Sharma MM, Georgiou G. 1994. Structural and immunological characterization of a biosurfactant produced by Bacillus licheniformis JF-2. Appl. Environ. Microbiol. 60:31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loo VG, et al. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449 [DOI] [PubMed] [Google Scholar]
- 32. Majerle A, Kidric J, Jerala R. 2003. Enhancement of antibacterial and lipopolysaccharide binding activities of a human lactoferrin peptide fragment by the addition of acyl chain. J. Antimicrob. Chemother. 51:1159–1165 [DOI] [PubMed] [Google Scholar]
- 33. Malina A, Shai Y. 2005. Conjugation of fatty acids with different lengths modulates the antibacterial and antifungal activity of a cationic biologically inactive peptide. Biochem. J. 390:695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin NI, et al. 2003. Isolation, structural characterization, and properties of mattacin (polymyxin M), a cyclic peptide antibiotic produced by Paenibacillus kobensis M. J. Biol. Chem. 278:13124–13132 [DOI] [PubMed] [Google Scholar]
- 35. Matsuzaki K, Murase O, Fujii N, Miyajima K. 1996. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry 35:11361–11368 [DOI] [PubMed] [Google Scholar]
- 36. McSpadden Gardener BB. 2004. Ecology of Bacillus and Paenibacillus spp. in agricultural systems. Phytopathology 94:1252–1258 [DOI] [PubMed] [Google Scholar]
- 37. Misumi S, Tsuruta M, Furuishi K, Shoji S. 1995. Determination of N-myristoyl peptide sequence both by MALDI TOF MASS and with an N-myristoyl cleaving enzyme (polymyxin acylase). Biochem. Biophys. Res. Commun. 217:632–639 [DOI] [PubMed] [Google Scholar]
- 38. Moran GJ, et al. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666–674 [DOI] [PubMed] [Google Scholar]
- 39. Ongena M, Jacques P. 2008. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16:115–125 [DOI] [PubMed] [Google Scholar]
- 40. Rodrigues L, Banat IM, Teixeira J, Oliveira R. 2006. Biosurfactants: potential applications in medicine. J. Antimicrob. Chemother. 57:609–618 [DOI] [PubMed] [Google Scholar]
- 41. Rodriguez-Palacios A, Lejeune JT. 2011. Moist-heat resistance, spore aging, and superdormancy in Clostridium difficile. Appl. Environ. Microbiol. 77:3085–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rosenthal KS, Swanson PE, Storm DR. 1976. Disruption of Escherichia coli outer membranes by EM 49. A new membrane active peptide. Biochemistry 15:5783–5792 [DOI] [PubMed] [Google Scholar]
- 43. Sklenar V, Piotto M, Leppik R, Saudek V. 1993. Gradient-tailored water suppression for 1H–15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. A 102:241–245 [Google Scholar]
- 44. Timmusk S, Wagner EG. 1999. The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: a possible connection between biotic and abiotic stress responses. Mol. Plant Microbe Interact. 12:951–959 [DOI] [PubMed] [Google Scholar]
- 45. Velkov T, Thompson PE, Nation RL, Li J. 2010. Structure-activity relationships of polymyxin antibiotics. J. Med. Chem. 53:1898–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. von der Weid I, Duarte GF, van Elsas JD, Seldin L. 2002. Paenibacillus brasilensis sp. nov., a novel nitrogen-fixing species isolated from the maize rhizosphere in Brazil. Int. J. Syst. Evol. Microbiol. 52:2147–2153 [DOI] [PubMed] [Google Scholar]
- 47. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu XC, et al. 2010. Paenimacrolidin, a novel macrolide antibiotic from Paenibacillus sp. F6–B70 active against methicillin-resistant Staphylococcus aureus. Microb. Biotechnol. 4:491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wüthrich K. 1986. NMR of proteins and nucleic acids. Wiley Interscience, New York, NY [Google Scholar]
- 50. Yakimov MM, Timmis KN, Wray V, Fredrickson HL. 1995. Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl. Environ. Microbiol. 61:1706–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60:1206–1215 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.