Abstract
The groundwater contaminant 1,4-dioxane (dioxane) is transformed by several monooxygenase-expressing microorganisms, but only a few of these, including Pseudonocardia dioxanivorans strain CB1190, can metabolize the compound as a sole carbon and energy source. However, nothing is yet known about the genetic basis of dioxane metabolism. In this study, we used a microarray to study differential expression of genes in strain CB1190 grown on dioxane, glycolate (a previously identified intermediate of dioxane degradation), or pyruvate. Of eight multicomponent monooxygenase gene clusters carried by the strain CB1190 genome, only the monooxygenase gene cluster located on plasmid pPSED02 was upregulated with dioxane relative to pyruvate. Plasmid-borne genes for putative aldehyde dehydrogenases, an aldehyde reductase, and an alcohol oxidoreductase were also induced during growth with dioxane. With both dioxane and glycolate, a chromosomal gene cluster encoding a putative glycolate oxidase was upregulated, as were chromosomal genes related to glyoxylate metabolism through the glyoxylate carboligase pathway. Glyoxylate carboligase activity in cell extracts from cells pregrown with dioxane and in Rhodococcus jostii strain RHA1 cells expressing the putative strain CB1190 glyoxylate carboligase gene further demonstrated the role of glyoxylate metabolism in the degradation of dioxane. Finally, we used 13C-labeled dioxane amino acid isotopomer analysis to provide additional evidence that metabolites of dioxane enter central metabolism as three-carbon compounds, likely as phosphoglycerate. The routing of dioxane metabolites via the glyoxylate carboligase pathway helps to explain how dioxane is metabolized as a sole carbon and energy source for strain CB1190.
INTRODUCTION
The cyclic ether 1,4-dioxane (dioxane) is a contaminant with an increasingly recognized wide environmental distribution (56). Dioxane is highly soluble in water and thus is easily transported in groundwater systems from point source contamination (56). Although the long-term effects of dioxane exposure on human health are not well understood, animal studies indicate that the compound is a potential carcinogen (42). Consequently, dioxane contamination has become the focus of remediation research, and microbial systems capable of dioxane degradation have been investigated (56). The degradation of dioxane under cometabolic conditions (where other substrates provide carbon and energy for growth) appears to be somewhat common (9, 23, 27, 41, 43, 48), but an increasing number of microorganisms have also been identified that can use dioxane as a sole source of carbon and energy (5, 15, 23, 27, 30).
Initial investigations of biological dioxane transformation were performed as part of toxicological studies, which identified hydroxyethoxyacetic acid (HEAA) and 1,4-dioxane-2-one as the major metabolites in humans (55) and rats (52), respectively. Recently, the pathways of dioxane biotransformation have been studied in a number of metabolizing and cometabolizing bacteria and fungi in an attempt to understand the fate of environmentally released dioxane. A common feature of dioxane transformation is the initial hydroxylation of the dioxane ring by monooxygenase enzyme systems (23, 41, 48), which is similar to the findings of the toxicological studies. The hydroxylated metabolite then undergoes uncharacterized biotic or abiotic processing, leading to an opening of the ring structure. Researchers have used a number of techniques to detect posthydroxylation metabolites in order to elucidate the dioxane catabolic pathway. In the dioxane-cometabolizing fungus Cordyceps sinensis, deuterated dioxane-d8 and gas chromatography-mass spectrometry (GC-MS) analysis were used to identify ethylene glycol, glycolic acid, and oxalic acid as metabolites (27). In Mycobacterium sp. strain PH-06, which grows using dioxane, deuterated dioxane and GC-MS analysis detected only two metabolites, 1,4-dioxane-2-ol and ethylene glycol (15). Meanwhile, 2-hydroxyethoxyacetic acid (2HEAA) was the sole metabolite detected with 14C-labeled dioxane in the cometabolizing bacterium Pseudonocardia strain ENV478 (48).
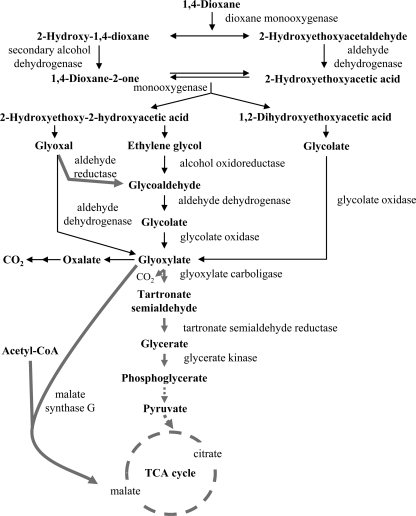
A more comprehensive analysis of dioxane metabolites was recently performed with the dioxane-metabolizing Pseudonocardia dioxanivorans strain CB1190 (designated strain CB1190) (24). By analyzing metabolites with Fourier transform ion cyclotron resonance mass spectrometry and tandem MS (MS/MS) and by tracking distribution of 14C-labeled dioxane metabolites, the previously identified metabolites were confirmed, and 2-hydroxyethoxyacetaldehyde, 1,2-dihydroxyethoxyacetic acid, 2-hydroxyethoxy-2-hydroxyacetic acid, glycoaldehyde, glyoxylic acid, and CO2 were also identified as dioxane metabolites (24). Based on these findings, a dioxane biodegradation pathway was proposed (Fig. 1, black arrows), whereby all metabolites feed through oxalic acid into central metabolism (24).
Fig 1.
Proposed pathways and enzymes involved in dioxane metabolism in strain CB1190. Black arrows indicate the transformations previously proposed by Mahendra et al. (24). Gray arrows indicate transformations supported by in silico or experimental results in the current work. Dashed arrows indicate multistep transformations.
These studies have improved our understanding of the fate of dioxane during microbial biodegradation, but there is a knowledge gap in terms of the biochemical and genetic bases for this pathway. While monooxygenase enzymes have been implicated in the initial activation of the dioxane ring, the further steps in dioxane metabolism have not been explored. In particular, the mechanism for incorporation of dioxane metabolites into biomass in microorganisms that can grow with dioxane as the sole carbon and energy source is unknown. In this work we used the recently determined genome of P. dioxanivorans strain CB1190 (38) to investigate how dioxane is assimilated into biomass to support growth. We present evidence highlighting the unique role of glyoxylate in bacterial dioxane metabolism.
MATERIALS AND METHODS
Culture growth.
Strain CB1190 was cultivated in ammonium mineral salts (AMS) medium (30). Replicate 250-ml screw-cap bottles were prepared with 50 ml of AMS medium and were amended with the appropriate carbon source. These were then inoculated with 0.1 ml (1/500 dilution) of stationary-phase dioxane-grown strain CB1190 culture and incubated at 30°C with shaking at 150 rpm. Sterile (uninoculated) controls were prepared in parallel. Bottles were amended with dioxane (4.7 mM final aqueous concentration), glycolate (2 mM), or pyruvate (45 mM). Dioxane and glycolate were used at low concentrations to avoid toxicity (data not shown). Glycolate was chosen since, of the known dioxane degradation intermediates, it best supported strain CB1190 growth. Glycolate bottles were neutralized with NaOH and were reamended with a second dose of glycolate after the first dose was consumed.
Cell harvesting and RNA extraction for transcription studies.
Strain CB1190 grows as clumps on the liquid surface, so standard optical density (OD) readings cannot be used to determine growth phase. Therefore, in order to generate highly active cultures, cells were harvested when approximately half of the added substrate had been consumed. For glycolate-grown cells, cell harvesting occurred 1 day after the second substrate amendment. Cells from replicate bottles were collected by filtration onto triplicate 0.45-μm-pore-size cellulose ester filters (Gelman Sciences, Ann Arbor, MI), scraped from the filters with a sterile scalpel, and transferred to 2-ml screw-top microcentrifuge tubes containing 1 g of 100-μm-diameter zirconia-silica beads (Biospec Products, Bartlesville, OK). Cells on each filter were collected from one, five, and six bottles, respectively, for pyruvate, dioxane, and glycolate treatments. Harvested cells were stored at −80°C until RNA extraction.
Nucleic acids were extracted using a modified version of the phenol method described previously (14). Briefly, each 2-ml microcentrifuge tube containing cells and zirconia-silica beads was filled with 250 μl of lysis buffer (50 mM sodium acetate, 10 mM EDTA [pH 5.1]), 100 μl of 10% sodium dodecyl sulfate, and 1.0 ml of phenol (pH 8; Sigma-Aldrich, St. Louis, MO). Cells were lysed by heating samples to 65°C for 2 min, followed by bead beating with a Mini Bead Beater (Biospec Products) for 2 min, incubation at 65°C for 8 min, and bead beating for an additional 2 min. Cellular debris was collected by centrifugation (5 min at 14,000 × g), and the aqueous lysate was transferred to a new microcentrifuge tube. The lysate was extracted twice with 1 volume of phenol-chloroform-isoamyl alcohol (pH 8) (24:24:1, vol/vol) and once with 1 volume of chloroform-isoamyl alcohol (24:1, vol/vol) (Sigma-Aldrich). Nucleic acids were precipitated by adding 0.1 volume of 3 M sodium acetate and 1 volume of ice-cold isopropanol and storing the tube at −20°C overnight. The precipitate was collected by centrifugation (30 min at 21,000 × g at 4°C), washed once with 70% ethanol, and resuspended in 100 μl of nuclease-free water.
To obtain RNA, resuspended nucleic acids were initially separated with an Allprep kit (Qiagen), and then the RNA was purified using an RNeasy kit (Qiagen). Following elution with 100 μl of RNase-free water, any contaminating DNA was removed by two successive DNase I treatments using a DNA-free kit (Ambion, Austin, TX) according to the manufacturer's instructions. Pure RNA was obtained with a final cleanup with an RNeasy kit. RNA yields ranged from 6.3 to 24.8 μg per microcentrifuge tube of harvested cells.
Microarray design and analyses.
A draft version of the strain CB1190 genome was obtained from the Joint Genome Institute and consisted of 6,896 protein-encoding sequences in 226 contigs. These were submitted to Affymetrix (Santa Clara, CA) for custom chip design. The final design consisted of a set of perfect-match-only 25-mer probes, with each sequence targeted by 11 to 13 probes, and also included a standard set of Affymetrix controls for prokaryotic gene expression microarrays. Subsequent to microarray production, a finished genome sequence for strain CB1190 was made available (38). The microarray probes were reannotated to reflect the finished genome sequence. Reannotated microarray probe sets targeted 6,391 (approximately 94%) of the coding sequences in the finished genome.
Five micrograms of total RNA was used as starting material for each microarray analysis. Triplicate microarrays were processed for each substrate treatment. cDNA was synthesized, fragmented, labeled, and hybridized to arrays according to the protocols outlined in section 3 of the Affymetrix GeneChip Expression Analysis technical manual (1), with the following changes: cDNA fragmentation was achieved by the addition of 0.07 U of DNase I/μg of cDNA, and the hybridization temperature was 52°C. Hybridized arrays were stained and washed according to the Affymetrix modified FlexMidi_euk2v3_450 fluidics protocol for the P. aeruginosa array and were scanned using an Affymetrix GeneChip Scanner 3000.
Microarray data were first processed with the RMAExpress software program (6) to compute gene expression summary values. Background adjustment and quantile normalization were performed across all microarrays. The average normalized expression ratio (dioxane or glycolate treatment/pyruvate control) was calculated for each gene. Ratios were considered significant if they were >2.0 or <0.5 and Student's t test indicated a P value (two-tailed) of <0.05.
Quantitative reverse transcription-PCR (qRT-PCR).
To verify microarray data, the relative transcription levels of 17 genes in the dioxane and glycolate treatments were compared with the pyruvate treatment. cDNA was synthesized from 500 ng of RNA using a TaqMan reverse transcription kit (Applied Biosystems, Foster City, CA) with random hexamers in 15-μl reaction mixtures according the manufacturer's protocol. cDNA was diluted 100-fold, and 2 μl was used in each subsequent quantitative PCR (qPCRs). All qPCRs were performed in triplicate with an Applied Biosystems StepOne Plus real-time PCR system. For most genes, SYBR green chemistry was used, and each 20-μl reaction mixture consisted of 1× Fast SYBR green master mix, 2 μl of diluted cDNA, and each primer at 0.5 mM. The PCR cycling conditions were as follows: 95°C for 20 s and then 40 cycles of 95°C for 3 s and annealing for 30 s, followed by melting curve analysis. The genes tpi, thiC, and rpoD were determined by the geNorm method (50) to be stably transcribed across all treatments and were thus used as internal references. TaqMan chemistry was used for these genes, and each qPCR mixture consisted of 1× Fast Universal Mix (Applied Biosystems), 2 μl of diluted cDNA, each primer at 0.5 mM, and the probe at 145 nM. The cycling conditions were as follows: 95°C for 20 s and then 40 cycles of 95°C for 1 s and annealing for 20 s. The primer sequences and dye and quencher chemistry for each probe are listed in Table S1 in the supplemental material. The efficiency of each qPCR assay was determined with a serial dilution of cDNA derived from a dioxane treatment replicate. Gene expression was normalized using the method of Vandesompele et al. (50).
Cloning and expression of strain CB1190 genes.
Strain CB1190 genes Psed_3889 and Psed_3890 were PCR amplified and ligated into linearized Rhodococcus expression plasmid pTip-QC2 (28) using the hetero-stagger cloning method (22). Rhodococcus jostii strain RHA1 was transformed with pTip-Psed_3889, pTip-Psed_3890, or pTip-QC2, and then expression of the inserted gene was induced by the addition of thiostrepton to the culture medium. Details of gene cloning and protein expression are presented in the supplemental material. Cell extracts from strain RHA1/pTip cells were prepared as described below for strain CB1190 cells.
Glyoxylate carboligase assay.
Strain CB1190 cells grown with either pyruvate or dioxane were harvested by filtration, and scraped cells were stored at −80°C. Cell extracts were prepared by resuspending the cells in 1 ml of sodium phosphate buffer (10 mM, pH 7.0), adding 1 g of 100-μm-diameter zirconia-silica beads, and bead beating for four cycles of 1 min, with cooling on ice for 1 min between cycles. Cell debris was collected by centrifugation for 10 min at 21, 000 × g at 4°C, and the supernatant (cell extract) was aliquoted and stored at −80°C until use. Protein was determined by the method of Bradford (8).
Glyoxylate carboligase activity was tested using the linked glyoxylate carboligase/tartronate semialdehyde reductase assay described by Cusa et al. (11). Briefly, 2-ml reaction mixtures, containing 100 mM sodium phosphate (pH 8.0), 5 mM MgCl2, 0.28 mM NADH, 0.5 mM thiamine pyrophosphate, and 200 μg of protein from cell extracts, were prepared in triplicate in 33-ml screw-top glass vials with Mininert caps. The headspace was purged for 5 min with N2, glyoxylate was added to 10 mM, and vials were incubated with shaking at 30°C for 16 h. Reactions were stopped, and CO2 was liberated by the addition of 0.2 ml of 10 N sulfuric acid and shaking for 15 min. CO2 was analyzed by gas chromatography-pulsed discharge detection (GC-PDD), and glyoxylate was analyzed by high-performance liquid chromatography (HPLC).
For assays with cell extracts from strain RHA1 carrying pTip plasmid constructs, the assay vials were prepared as above for the following treatments: (i) pTip-Psed_3889, 8 μg of protein; (ii) pTip-Psed_3890, 7 μg of protein; (iii) pTip-Psed_3889 and pTip-Psed_3890, 8 μg and 7 μg of protein of each, respectively; (iv) pTip-QC2, 10 μg of protein; (v) buffer control. CO2 production was determined by GC-PDD, and glyoxylate consumption and glycerate production were determined by HPLC (see below).
Growth with 1,4-[U-13C]dioxane.
A series of strain CB1190 cultures was prepared with 1,4-[U-13C]dioxane (99% pure; Sigma-Aldrich). Single serum bottles (160 ml; Wheaton Science Products, Millville, NJ) with black butyl rubber stoppers and containing 30 ml of AMS medium were amended with 1.6 mM [13C]dioxane, or 1.6 mM [13C]dioxane plus 3% CO2. These bottles were inoculated with 0.1 ml (1/300 dilution) of [13C]dioxane-grown strain CB1190 culture and were incubated at 30°C with shaking at 150 rpm. Following complete removal of the dioxane, cells (0.1 ml) from each treatment were transferred to fresh triplicate bottles with the same amendments and were incubated with shaking until dioxane removal was complete. Cells were then harvested by filtration, scraped from the filter, and stored at −80°C in 2-ml screw-top vials until isotopomer amino acid analysis.
Analytical methods.
Pyruvate, glycolate, and glyoxylate consumption and glycerate production were monitored with a Waters (Milford, MA) model 2695 HPLC equipped with a model 2996 photodiode array detector and an Amidex HPX-87H column (Bio-Rad, Hercules, CA). Samples (50 μl) were analyzed isocratically with 5 mM H2SO4 at a flow rate of 0.6 ml/min, and the column was maintained at 30°C. Dioxane consumption was monitored by direct injection of 5-μl samples onto a Varian 3400 GC equipped with a flame ionization detector (FID) and a 1% AT-1000 on carbograph 1-packed column (Grace, Columbia, MD). Samples were analyzed isothermally at 170°C, and the injector and detector temperatures were 230°C and 250°C, respectively. CO2 production in cell extracts was determined by injecting headspace samples (2 ml) onto a HP 6890 GC equipped for PDD, a 1.5-m Hayesep-DB 100/120 precolumn, and a 2-m Hayesep-DB 120/140 column. The oven temperature was maintained at 90°C. The flow rate of helium carrier gas was set at 10 ml/min.
The preparation and isotopomeric analysis of proteinogenic amino acids were performed as previously described (33, 44). In brief, biomass was hydrolyzed in 6 M HCl at 100°C for 24 h. During the hydrolysis, tryptophan and cysteine are degraded, while asparagine and glutamine are converted into aspartate and glutamate, respectively. The amino acid solution was dried under air flush overnight. Amino acid samples were derivatized in tetrahydrofuran (THF) and N-(tert-butyl dimethylsilyl)-N-methyl-trifluoroacetamide (Sigma-Aldrich, St. Louis, MO) at 70°C for 1 h. A GC (Hewlett-Packard model 6890; Agilent technologies, Palo Alto, CA) equipped with a DB5-MS column (J&W Scientific, Folsom, CA) and a mass spectrometer (MS) (5975; Agilent Technologies, Palo Alto, CA) was used for isotopomer analysis. Two types of charged fragments were clearly detected by GC-MS for the derivatized amino acids: the [M-57]+ which contains the entire amino acid and the [M-159]+, which contains the amino acid without the first carbon (α carboxyl group). The [M-57]+ peaks in leucine, isoleucine, and proline overlap other peaks, so the [M-159]+ group was used to obtain the isotopomer labeling information of those amino acids. The final isotopomer labeling fractions were indicated as M0 (unlabeled fraction), M1 (single labeled carbon fraction), M2 (fraction with two labeled carbons), M3 (fraction with three labeled carbons), and so forth.
Microarray data accession number.
Details of the microarray design, transcriptomic experimental design, and transcriptomic data have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE33197.
RESULTS AND DISCUSSION
In silico analysis of dioxane degradation pathway genes.
The pathway for dioxane degradation that was proposed by Mahendra et al. (24) (Fig. 1, black arrows) was the starting point for this study. We searched the literature for descriptions of enzymatic activities catalyzing the proposed dioxane metabolic transformations and used the amino acid sequence of characterized enzymes to identify homologues in the strain CB1190 genome sequence (See Table S2 in the supplemental material) using blastp (2).
Dioxane degradation is initiated by the activity of a monooxygenase (23, 48). The strain CB1190 genome carries eight bacterial multicomponent monooxygenase gene clusters, seven of which are carried on the chromosome and one that is on plasmid pPSED02 (38).
The transformation of the initial hydroxylated dioxane metabolite is proposed to involve spontaneous ether cleavage and/or aldehyde/carboxylic acid and/or secondary alcohol/ketone conversions (24). Strain CB1190 has 13 genes encoding proteins with at least 30% amino acid identity with characterized bacterial aldehyde dehydrogenases (Escherichia coli AldA [accession number NP_415933], E. coli AldB [AAC76612], Rhodococcus erythropolis AldH [AAZ14956]), which catalyze the NAD(P)+-dependent oxidation of aldehydes to carboxylic acids. Eleven of these genes are located on the chromosome, while the remaining two are on plasmid pPSED02 (see Table S2 in the supplemental material). Three genes encoding proteins with 29 to 36% amino acid identity to a secondary alcohol dehydrogenase (accession number CAD36475) of the actinomycete Rhodococcus ruber are on the chromosome; this enzyme catalyzes the NAD+-dependent transformation of secondary alcohols to ketones (23, 48).
The second proposed hydroxylation reaction generating single ether-containing 2-hydroxyethoxy-2-hydroxyacetic acid or 1,2-dihydroxyethoxyacetic acid has also been postulated to be catalyzed by a monooxygenase (24) (Fig. 1).
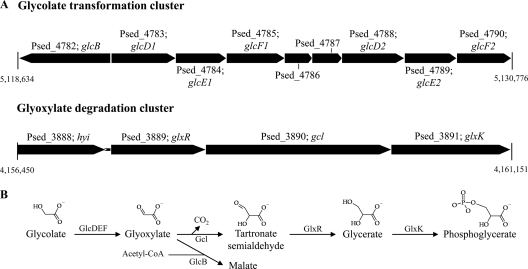
The subsequent cleavage of the second ether bond leads to the production of two-carbon intermediates, of which glyoxal, ethylene glycol, glycoaldehyde, glycolate, glyoxylate, and oxalate have been identified (24). The strain CB1190 genome has three genes encoding proteins with 27 to 34% amino acid identity to E. coli 1,2-propanediol oxidoreductase (accession number AP_003365), which transforms ethylene glycol to glycoaldehyde (10). Glycoaldehyde is transformed to glycolate by aldehyde dehydrogenases (see above), while glycolate is converted to glyoxylate by glycolate oxidase (18). In E. coli, glycolate oxidase is encoded by glcDEF (31); strain CB1190 has two adjacent homologues, glcD1E1F1 and glcD2E2F2 (Fig. 2). These homologues share 72%, 47%, and 64% amino acid identity for GlcD, GlcE, and GlcF, respectively.
Fig 2.
Strain CB1190 chromosomal regions of interest implicated in glycolate and glyoxylate transformations during dioxane metabolism. (A) Gene clusters implicated in glycolate and glyoxylate metabolism. (B) Proposed transformations of glycolate and glyoxylate catalyzed by enzymes putatively encoded by strain CB1190 gene clusters.
Mahendra et al. (24) proposed that glyoxal is transformed to glyoxylate, and although this enzymatic activity has not been previously described in the literature, the numerous aldehyde dehydrogenases encoded in the strain CB1190 genome could potentially catalyze this reaction. Alternatively, genes encoding aldehyde reductase homologues (E. coli YqhD [accession number Q46856] and Bacillus subtilis YvgN [accession number O32210]) that catalyze the transformation of glyoxal to glycoaldehyde were identified. Furthermore, in E. coli the glyoxalase I/II system, encoded by the genes gloA and gloB, uses S-glutathione to transform glyoxal to glycolate (47); however, no homologous genes were identified in the strain CB1190 genome.
Since glyoxylate has been identified as a metabolite of dioxane (24, 27), we examined the strain CB1190 genome for genes involved in known glyoxylate assimilation pathways. Strain CB1190 has a cluster of genes (Fig. 2) homologous to those encoding the glyoxylate carboligase pathway that converts glyoxylate through tartronate semialdehyde and glycerate to phosphoglycerate in order to provide both carbon and energy for growth in E. coli (20). Strain CB1190 also has a gene (Psed_4782) that encodes malate synthase G, which in E. coli (and in strain CB1190) (Fig. 2) is linked to the locus encoding glycolate oxidase and which assimilates glyoxylate directly into the tricarboxylic acid cycle (TCA) cycle via malate (19, 26). Malate synthase G is nonessential for the growth of E. coli on glyoxylate and is therefore considered to play an anaplerotic role (29).
While the transformation of glyoxylate to oxalate has been reported in Pseudomonas fluorescens (40), the gene sequence for the enzyme catalyzing this reaction has not yet been determined in bacteria. Finally, aerobic bacterial growth on oxalate was shown to be dependent on the oxalate decarboxylase OxdC, which converts oxalate to formate (45). However, the strain CB1190 genome lacks a homologue for this enzyme.
Gene expression during growth on dioxane, glycolate, and pyruvate.
We used gene expression microarrays targeting the strain CB1190 genome to determine the genes that are differentially regulated during growth on dioxane or glycolate relative to growth on pyruvate. The expression of 383 genes differed significantly; 97 genes were upregulated with dioxane relative to the pyruvate control, whereas 286 genes were downregulated (see Table S3 in the supplemental material). When strain CB1190 was grown with glycolate, the expression of 506 genes differed significantly relative to pyruvate-grown cells, with 203 genes upregulated and 303 genes downregulated (see Table S4). A comparison of genes differentially regulated with either dioxane or glycolate relative to pyruvate identified 36 genes that were upregulated with both substrates, whereas 121 genes were downregulated with both substrates. Table 1 provides a list of genes that were upregulated with dioxane alone or with both dioxane and glycolate.
Table 1.
Strain CB1190 genes on microarray upregulated during growth with dioxanea
| Gene ID | Gene name | Gene product | Repliconb | Expression ratioc |
|
|---|---|---|---|---|---|
| Dioxane/pyruvate | Glycolate/pyruvate | ||||
| Psed_0038 | Regulatory protein ArsR | c | 2.0 | 0.37 | |
| Psed_0350 | Bile acid-sodium symporter | c | 2.9 | 13 | |
| Psed_1076 | Short-chain dehydrogenase/reductase SDR | c | 2.6 | 1.7 | |
| Psed_1302 | Protein of unknown function DUF156 | c | 3.2 | 11 | |
| Psed_1303 | Heavy metal transport/detoxification protein | c | 2.7 | 8.0 | |
| Psed_1304 | Heavy metal translocating P-type ATPase | c | 2.5 | 9.7 | |
| Psed_1584 | 6-Phosphofructokinase | c | 3.3 | 3.9 | |
| Psed_1594 | Protein of unknown function UPF0016 | c | 2.3 | 1.2 | |
| Psed_1658 | Trimethylamine-N-oxide reductase (cytochrome c) | c | 5.3 | 0.63 | |
| Psed_2030 | Linalool 8-monooxygenase | c | 2.2 | 2.4 | |
| Psed_2371 | ABC-type transporter, periplasmic subunit | c | 2.0 | 3.1 | |
| Psed_3041 | NADH dehydrogenase (ubiquinone) 24-kDa subunit | c | 2.0 | 1.1 | |
| Psed_3522 | Response regulator receiver | c | 3.5 | 3.9 | |
| Psed_3564 | Aromatic-amino-acid transaminase | c | 3.0 | 1.2 | |
| Psed_3888 | hyi | Hydroxypyruvate isomerase | c | 106 | 112 |
| Psed_3889 | glxR | Tartronate semialdehyde reductase | c | 114 | 114 |
| Psed_3890 | gcl | Glyoxylate carboligase | c | 98 | 108 |
| Psed_3891 | glxK | Glycerate kinase | c | 49 | 56 |
| Psed_3934 | Major facilitator superfamily MFS_1 | c | 2.4 | 1.6 | |
| Psed_3935 | GntR domain protein | c | 2.5 | 1.1 | |
| Psed_4031 | Iron-sulfur cluster binding protein | c | 2.1 | 2.8 | |
| Psed_4146 | Potassium-transporting ATPase B chain | c | 2.6 | 1.2 | |
| Psed_4147 | Potassium-transporting ATPase A chain | c | 2.6 | 1.2 | |
| Psed_4148 | K+-transporting ATPase, F subunit | c | 3.0 | 1.3 | |
| Psed_4512 | ABC-type transporter, integral membrane subunit | c | 2.2 | 2.7 | |
| Psed_4513 | Cobalamin (vitamin B12) biosynthesis CbiX protein | c | 2.1 | 2.4 | |
| Psed_4577 | Cold-shock protein DNA-binding | c | 2.2 | 3.7 | |
| Psed_4755 | Ammonium transporter | c | 2.2 | 9.3 | |
| Psed_4782 | glcB | Malate synthase | c | 10 | 11 |
| Psed_4788 | glcD2 | d-Lactate dehydrogenase (cytochrome) | c | 43 | 44 |
| Psed_4789 | glcE2 | FAD linked oxidase domain protein | c | 24 | 24 |
| Psed_4790 | glcF2 | Protein of unknown function DUF224 cysteine-rich region domain protein | c | 27 | 26 |
| Psed_5025 | 4-Hydroxyacetophenone monooxygenase | c | 5.8 | 3.6 | |
| Psed_5026 | Regulatory protein TetR | c | 2.4 | 2.8 | |
| Psed_5135 | Malic protein NAD-binding | c | 2.6 | 1.4 | |
| Psed_5406 | NAD(P)+ transhydrogenase (AB-specific) | c | 2.1 | 2.2 | |
| Psed_5524 | Hydrolase | c | 2.1 | 1.3 | |
| Psed_5736 | Transglycosylase-like domain protein | c | 2.8 | 8.4 | |
| Psed_5938 | Glycoside hydrolase family 13 domain-containing protein | c | 2.3 | 1.5 | |
| Psed_6175 | Regulatory protein TetR | c | 3.5 | 4.0 | |
| Psed_6259 | FMN-dependent oxidoreductase, nitrilotriacetate monooxygenase family | c | 2.5 | 1.2 | |
| Psed_6261 | ABC-type transporter, integral membrane subunit | c | 2.3 | 1.1 | |
| Psed_6262 | ABC-type transporter, integral membrane subunit | c | 2.2 | 0.97 | |
| Psed_6263 | Extracellular ligand-binding receptor | c | 2.2 | 0.92 | |
| Psed_6600 | Acyl-CoA dehydrogenase domain-containing protein | c | 4.2 | 0.95 | |
| Psed_6728 | Amidase | p1 | 2.6 | 0.99 | |
| Psed_6730 | Luciferase-like, subgroup | p1 | 2.8 | 0.72 | |
| Psed_6732 | MaoC domain protein dehydratase | p1 | 2.2 | 1.0 | |
| Psed_6735 | Flavoprotein WrbA | p1 | 2.1 | 1.1 | |
| Psed_6742 | NLP/P60 protein | p1 | 2.7 | 1.1 | |
| Psed_6745 | ATP-binding protein | p1 | 3.7 | 0.72 | |
| Psed_6751 | Transglycosylase-like domain protein | p1 | 5.0 | 0.60 | |
| Psed_6779 | Integrase catalytic region | p1 | 6.9 | 5.4 | |
| Psed_6782 | Hydroxyacid-oxoacid transhydrogenase | p1 | 4.6 | 4.3 | |
| Psed_6784 | Alkylglycerone-phosphate synthase | p1 | 3.5 | 3.5 | |
| Psed_6787 | Formyl-CoA transferase | p1 | 13 | 11.3 | |
| Psed_6791 | IstB domain protein ATP-binding protein | p1 | 3.5 | 2.4 | |
| Psed_6799 | Transposase IS4 family protein | p1 | 2.0 | 1.7 | |
| Psed_6970 | d-Lactate dehydrogenase (cytochrome) | p2 | 6.3 | 0.67 | |
| Psed_6971 | Hydroxyacid-oxoacid transhydrogenase | p2 | 5.1 | 0.69 | |
| Psed_6972 | GntR domain protein | p2 | 3.4 | 0.62 | |
| Psed_6974 | Ethyl tert-butyl ether degradation EthD | p2 | 3.2 | 0.71 | |
| Psed_6975 | Betaine-aldehyde dehydrogenase | p2 | 2.6 | 0.74 | |
| Psed_6977 | thmD | Ferredoxin-NAD(+) reductase | p2 | 4.2 | 0.72 |
| Psed_6978 | thmB | Methane/phenol/toluene hydroxylase | p2 | 2.7 | 0.80 |
| Psed_6979 | thmC | Monooxygenase component MmoB/DmpM | p2 | 3.7 | 0.76 |
| Psed_6981 | Aldehyde dehydrogenase | p2 | 9.5 | 0.83 | |
| Psed_6982 | Mn2+/Fe2+ transporter, NRAMP family | p2 | 14 | 1.0 | |
| Psed_7002 | Transcription factor WhiB | p2 | 2.7 | 0.67 | |
Only annotated genes with predicted functions are shown. The following genes (Psed_XXXX) were automatically annotated as hypothetical proteins: chromosome, 2752, 3653, 3669, 4149, 4306, 4424, and 5371; plasmid pPSED01, 6729, 6743, 6744, 6746, 6747, 6748, 6749, 6750, 6752, 6753, 6754, 6755, 6757, 6758, and 6780; plasmid pPSED02, 6913, 6973, 6980, 7003, 7007, and 7008.
c, chromosome; p1, plasmid pPSED01; p2, plasmid pPSED02.
Genes upregulated with dioxane but not with glycolate are indicated in boldface.
The replicon locations of the dioxane- and glycolate-induced genes were examined. For dioxane-grown cells, 27 and 17 genes were induced from plasmids pPSED01 and pPSED02, respectively, with the remaining induced genes on the chromosome. For glycolate-grown cells, 15 induced genes were from plasmid pPSED01, while the rest were on the chromosome; no genes were induced from plasmid pPSED02.
Sixty-one genes were upregulated with dioxane but not with glycolate, relative to pyruvate (Table 1). Of these genes, 21 were from plasmid pPSED01, while 17 genes were from plasmid pPSED02. Of the dioxane-induced plasmid pPSED01 genes, 14 of these encode hypothetical proteins.
Upregulation of a multicomponent monooxygenase gene cluster during dioxane metabolism.
While the strain CB1190 genome encodes eight monooxygenase gene clusters, microarray analysis of dioxane-induced transcription revealed that only the plasmid pPSED02-encoded monooxygenase cluster was upregulated compared with growth with pyruvate (Table 1). Upregulation of this cluster was verified with qRT-PCR using primers targeting Psed_6976, encoding a homologue of THF monooxygenase α-subunit (Table 2). Three chromosomally encoded monooxygenase gene clusters were missing from the microarray, so their potential involvement in dioxane metabolism was tested using qRT-PCR with primers targeting the genes encoding the α-subunits. As Table 2 shows, no significant differential transcription was observed for these three monooxygenase genes (Psed_0629, Psed_0768, and Psed_0815) in cDNA from dioxane-, glycolate-, or pyruvate-grown cells.
Table 2.
Normalized relative expression levels for selected strain CB1190 genes analyzed by qRT-PCR
| Gene group | Gene ID | Gene name | Gene product | Replicona | Expression ratiob |
|
|---|---|---|---|---|---|---|
| Dioxane/pyruvate | Glycolate/pyruvate | |||||
| Monooxygenase genes lacking probes on microarray | Psed_6976 | thmA | THF monooxygenase α-subunit | p2 | 15 ± 0.2 | 1.0 ± 0.5 |
| Psed_0629 | prmA | Methane monooxygenase | c | 0.4 ± 0.1 | 0.8 ± 0.5 | |
| Psed_0768 | Phenol 2-monooxygenase | c | 2 ± 2 | 0.9 ± 0.4 | ||
| Psed_0815 | Methane/phenol/toluene hydroxylase | c | 1.1 ± 0.5 | 0.3 + 0.4 | ||
| Dioxane metabolism intermediate-related genes | Psed_6782 | Alcohol dehydrogenase | p1 | 7.3 ± 0.8 | 3.3 ± 0.4 | |
| Psed_6783 | Fatty-acid-CoA ligase | p1 | 24 ± 2 | 7 ± 5 | ||
| Psed_6970 | FAD/FMN-dependent dehydrogenase | p2 | 6.1 ± 0.8 | 1.5 ± 0.9 | ||
| Psed_6971 | Alcohol dehydrogenase | p2 | 8.7 ± 0.8 | 1.3 ± 0.4 | ||
| Psed_6975 | Aldehyde dehydrogenase | p2 | 23 ± 2 | 0.9 ± 0.3 | ||
| Psed_6981 | Aldehyde dehydrogenase | p2 | 11 ± 1 | 1.5 ± 0.9 | ||
| Glycolate and glyoxylate metabolism-related genes | Psed_4783 | glcD1 | Glycolate oxidase | c | 0.3 ± 0.1 | 0.7 ± 0.5 |
| Psed_4788 | glcD2 | Glycolate oxidase | c | 3.6 ± 0.3 | 2.8 ± 0.5 | |
| Psed_4782 | glcB | Malate synthase G | c | 5.4 ± 0.2 | 2 ± 1 | |
| Psed_3888 | hyi | Hydroxypyruvate isomerase | c | 5 ± 1 | 3.1 ± 0.3 | |
| Psed_3889 | glxR | Tartronate semialdehyde reductase | c | 13 ± 1 | 6 ± 2 | |
| Psed_3890 | gcl | Glyoxylate carboligase | c | 18 ± 2 | 10 ± 4 | |
| Psed_3891 | glxK | Glycerate kinase | c | 1.9 ± 0.6 | 1.9 ± 0.7 | |
c, chromosome; p1, plasmid pPSED01; p2, plasmid pPSED02.
Genes upregulated with dioxane but not with glycolate are indicated in boldface.
The dioxane-induced, plasmid pPSED02-borne monooxygenase gene cluster is homologous to the thm gene cluster involved in THF utilization in Pseudonocardia tetrahydrofuranoxydans strain K1 (46). This gene cluster has also been identified in DNA extracted from Pseudonocardia strain ENV478 (25), and in both strains K1 and ENV478 growth with THF induces transcription of genes in this cluster (25, 46). Although strains K1 and ENV478 degrade dioxane cometabolically when induced with THF (23, 48), the published results did not directly link the THF-induced monooxygenase gene cluster to biochemical dioxane transformation activity. However, the upregulation of the homologous thm monooxygenase gene cluster in strain CB1190 during dioxane metabolism reported here supports the involvement of the thm monooxygenase gene clusters in dioxane degradation in strain K1 and ENV478.
Genes potentially contributing to transformation of C4 dioxane metabolites.
After the initial monooxygenase-catalyzed hydroxylation, the C4 dioxane metabolites undergo a series of transformations that ultimately yield two C2 compounds (Fig. 1). The set of genes on plasmid pPSED02 that are induced by dioxane but not by glycolate could potentially be involved in these transformations (Table 1). The operon containing the thm monooxygenase-encoding gene cluster also includes genes encoding two aldehyde dehydrogenases (Psed_6975 and Psed_6981), and upstream of the monooxygenase cluster and in the reverse orientation is an upregulated gene cluster encoding an alcohol dehydrogenase and a FAD/FMN-dependent dehydrogenase, as well as a transcriptional regulator (Psed_6970–6972). Dioxane-dependent upregulation of these genes was verified by qRT-PCR (Table 2). The aldehyde-carboxylic acid conversion of C4 dioxane metabolites could be catalyzed by these dioxane-induced aldehyde dehydrogenases. In contrast, a candidate gene encoding an enzyme for the secondary alcohol-ketone conversion of C4 metabolites (i.e., 2-hydroxy-1,4-dioxane to 1,4-dioxane-2-one) is not immediately clear, since none of the secondary alcohol dehydrogenase genes identified by in silico analysis (see Table S2 in the supplemental material) were specifically upregulated with dioxane. The alcohol dehydrogenase-encoding Psed_6971 or constitutively expressed secondary alcohol dehydrogenases (not identified in the differential transcription analyses reported here) could be involved in this reaction. We are currently cloning the upregulated dehydrogenases and reductases to functionally test their role in the transformation of C4 dioxane metabolites.
The dioxane metabolic pathway also requires a second hydroxylation and two ether cleavage steps. Ether cleavage may occur spontaneously following hydroxylation, or it may occur enzymatically (51). A heterogeneous group of ether-cleaving enzymes have been identified (reviewed in reference 51), and an ether bond-cleaving diglycolic acid dehydrogenase has been purified from P. tetrahydrofuranoxydans strain K1 (54). However, genomic and transcriptomic analyses of dioxane metabolism did not identify such enzymes in strain CB1190.
Genes potentially contributing to transformation of C2 dioxane metabolites to glyoxylate.
The plasmid pPSED02-borne putative aldehyde and alcohol dehydrogenases that were upregulated during growth on dioxane but not on glycolate could also contribute to the transformation of the dioxane metabolites glyoxal, glycoaldehyde, and ethylene glycol. In E. coli the aldehyde reductase YqhD catalyzes the NADPH-dependent reduction of glyoxal to glycoaldehyde (21), while in Bacillus subtilis the nonhomologous YvgN catalyzes the same reaction (36). The upregulated aldehyde dehydrogenase gene products share only low amino acid sequence identity with these proteins, but Psed_6975 has 35% identity to E. coli AldA, which catalyzes the NAD+-dependent oxidation of glycoaldehyde to glycolate (13). Boronat et al. (7) identified an E. coli mutant that employed the activity of propanediol oxidoreductase (fucO [10]) to transform ethylene glycol to glycoaldehyde, which was further transformed to glycolate. The putative alcohol dehydrogenase Psed_6971 that was upregulated with dioxane shares 42% amino acid similarity to E. coli 1,2-propanediol oxidoreductase, so the gene product may catalyze the conversion of ethylene glycol to glycoaldehyde during dioxane metabolism. It should be noted that the product of the glycolate- and dioxane-induced alcohol dehydrogenase gene Psed_6782 has similar amino acid identity to E. coli propanediol oxidoreductase although its role in glycolate metabolism is not clear.
The set of genes upregulated with both dioxane and glycolate relative to pyruvate included the putative glycolate oxidase-encoding chromosomal gene cluster glcD2E2F2 (Psed_4788–4790) (Table 1). In contrast, the adjacent homologous glcD1E1F1 gene cluster was not upregulated with either dioxane or glycolate. This lack of upregulation was confirmed with qRT-PCR (Table 2). Therefore, the enzyme encoded by glcD2E2F2 likely catalyzes the transformation of glycolate to glyoxylate (31). In E. coli, genes for malate synthase G (glcB) and glcDEF are proximal, oriented in the same direction, and are cotranscribed during growth with glycolate (32). While glcB (Psed_4782) and glcD2E2F2 are in close proximity in strain CB1190, they have opposing orientation and are separated by the homologous but nonidentical glcD1E1F1 (Psed_4783–4785) cluster that was not upregulated during growth with either dioxane or glycolate (Fig. 2). Between glcD2E2F2 and glcD1E1F1 is a gene (Psed_4787) encoding a putative GntR family transcriptional regulator, which may be involved in regulation of this glycolate oxidase homologue.
Glyoxylate metabolism during dioxane degradation by strain CB1190.
Genes encoding two divergent routes for glyoxylate assimilation were upregulated with both dioxane and glycolate, as determined by microarray analysis. The first route was through the glcB-encoded malate synthase G (Psed_4782) (Table 1) although qRT-PCR did not confirm this upregulation (Table 2). The upregulated gene cluster encoding glyoxylate carboligase, tartronate semialdehyde reductase, hydroxypyruvate isomerase, and glycerate kinase (Psed_3888–3891) (Fig. 2) represents the second route for glyoxylate assimilation. The upregulation of three genes from this cluster was verified by qRT-PCR (Table 1).
These transcriptional results point to glyoxylate, rather than oxalate (Fig. 1), as the key intermediate in the assimilation of carbon into central metabolism during dioxane degradation by strain CB1190. In the glyoxylate carboligase pathway, two glyoxylate molecules are combined by glyoxylate carboligase, producing a C3 compound that is eventually incorporated into glycolysis at phosphoglycerate (20) (Fig. 1 and 2). In the typical malate synthase G pathway, glyoxylate and acetyl-coenzyme A (CoA) are condensed to form malate, which is incorporated in the TCA cycle (19) (Fig. 1 and 2). As noted above, the glyoxylate carboligase pathway supports growth through carbon and energy conservation, whereas malate synthase G is involved in anaplerotic reactions but is not known to support growth in the absence of glyoxylate carboligase with glyoxylate as a sole substrate (29). Therefore, the growth of strain CB1190 on dioxane is likely dependent on the glyoxylate carboligase pathway and is potentially enhanced by malate synthase G.
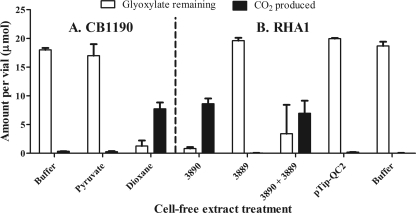
Since strain CB1190 can grow with dioxane, the bacterium must derive energy from the compound's transformation. The glyoxylate carboligase pathway represents a potential route to energy generation during dioxane metabolism via glyoxylate, so we sought to confirm the activity of this pathway in dioxane-grown cells. Glyoxylate carboligase (Gcl) activity (2 glyoxylate → CO2 + tartronate semialdehyde) (Fig. 2) was tested in cell extracts prepared from strain CB1190 cells. As Fig. 3A shows, pregrowth on dioxane resulted in significantly greater consumption of glyoxylate than pregrowth on pyruvate. Glyoxylate disappearance was accompanied by a stoichiometric production of CO2, a specific product of the glyoxylate carboligase reaction. Heating of the dioxane cell extracts for 10 min at 80°C reduced CO2 production to that observed with the buffer treatment (data not shown). No CO2 was detected in dioxane cell extracts when glyoxylate was omitted.
Fig 3.
Glyoxylate carboligase activity in strain CB1190 and strain RHA1/pTip cell extracts. (A) Phosphate buffer or cell extracts from pyruvate- or dioxane-grown strain CB1190 cells were amended with 10 mM glyoxylate, and then the amount of glyoxylate remaining and CO2 production were determined. (B) The same assay as described for panel A was performed with cell extracts prepared from strain RHA1 cells with pTip-Psed_3890 (3890; putative glyoxylate carboligase), pTip-3889 (3889; putative tartronate semialdehyde reductase), a mix of the two (3890 + 3889), pTip-QC2 (empty vector), or buffer.
To verify the putative roles of strain CB1190 genes in the dioxane-induced glyoxylate carboligase pathway, we cloned and overexpressed Psed_3890 (putative glyoxylate carboligase gene) and Psed_3889 (putative tartronate semialdehyde reductase gene) (Fig. 2) in Rhodococcus jostii strain RHA1 using the thiostrepton-inducible plasmid pTip-QC2 (28). As expected, glyoxylate was consumed and CO2 was produced only in cell extracts from Psed_3890-expressing strain RHA1 or when these extracts were mixed with extracts from Psed_3889-expressing RHA1 (Fig. 3B). The mixture of cell extracts from RHA1/pTip-Psed_3890 and RHA1/pTip-Psed_3889 also produced a small amount of glycerate (0.71 ± 0.23 μmol per vial). No glycerate was produced with cell extracts from either construct alone or with cell extracts from strain RHA1 with empty pTip-QC2. The amount of glycerate detected with the mix of RHA1/pTip-Psed_3890 and RHA1/pTip-Psed_3889 cell extracts represented ∼7% of that expected if tartronate semialdehyde was stoichiometrically transformed to glycerate (Fig. 2) as a dead-end product. Tartronate semialdehyde was not detectable with the HPLC method used, so it is unclear whether the small amount of glycerate detected was due to only a minor transformation of tartronate semialdehyde or to the further transformation of the generated glycerate to unspecified products. Regardless, these results support the annotation of Psed_3890 as a glyoxylate carboligase gene and Psed_3889 as a tartronate semialdehyde reductase gene.
Amino acid isotopomer analysis to identify routes for dioxane carbon assimilation.
The glyoxylate carboligase pathway ultimately leads to the assimilation of carbon as three-carbon metabolites in glycolysis in the form of 3-phosphoglycerate and later pyruvate (12). In order to obtain further support for the hypothesis that carbon from dioxane enters central metabolism via the glyoxylate carboligase pathway as three-carbon compounds, we performed isotopomer amino acid analysis with 13C fully labeled dioxane. Strain CB1190 was grown either with [13C]dioxane as the sole carbon source or with [13C]dioxane and elevated (3%) CO2. With [13C]dioxane as the sole carbon source, all of the detected amino acids were heavily labeled (see Table S5 in the supplemental material), which confirmed, as expected, that strain CB1190 can directly use dioxane as a sole carbon source.
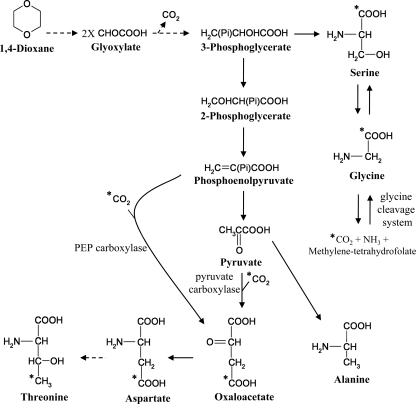
Pyruvate-derived alanine, valine, phenylalanine, and tyrosine were predominantly 13C-labeled when strain CB1190 was grown with [13C]dioxane as the sole carbon source (see Table S5 in the supplemental material), and the addition of elevated unlabeled CO2 did not affect these labeling patterns (see Table S6). These results are in contrast to aspartate, threonine, serine, and glycine; for these, a small proportion of unlabeled carbon was integrated when the cells were grown with [13C]dioxane as the sole carbon source, and this proportion increased greatly when strain CB1190 was grown with [13C]dioxane and elevated unlabeled CO2. These results indicate that carbon from CO2 was assimilated during the synthesis of these amino acids (Fig. 4).
Fig 4.
Proposed assimilation of unlabeled carbon during growth of strain CB1190 with uniformly 13C-labeled dioxane. Unlabeled carbons are indicated with an asterisk (*); all other carbons are considered 13C. The dashed arrow indicates a multistep reaction. The amino acids valine, phenylalanine, and tyrosine were omitted for simplicity.
For the oxaloacetate-derived aspartate and threonine, the unlabeled carbon was present at a non-first-carbon position (see M-159 results in Tables S5 and S6 in the supplemental material) under both experimental conditions. These results indicate that the anaplerotic pathway transforming either phosphoenolpyruvate (PEP) or pyruvate to oxaloacetate through the addition of CO2 (17, 53) was active during dioxane metabolism (Fig. 4). Strain CB1190 has homologous genes for both PEP carboxylase (Psed_6164) and pyruvate carboxylase (Psed_3032), which catalyze these anaplerotic reactions. The enzyme that catalyzes the reverse of the PEP-utilizing process, PEP carboxykinase, is putatively encoded by Psed_6619, and this gene was downregulated during growth with both dioxane and glycolate, relative to pyruvate (see Tables S3 and S4). Taken together, these results indicate that with dioxane, carbon flows predominantly from the direction of PEP toward the TCA cycle and support the proposed role of the three-carbon compound-generating glyoxylate carboligase pathway in dioxane metabolism. In contrast, when grown with pyruvate, strain CB1190 needs to generate PEP and further gluconeogenic compounds necessary for amino acid synthesis, which explains the relative upregulation of the gene (Psed_6619) encoding the PEP-generating PEP carboxykinase.
The inconsistency between the labeling patterns of 3-phosphoglycerate-derived serine and glycine and pyruvate-derived alanine, valine, phenylalanine, and tyrosine is noteworthy since 3-phosphoglycerate is a precursor of pyruvate in glycolysis. An explanation for this inconsistency is an alternative pathway of serine and glycine biosynthesis, specifically, by the reverse activity of the glycine cleavage system (GCS). The GCS converts glycine and tetrahydrofolate to CO2, NH3, and methylene-tetrahydrofolate (35), and it is putatively encoded by the strain CB1190 genome (37). The GCS is reversible (16), and reverse GCS activity in the presence of elevated unlabeled CO2 would result in increased unlabeled carbon in the glycine pool and, consequently, in the serine pool as well (Fig. 4). Thus, during dioxane metabolism, it is possible that serine and glycine are synthesized from both the glyoxylate carboligase pathway via 3-phosphoglycerate and the reverse GCS.
Glyoxylate could contribute to the serine pool in yet another way. The strain CB1190 glyoxylate carboligase cluster has a gene (hyi) for hydroxypyruvate isomerase, which catalyzes the isomerization of hydroxypyruvate to tartronate semialdehyde (3) and which is upregulated in strain CB1190 during growth with dioxane (Tables 1 and 2). Hydroxypyruvate can be used as a precursor for serine biosynthesis by the activity of transaminases (39). However, no genes with significant homology to known amino acid-glyoxylate aminotransferase or amino acid-hydroxypyruvate transaminase genes were upregulated in strain CB1190 during growth with dioxane or glycolate, so the role of hydroxypyruvate isomerase in dioxane metabolism remains unclear.
Energy generation in strain CB1190 dioxane metabolism.
Based on the current work, we present an updated proposed dioxane degradation pathway for strain CB1190, linking dioxane metabolites to central metabolism (Fig. 1). The ability of strain CB1190 to grow on dioxane necessitates energy generation, and the employment of the glyoxylate carboligase pathway during dioxane degradation conceptually provides the basis for energy production since this pathway results in the pyruvate precursors that can then be metabolized to yield NADH and ATP (20). An analysis (see Table S7 in the supplemental material) of the reducing equivalents consumed or produced in the individual steps proposed for dioxane metabolism in Fig. 1 reveals a maximum net yield of 6 mol NADH and 1 mol GTP per mol dioxane. Assuming an equivalence of 3 mol NADH per mol ATP, this means the theoretical maximum yield of ATP generated per dioxane is 19. Mahendra and Alvarez-Cohen (23) previously reported a yield of 0.09 g of protein per g of dioxane for strain CB1190, so assuming that protein constitutes 50% of dry cell weight and integrating this cell yield with the theoretical maximum yield of ATP from dioxane, this results in a yield coefficient of ATP (YATP) (4) of 0.83 g of dry cell weight per mol of ATP. In comparison, Lactobacillus casei growing with glucose (34) and Actinomyces strains growing with lactate (49) had YATP values of 24 and 10 g of dry cell weight per mol of ATP, respectively.
This work represents the first insights into the genetic basis for microbial dioxane metabolism and provides a more complete picture of the pathway leading to carbon assimilation and energy generation during growth on dioxane. It will be useful to determine if dioxane supports growth through a similar pathway in the other reported dioxane-metabolizing microorganisms, including Pseudonocardia benzenivorans strain B5 (23), Mycobacterium sp. PH-06 (15), C. sinensis (27), and Rhodococcus ruber strain 219 (5).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by Strategic Environmental Research and Development Program grant ER-1417.
We thank Ping Hu (Lawrence Berkeley National Lab) for help in microarray design and problem solving. Vanessa Brisson reannotated the microarray probes, and Charles Lee helped with primer development. We also appreciate the help of Donald Hermann in the lab of Mary Firestone, University of California, Berkeley, and Xueyang Feng in the lab of Yinjie Tang, Washington University in St. Louis, for their technical assistance in CO2 and amino acid isotopomer analyses, respectively, and Rebecca Parales, University of California, Davis, for constructive comments on the manuscript. Plasmid pTip-QC2 was obtained from the National Institute of Advanced Industrial Science and Technology (Japan).
Footnotes
Published ahead of print 10 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Affymetrix 2009. GeneChip expression analysis technical manual. Affymetrix, Santa Clara, CA: http://media.affymetrix.com/support/downloads/manuals/expression_analysis_manual.pdf [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Ashiuchi M, Misono H. 1999. Biochemical evidence that Escherichia coli hyi (orf b0508, gip) gene encodes hydroxypyruvate isomerase. Biochim. Biophys. Acta 1435:153–159 [DOI] [PubMed] [Google Scholar]
- 4. Bauchop T, Elsden SR. 1960. The growth of micro-organisms in relation to their energy supply. J. Gen. Microbiol. 23:457–469 [DOI] [PubMed] [Google Scholar]
- 5. Bernhardt D, Diekmann H. 1991. Degradation of dioxane, tetrahydrofuran and other cyclic ethers by an environmental Rhodococcus strain. Appl. Microbiol. Biotech. 36:120–123 [DOI] [PubMed] [Google Scholar]
- 6. Bolstad BM, Irizarry RA, Åstrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 7. Boronat A, Caballero E, Aguilar J. 1983. Experimental evolution of a metabolic pathway for ethylene glycol utilization by Escherichia coli. J. Bacteriol. 153:134–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 9. Burback BL, Perry JJ. 1993. Biodegradation and biotransformation of groundwater pollutant mixtures by Mycobacterium vaccae. Appl. Environ. Microbiol. 59:1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conway T, Ingram LO. 1989. Similarity of Escherichia coli propanediol oxidoreductase (fucO product) and an unusual alcohol dehydrogenase from Zymomonas mobilis and Saccharomyces cerevisiae. J. Bacteriol. 171:3754–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cusa E, Obradors N, Baldoma L, Badia J, Aguilar J. 1999. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 181:7479–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hansen RW, Hayashi JA. 1962. Glycolate metabolism in Escherichia coli. J. Bacteriol. 83:679–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hidalgo E, Chen YM, Lin EC, Aguilar J. 1991. Molecular cloning and DNA sequencing of the Escherichia coli K-12 ald gene encoding aldehyde dehydrogenase. J. Bacteriol. 173:6118–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson DR, et al. 2008. Temporal transcriptomic microarray analysis of “Dehalococcoides ethenogenes” strain 195 during the transition into stationary phase. Appl. Environ. Microbiol. 74:2864–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim Y-M, Jeon J-R, Murugesan K, Kim E-J, Chang Y-S. 2009. Biodegradation of 1,4-dioxane and transformation of related cyclic compounds by a newly isolated Mycobacterium sp. PH-06. Biodegradation 20:511–519 [DOI] [PubMed] [Google Scholar]
- 16. Kochi H, Kikuchi G. 1969. Reactions of glycine synthesis and glycine cleavage catalyzed by extracts of Arthrobacter globiformis grown on glycine. Arch. Biochem. Biophys. 132:359–369 [DOI] [PubMed] [Google Scholar]
- 17. Kornberg HL. 1966. Anaplerotic sequences and their role in metabolism, p 1–31 In Campbell PN, Greville GD. (ed), Essays in biochemistry, vol. 2 Academic Press, New York NY [Google Scholar]
- 18. Kornberg HL, Elsden SR. 1961. The metabolism of 2-carbon compounds by microorganisms. Adv. Enzymol. Relat. Subj. Biochem. 23:401–470 [DOI] [PubMed] [Google Scholar]
- 19. Kornberg HL, Krebs HA. 1957. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179:988–991 [DOI] [PubMed] [Google Scholar]
- 20. Krakow G, Barkulis SS, Hayashi JA. 1961. Glyoxylic acid carboligase: an enzyme present in glycolate-grown Escherichia coli J. Bacteriol. 81:509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee C, et al. 2010. Transcriptional activation of the aldehyde reductase YqhD by YqhC and its implication in glyoxal metabolism of Escherichia coli K-12. J. Bacteriol. 192:4205–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Z. 1996. Hetero-stagger cloning: efficient and rapid cloning of PCR products. Nucleic Acids Res. 24:2458–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mahendra S, Alvarez-Cohen L. 2006. Kinetics of 1,4-dioxane biodegradation by monooxygenase-expressing bacteria. Environ. Sci. Technol. 40:5435–5442 [DOI] [PubMed] [Google Scholar]
- 24. Mahendra S, Petzold CJ, Baidoo EE, Keasling JD, Alvarez-Cohen L. 2007. Identification of the intermediates of in vivo oxidation of 1,4-dioxane by monooxygenase-containing bacteria. Environ. Sci. Technol. 41:7330–7336 [DOI] [PubMed] [Google Scholar]
- 25. Masuda H. 2009. Identification and characterization of monooxygenase enzymes involved in 1,4-dioxane degradation in Pseudonocardia sp. strain ENV478, Mycobacterium sp. strain ENV421, and Nocardia sp. strain ENV425. Ph.D. dissertation Rutgers University, New Brunswick, NJ [Google Scholar]
- 26. Molina I, Pellicer M-T, Badia J, Aguilar J, Baldoma L. 1994. Molecular characterization of Escherichia coli malate synthase G. Eur. J. Biochem. 224:541–548 [DOI] [PubMed] [Google Scholar]
- 27. Nakamiya K, Hashimoto S, Ito H, Edmonds JS, Morita M. 2005. Degradation of 1,4-dioxane and cyclic ethers by an isolated fungus. Appl. Environ. Microbiol. 71:1254–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakashima N, Tamura T. 2004. Isolation and characterization of a rolling-circle-type plasmid from Rhodococcus erythropolis and application of the plasmid to multiple-recombinant-protein expression. Appl. Environ. Microbiol. 70:5557–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ornston LN, Ornston MK. 1969. Regulation of glyoxylate metabolism in Escherichia coli K-12. J. Bacteriol. 98:1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parales RE, Adamus JE, White N, May HD. 1994. Degradation of 1,4-dioxane by an actinomycete in pure culture. Appl. Environ. Microbiol. 60:4527–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pellicer MT, Badia J, Aguilar J, Baldoma L. 1996. glc locus of Escherichia coli: characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J. Bacteriol. 178:2051–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pellicer MT, et al. 1999. Cross-induction of glc and ace operons of Escherichia coli attributable to pathway intersection. J. Biol. Chem. 274:1745–1752 [DOI] [PubMed] [Google Scholar]
- 33. Pingitore F, Tang Y, Kruppa GH, Keasling JD. 2007. Analysis of amino acid isotopomers using FT-ICR MS. Anal. Chem. 79:2483–2490 [DOI] [PubMed] [Google Scholar]
- 34. Russell JB, Cook GM. 1995. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 59:48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sagers RD, Gunsalus IC. 1961. Intermediary metabolism of Diplococcus glycinophilus. I. Glycine cleavage and one-carbon interconversions. J. Bacteriol. 81:541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakai A, Katayama K, Katsuragi T, Tani Y. 2001. Glycolaldehyde-forming route in Bacillus subtilis in relation to vitamin B6 biosynthesis. J. Biosci. Bioeng. 91:147–152 [DOI] [PubMed] [Google Scholar]
- 37. Sales CM. 2012. Functional genomics of bacterial degradation of the emerging contaminants 1,4-dioxane and N-nitrosodimethylamine (NDMA). Ph.D. thesis University of California, Berkeley, Berkeley, CA [Google Scholar]
- 38. Sales CM, et al. 2011. Genome sequence of the 1,4-dioxane-degrading Pseudonocardia dioxanivorans strain CB1190. J. Bacteriol. 193:4549–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sallach HJ. 1956. Formation of serine from hydroxypyruvate and l-alanine. J. Biol. Chem. 223:1101–1108 [PubMed] [Google Scholar]
- 40. Singh R, et al. 2009. An ATP and oxalate generating variant tricarboxylic acid cycle counters aluminum toxicity in Pseudomonas fluorescens. PLoS One 4:e7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Skinner K, Cuiffetti L, Hyman M. 2009. Metabolism and cometabolism of cyclic ethers by a filamentous fungus, a Graphium sp. Appl. Environ. Microbiol. 75:5514–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stickney JA, et al. 2003. An updated evaluation of the carcinogenic potential of 1,4-dioxane. Regul. Toxicol. Pharmacol. 38:183–195 [DOI] [PubMed] [Google Scholar]
- 43. Sun B, Ko K, Ramsay JA. 2011. Biodegradation of 1,4-dioxane by a Flavobacterium. Biodegradation 22:651–659 [DOI] [PubMed] [Google Scholar]
- 44. Tang Y, et al. 2007. Pathway confirmation and flux analysis of central metabolic pathways in Desulfovibrio vulgaris Hildenborough using gas chromatography-mass spectrometry and Fourier transform-ion cyclotron resonance mass spectrometry. J. Bacteriol. 189:940–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanner A, Bornemann S. 2000. Bacillus subtilis YvrK is an acid-induced oxalate decarboxylase. J. Bacteriol. 182:5271–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thiemer B, Andreesen JR, Schräder T. 2003. Cloning and characterization of a gene cluster involved in tetrahydrofuran degradation in Pseudonocardia sp. strain K1. Arch. Microbiol. 179:266–277 [DOI] [PubMed] [Google Scholar]
- 47. Thornalley PJ. 1998. Glutathione-dependent detoxification of alpha-oxoaldehydes by the glyoxalase system: involvement in disease mechanisms and antiproliferative activity of glyoxalase I inhibitors. Chem. Biol. Interact. 111–112:137–151 [DOI] [PubMed] [Google Scholar]
- 48. Vainberg S, et al. 2006. Biodegradation of ether pollutants by Pseudonocardia sp. strain ENV478. Appl. Environ. Microbiol. 72:5218–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van der Hoeven JS, van den Kieboom CWA. 1990. Oxygen-dependent lactate utilization by Actinomyces viscosus and Actinomyces naeslundii. Oral Microbiol. Immunol. 5:223–225 [DOI] [PubMed] [Google Scholar]
- 50. Vandesompele J, et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034. http://genomebiology.com/content/3/7/RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. White GF, Russell NJ, Tidswell EC. 1996. Bacterial scission of ether bonds. Microbiol. Rev. 60:216–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woo Y-T, Arcos JC, Argus MF, Griffin GW, Nishiyama K. 1977. Metabolism of dioxane: identification of ρ-dioxane-2-one as the major urinary metabolite. Biochem. Pharmacol. 26:1535–1538 [DOI] [PubMed] [Google Scholar]
- 53. Wood HG, Utter MF. 1965. The role of CO2 fixation in metabolism. Essays Biochem. 1:1–27 [PubMed] [Google Scholar]
- 54. Yamashita M, Tani A, Kawai F. 2004. Cloning and expression of an ether-bond-cleaving enzyme involved in the metabolism of polyethylene glycol. J. Biosci. Bioeng. 98:313–315 [DOI] [PubMed] [Google Scholar]
- 55. Young JD, Braun WH, Gehring PJ, Horvath BS, Daniel RL. 1976. 1,4-Dioxane and β-hydroxyethoxyacetic acid excretion in urine of humans exposed to dioxane vapors. Toxicol. Appl. Pharmacol. 38:643–646 [DOI] [PubMed] [Google Scholar]
- 56. Zenker MJ, Borden RC, Barlaz MA. 2003. Occurrence and treatment of 1,4-dioxane in aqueous environments. Environ. Eng. Sci. 20:423–432 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.